Abstract
Hereditary sensory and autonomic neuropathies (HSAN) are clinically and genetically heterogeneous disorders characterized by axonal atrophy and degeneration, exclusively or predominantly affecting the sensory and autonomic neurons. So far, disease-associated mutations have been identified in seven genes: two genes for autosomal dominant (SPTLC1 and RAB7) and five genes for autosomal recessive forms of HSAN (WNK1/HSN2, NTRK1, NGFB, CCT5 and IKBKAP). We performed a systematic mutation screening of the coding sequences of six of these genes on a cohort of 100 familial and isolated patients diagnosed with HSAN. In addition, we screened the functional candidate gene NGFR (p75/NTR) encoding the nerve growth factor receptor. We identified disease-causing mutations in SPTLC1, RAB7, WNK1/HSN2 and NTRK1 in 19 patients, of which three mutations have not previously been reported. The phenotypes associated with mutations in NTRK1 and WNK1/HSN2 typically consisted of congenital insensitivity to pain and anhidrosis, and early-onset ulcero-mutilating sensory neuropathy, respectively. RAB7 mutations were only found in patients with a Charcot-Marie-Tooth type 2B (CMT2B) phenotype, an axonal sensory-motor neuropathy with pronounced ulcero-mutilations. In SPTLC1, we detected a novel mutation (S331F) corresponding to a previously unknown severe and early-onset HSAN phenotype. No mutations were found in NGFB, CCT5 and NGFR. Overall disease-associated mutations were found in 19% of the studied patient group, suggesting that additional genes are associated with HSAN. Our genotype–phenotype correlation study broadens the spectrum of HSAN and provides additional insights for molecular and clinical diagnosis.
Keywords: HSAN, SPTLC1, RAB7, WNK1/HSN2, NTRK1
Introduction
Hereditary sensory and autonomic neuropathies (HSAN) are a clinically and genetically heterogeneous group of inherited peripheral neuropathies, primarily affecting the peripheral sensory and autonomic neurons (Dyck, 1993). Patients usually exhibit prominent distal sensory loss with manifest insensitivity to pain in some. The prominent distal sensory loss frequently leads to chronic ulcerations in feet and hands, sometimes resulting in severe complications such as extensive soft tissue infections, osteomyelitis necessitating amputations of toes and fingers or, in rare instances, even of more proximal parts of the extremities (Dyck, 1993). Autonomic dysfunction, such as anhidrosis, fever, blood pressure fluctuations and gastro-intestinal disturbances are present in some patients. Electrophysiologically, axonal nerve damage of the sensory neurons is often found, but additional demyelination may also be present (Auer-Grumbach et al., 2003).
HSAN can be transmitted as an autosomal dominant (AD) or autosomal recessive (AR) trait. Isolated patients have also been described (Dyck, 1993; Auer-Grumbach, 2004). The AD types of HSAN usually present in the second or third decade of life with marked sensory involvement and minimal autonomic and variable motor involvement, while AR HSAN present either as congenital syndromes with striking sensory and autonomic abnormalities or as almost pure autonomic disorders (Verpoorten et al., 2006a).
A classification of the hereditary sensory neuropathies into types HSAN I–V (Dyck, 1993) was made based on age at onset, inheritance pattern and additional features. Although the clinical classification of these HSAN types is based on a small number of individuals, it still stands after the molecular characterization of the subtypes in recent years. There is variable motor involvement in the AD form of HSAN, making the distinction with hereditary motor and sensory neuropathies (HMSN) or Charcot-Marie-Tooth disease (CMT) difficult. In CMT2B, sensory loss and the associated ulcerations are such prominent phenotypic features that inclusion within the HSAN-spectrum is justified (Vance et al., 1996; Verpoorten et al., 2006a). However, due to the concomitant motor involvement with distal muscle atrophy and weakness, this phenotype was originally classified as HMSN (Kwon et al., 1995).
So far, seven genes have been identified for the different types of HSAN (http://www.molgen.ua.ac.be/CMTMutations/). Two genes have been associated with AD HSAN: missense mutations in serine palmitoyltransferase long chain subunit 1 (SPTLC1) are found in families and individuals with HSAN type I, an adult-onset sensory neuropathy (Bejaoui et al., 2001; Dawkins et al., 2001). Mutations in the small GPTase late endosomal protein RAB7, cause CMT2B (Verhoeven et al., 2003; Meggouh et al., 2006). Mutations in the WNK1/HSN2 gene [protein kinase with-no-lysine(K)-1/hereditary sensory neuropathy type 2] cause AR HSAN type II, an early-onset ulcero-mutilating sensory neuropathy (Lafreniere et al., 2004). HSAN type III, also known as Familial Dysautonomia or Riley–Day syndrome, presents with typical prominent autonomic manifestations early in life and is caused by mutations in the inhibitor of kappa-light polypeptide gene enhancer in B cells, kinase complex associated protein (IKBKAP) (Slaugenhaupt et al., 2001). Mutations in neurotrophic tyrosine kinase, receptor type 1 (NTRK1) are reported in families with congenital insensitivity to pain, anhidrosis and mental retardation (CIPA or HSAN type IV) (Indo et al., 1996). HSAN type V, a phenotype closely related to CIPA but with normal mental development and less pronounced anhidrosis, can be caused by mutations in nerve growth factor beta (NGFB) (Einarsdottir et al., 2004) but also by NTRK1-mutations (Houlden et al., 2001; Einarsdottir et al., 2004). Apart from these six HSAN subtypes other forms with distinct additional features exist, e.g. HSAN with gastroesophageal reflux and cough (Kok et al., 2003) and HSAN with spastic paraplegia (Bouhouche et al., 2006b). Recently, the gene for this last form has been identified as cytosolic chaperonin-containing t-complex peptide-1 (CCT5) (Bouhouche et al., 2006a). The identification of causative genes for the HSAN forms in recent years has provided preliminary insights in the pathogenesis of these rare neuropathies although the fundamental underlying pathomechanisms still remain to be unveiled (Verhoeven et al., 2006).
In this study, we investigated a cohort of 100 familial and isolated patients who had a clinical diagnosis compatible with any of the subtypes of HSAN listed above, and we determined the contribution of mutations in the known genes associated to the distinct phenotypes. IKBKAP was not screened since our cohort did not contain patients with familial dysautonomia. The cohort included 16 index patients of families that have previously been reported in manuscripts describing novel genes and related phenotypes. We broadened the screening of the individual genes to non-associated phenotypes in order to establish potential new genotype–phenotype correlations. Furthermore, we performed the first large-scale mutation screening of WNK1 and CCT5. Additionally, we screened the functional candidate gene NGFR (p75/NTR) because of its importance in development and function of sensory neurons (Lee et al., 1992).
Patients and Methods
Selection criteria
For this study, we selected a group of 100 individuals from our patient database. These were individuals presenting with a clinical phenotype compatible with any of the HSAN subforms described earlier (Dyck, 1993; Auer-Grumbach et al., 2006). The majority of patients presented with progressive distal sensory loss, often associated with one or several of the following additional features: skin changes (e.g. hyperkeratosis, ulcerations), spontaneous fractures, amputations and autonomic features. Because variable motor involvement under the form of distal muscle wasting and weakness can be present in some subtypes of sensory neuropathies, we also included a group of patients diagnosed with CMT2B, a variant of axonal motor and sensory neuropathy (HMSN II), with prominent ulcero-mutilations (Vance et al., 1996; Verpoorten et al., 2006a). To avoid inclusion of classic axonal CMT variants unrelated to the HSAN spectrum, we only included patients with motor and sensory neuropathies if their clinical presentation was complicated by the development of ulcerations. Overall, the cohort in this study could be described as a group of hereditary ulcero-mutilating and sensory neuropathies. Autonomic symptoms were only seen in patients who also presented with sensory abnormalities. Our cohort did not contain patients with predominant or pure dysautonomia, the hallmark feature of HSAN type III or Riley–Day syndrome. Diagnosis was based on clinical presentation, complemented with nerve conduction velocity (NCV) measurements and EMG. No strict electrophysiological selection criteria were applied to our cohort, given the broad range of electrophysiological features associated with the various HSAN phenotypes. Typically, a predominantly sensory axonal neuropathy was found, which was often more severe in the lower limbs. Occasional electrophysiological signs of demyelination can also be found in HSAN. Electrophysiological abnormalities in motor nerves such as reduced amplitudes of compound muscle action potentials and slightly reduced motor NCV can be found, illustrating the overlap between HSAN and HMSN. In CIPA patients, nerve conduction studies can be within normal range (Shatzky et al., 2000; Auer-Grumbach et al., 2003; Axelrod and Gold-von Simson, 2007). In several patients, nerve and skin biopsies were performed.
Patient cohort
The cohort consisted of 100 index patients who were referred to our laboratory for molecular genetic testing in the context of HSAN. Genomic DNA samples were provided through Neurologic and Paediatric Departments and Neuromuscular Centres worldwide. The majority of samples were of European origin. In 43 patients, autonomic features were noted, 44 had a pure sensory neuropathy and the remaining 13 were diagnosed as sensory-motor neuropathy with ulcero-mutilations. In two patients, the HSAN phenotype presented with an associated spastic paraplegia. For 21 out of 100 index patients, a dominant inheritance pattern, based on a parent to child transmission, could be determined. For eight index patients, a recessive inheritance pattern characterized by the presence of affected siblings in the pedigree could be determined. Twenty-four patients were referred as ‘isolated’ since they did not have a familial history of neuropathy in the first- and second-degree relatives. No family history was available for the remaining 47 patients. In five index patients, there was a clear indication of consanguinity of the parents. For 44 patients, detailed information about the age at onset was available; in nine patients, first symptoms occurred in the first year of life, in 11 patients the disease started in the first decade and in 12 patients in the second decade. In the remaining 12 patients, onset was after the age of 20 years. In 35 patients, ulcerations were present and 17 out of them displayed additional complications such as osteomyelitis or amputations. Detailed electrophysiological data were available for 37 patients, the remaining patients had an electrophysiological evaluation by their referring physician but detailed information was not available. In 17 patients, a nerve biopsy or a combined sural nerve/muscle biopsy was performed showing abnormalities compatible with a diagnosis of HSAN. All referring clinicians were neurologists, orthopaedic surgeons or paediatricians active in the field of rare neuromuscular diseases and well acquainted with the clinical presentation of HSAN and related phenotypes. The referring clinicians obtained informed consent from all patients or their legal representatives prior to enrolment in this study.
Molecular genetic analysis
All DNA samples were amplified using the whole genome amplification kit ‘GenomiPhi V2 DNA Amplication Kit’ (GE Healthcare, Waukesha, USA). The protocol was performed according to the manufacturer's instruction.
The coding regions and exon–intron boundaries up to 100 bp up- and downstream of the exons of SPTLC1, RAB7, WNK1/HSN2, NTRK1, NGFB, CCT5 and NGFR were PCR-amplified using primer oligonucleotides designed with the Primer3 and SNPbox software tools (Rozen and Skaletsky, 2000; Weckx et al., 2004). PCR conditions are available upon request. PCR products were cleaned up using the Exonuclease I-Shrimp Alkaline Phosphatase enzyme (USB, Cleveland, USA). Mutation screening was performed by direct sequencing of the purified PCR fragments using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, USA). Fragments were separated on an ABI3730xl DNA Analyser (Applied Biosystems, Foster City, USA). The resulting sequences were aligned and analysed with the novoSNP (Weckx et al., 2005) and SeqMan™II (DNASTAR Inc., Madison, USA) programs. The nucleotide numbering of the genes is relative to the ATG translation initiation site with A as +1 of the corresponding cDNA sequences (SPTLC1: NM_006415.2; RAB7: NM_004637.5; NTRK1: NM_002529.3; NGFB: NM_002506.2; CCT5: NM_012073.3; NGFR: NM_002507). The nucleotide numbering of WNK1/HSN2 is relative to the first nucleotide of the HSN2-specific exon of WNK1 (NM_213665.1). Mutations are described according to the latest conventions on the nomenclature of DNA sequence variants (http://www.hgvs.org/mutnomen). Sequence variants were confirmed by repeated PCR on original DNA samples and bidirectional sequencing. Where possible, segregation of the mutation with the disease phenotype was analysed in the family.
Genotyping and paternity testing
Paternity was tested using 15 highly informative short tandem repeats (STRs) distributed throughout the genome (ATA38A05, D1S1646, D1S1653, D1S1360, D2S2256, D3S3037, D4S2382, D4S3240, D7S509, D8S1759, D9S1118, D12S1056, D12S2082, D16S2619 and GATA152H04). STRs were PCR-amplified and PCR fragments were loaded on an ABI3730xl DNA Analyser. Genotypes were analysed using Local Genotype Viewer, a software program developed in-house (http://www.vibgeneticservicefacility.be/).
Analysis of exon skipping
Analysis of exon skipping was performed by RT-PCR on mRNA isolated from lymphoblast cell lines of patients CMT-841.01 and CMT-886.01. mRNA was first purified from peripheral blood lymphoblasts using the RNeasy mini kit (Qiagen, Hilden, Germany). DNA inactivation was performed using the Turbo DNA free kit (Ambion, Austin, USA) and subsequently analysed for splicing defects by RT-PCR and sequencing using exonic primers located in exons 1 and 5 for patient CMT-886.01 (forward: 5′-CTGCTGGCTTGGCTGATACT-3′, reverse: 5′-CACTGCAGCTTCTGTTCAGG-3′) and exonic primers in exons 12 and 16 for CMT-841.01 (forward: 5′-CCTTGTGCTCAACAAATGTGG-3′, reverse: 5′-AGCCAGCAGCTTGGCAT-3′) (Superscript III First-Strand Synthesis System for RT-PCR; Life Technologies, San Diego, USA).
Results
In 19 index patients, out of a cohort of 100, pathogenic mutations were found in four HSAN disease associated genes: SPTLC1, RAB7, NTRK1 and WNK1/HSN2. These mutations were absent from 600 European control chromosomes. No pathogenic variations could be detected in NGFB, CCT5 and NGFR. Clinical and electrophysiological data on these 19 index patients are summarized in Tables 1 and 2.
Table 1.
Clinical features of HSAN patients with proven mutation
| Patient | Gene | AA change | Origin | Diagnosis | Inheritance | AAO | SAO | ALE | Sensory loss | Skin changes | Amputations | Bone complications | Autonomic symptoms |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CMT-186.05 | SPTLC1 | Ala352Val | Austria | HSAN1 | IC | 16y | Sensory abnormalities | 46y | Severe LL | – | − | − | − |
| CMT-791.01 | SPTLC1 | Ser331Phe | France (Gypsy) | HSAN/CIPA | IC | cong | Insensitivity to pain | 7y | Severe loss of superficial touch distal LL, insensitivity to pain | eschar and ulceration foot | – | – | Gastro- oesophageal reflux |
| PN-626.01 | RAB7 | Val162Met | Belgium | CMT2B | AD | 37y | Osteomyelitis toes | 54y | Diminished L>R | Ulcerations toes | + (toes) | Osteomyelitis in the toes | – |
| CMT-90.01 | RAB7 | Val162Met | UK | CMT2B | AD | 28y | Ulcerations toes | 64y | Severe in distal LL, position sense conserved, UL less pronounced | Ulcerations toes | + (all toes R foot) | Unk | – |
| CMT-126.01 | RAB7 | Leu129Phe | Austria | CMT2B | AD | 15y | Weakness in LL | 75y | Severe | Ulcerations feet | – | Osteomyelitis and -necrosis | – |
| CMT-140.01 | RAB7 | Leu129Phe | Austria | CMT2B | AD | 13y | Ulcerations toes | 46y | Moderate | Ulcerations and foot callus | + (toes) | Osteomyelitis and -necrosis | – |
| CMT-186.26 | RAB7 | Leu129Phe | Austria | CMT2B | AD | 20y | Ulcerations toes | 49y | Severe in LL (toes and feet) | Ulcerations toes | – | Osteomyelitis and -necrosis | – |
| CMT-186.28 | RAB7 | Val162Met | Austria | CMT2B | AD | 15y | Steppage gait | 34y | No clinical sensory loss | Multiple ulceration toes | + (lower leg L, several toes R) | Osteomyelitis | – |
| CMT-195.01 | RAB7 | Val162Met | USA | CMT2B | AD | ad | Gait disturbances | Unk | Distal LL | Ulcerations feet | Unk | Unk | – |
| CMT-178.01 | HSN2 | Ile355AsnfsX7 | Belgium | HSAN2 | IC | <2y | Ulcerations hand and feet, poor healing | 5y | Distally severely reduced for all modalities, LL>UL | Ulcerations toes and fingers | – | Spontaneous fractures LL + Charcot atropathy | – |
| CMT-260.01 | HSN2 | Gln184X | Austria | HSAN2 | IC | inf | Clumsiness hands, osteomyelitis in the foot | 50y | Distally severely reduced for all modalities, LL>UL | Ulcerations | + (fingers and toes) | Osteomyelitis in the foot | – |
| CMT-451.04 | HSN2 | Pro85ProfsX14 + Gln364SerfsX16 | Italy | HSAN2 | IC | <1y | Difficulties in hand manipulation | 33y | Distally severely reduced for all modalities, LL>UL | Ulcerations in hands and feet | + (progressive spontaneous amputations hands, and LL to knees) | Painless pathologic fractures LL | Unk |
| CMT-179.01 | NTRK1 | Gly181GlyfsX16 | Belgium | HSAN4/CIPA | IC | cong | Delayed motor milestones? | 12y | Insensitivity to pain | Thickening and ulcerations LL | – | – | Anhidrosis |
| CMT-197.01 | NTRK1 | c.359+ 5G>T aberrant splicing | Belgium | HSAN4/CIPA | IC | <8m | Fever of unknown cause | 14y | Insensitivity to pain | Unk | – | – | Anhidrosis, fever episodes |
| CMT-366.01 | NTRK1 | Arg761Trp | The Netherlands | HSAN4/CIPA | IC | cong | Anhidrosis, hyperthermia, hypotonia | 3y | Insensitivity to pain | Unk | – | Painless fracture right calcaneus | Anhidrosis, fever episodes |
| CMT-826.01 | NTRK1 | c.2046+ 3 A>C aberrant splicing | Spain | HSAN4/CIPA | AR | 5m | Skin ulcerations | 20y | Insensitivity to pain and temperature | Hyperkeratosis, ulcerations | + (toes) | Osteomyelitis | Anhidrosis, fever episodes |
| CMT-841.01 | NTRK1 | Arg565Gln | The Netherlands (Moroccan origin) | HSAN4/CIPA | AR (cons) | 4 yrs | Painless tibial fracture, poor healing | 35y | Insensitivity to pain | – | – | Multiple factures (pelvic bone, upper and lower limbs and foot bones), osteomyelitis | Anhidrosis |
| PN-1192.03 | NTRK1 | Gln626GlnfsX7 | The Netherlands (Moroccan origin) | HSAN4/CIPA | IC (cons) | cong | Hypotonia, recurrent episodes of fever | 2y | Insensitivity to pain | Keratodermatitis, xerodermia, selfmutilation of fingers and tongue, necrotizing fasciitis of right hand | – | – | Anhidrosis, fevers |
| CMT-886.01 | NTRK1 | c.354_359+3 delTCGCCTGAA aberront splicing | Turkey | HSAN4/CIPA | IC | 5m | Fever, recurrent infections | 7y | Insensitivity to pain | Hyperkeratosis, ulcerations, nail dystrophy, automutilations, neck abscess | – | Avascular necrosis left talus | Anhidrosis |
AA = amino acid; AAO = age at onset (years); SAO = symptoms at onset; ALE = age at last exam (y); cong = congenital; inf = infancy; ad = adulthood; IC = isolated case; AD = autosomal dominant; AR = autosomal recessive; Fam = familial; cons = consanguineous; LL = lower limb; UL = upper limb; + = present; − = absent; unk = unknown; dist = distal; prox = proximal; R = right; L = left.; y = year; m = month.
Table 2.
Clinical features of HSAN patients with proven mutation
| Patient | Gene/AA change | Foot deformities | Walking difficulties | Weakness | Atrophy | Reflexes | Mental retardation | Nerve conduction studies + electromyography |
Additional features | Reference | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensory | Motor | ||||||||||
| CMT-186.05 | SPTLC1 Ala352Val | Mild pes cavus | Steppage | Distal LL | Peroneal | +, Ach absent | – | Axonal loss UL, absent responses LL | Absent responses LL | Severe hypesthesia and spontaneaous lancinating pain LL | Novel family |
| CMT-791.01 | SPTLC1 Ser331Phe | Pes cavus/equinovarus | + | + (global) | Global amyotrophy UL and LL | ↓/− | +, microcephaly | Absent responses UL/LL | Absent responses UL/LL | Severe growth retardation, hypotonia, joint hyperlaxity, vocal cord paralysis, bilateral cataract, respiratory involvement with sleep apnea requiring non-invasive ventilation | Novel family |
| PN-626.01 | RAB7 Val162Met | – | Steppage L>R | Distal and proximal LL | Unk | ↓ (L<R) | – | Axonal loss UL, absent responses LL | Normal UL, absent responses LL | Brown-Séquard syndrome and quadriplegia due to bleeding cervical spinal angioma, IgA nephropathy | (Verhoeven et al., 2003) |
| CMT-90.01 | RAB7 Val162Met | Multiple toe amputations | Steppage | Distal in LL | Peroneal | ↓ to − | – | Axonal loss UL, absent responses LL | UL normal, absent responses LL | – | (Verhoeven et al., 2003) |
| CMT-126.01 | RAB7 Leu129Phe | Pes cavus | + (severe) | Distal LL>UL | Peroneal | ↓ | – | Axonal loss UL, absent responses LL | Axonal loss UL, absent responses LL | – | (Verhoeven et al., 2003) |
| CMT-140.01 | RAB7 Leu129Phe | Pes cavus | Mild steppage gait | Distal UL and LL, mild | Mild in UL and in LL | ↓ | – | Normal UL, axonal loss LL | Axonal- demyelinating NCVs in UL and LL | – | (Verhoeven et al., 2003) |
| CMT-186.26 | RAB7 Leu129Phe | Charcot foot deformity | Mild steppage gait | Distal LL, mild | Distal LL, small foot muscles | ↑ | – | Normal UL, axonal loss LL | Normal UL, axonal loss LL | – | (Verhoeven et al., 2003) |
| CMT-186.28 | RAB7 Val162Met | Toe deformities | Steppage, after amputation, wheelchair dependent | Distal LL>UL | Peroneal and hands | + to ↑ | – | Absent responses LL | Absent responses LL | – | (Verhoeven et al., 2003) |
| CMT-195.01 | RAB7 Val162Met | Pes cavus, hammer toes | Steppage | Distal LL | Unk | ↓ | – | Axonal loss | Axonal loss | – | (Verhoeven et al., 2003) |
| CMT-178.01 | HSN2 Ile355AsnfsX7 | – | – | – | – | – | – | Absent responses UL/LL | Normal | – | (Coen et al., 2006) |
| CMT-260.01 | HSN2 Gln184X | Deformed due to recurrent infections | Disturbed due to amputations | – | – | ↓ to − | – | Absent responses UL/LL | UL normal, LL axonal loss | – | (Coen et al., 2006) |
| CMT-451.04 | HSN2 Pro85HisfsX14 + Gln364SerfsX16 | – | Disturbed due to amputations | – | – | – | – | Absent responses UL/LL | Normal | Sural nerve biopsy: complete loss of myelinated fibers, endoneuronal fibrosis | (Coen et al., 2006) |
| CMT-179.01 | NTRK1 Gly181GlyfsX16 | – | – | – | – | + | + | Unk | Unk | Henoch-Schönlein vasculitis, pseudotumor cerebri, tooth abscesses | (Verpoorten et al., 2006b) |
| CMT-197.01 | NTRK1 c.359+5G>T | Unk | Unk | – | – | ↓ | + | Unk | Unk | Skin biopsy: innervation of blood vessels and eccrine sweat glands absent | (Verpoorten et al., 2006b) |
| CMT-366.01 | NTRK1 Arg761Trp | Charcot joint on the right ankle | Normal | – | – | + | Unk | Unk | Unk | – | (Verpoorten et al., 2006b) |
| CMT-826.01 | NTRK1 c.2046+3A>C | Charcot joint on both ankles and knees | Disturbed due to Charcot joint | – | – | ↓ | + (mild) | Axonal loss LL | Axonal loss UL/LL | Corneal opacities | Novel family |
| CMT-841.01 | NTRK1 Arg565Gln | Charcot deformity right ankle and knee | Walks with rollator | – | – | ↓ to − | − (younger affected sister has learning difficulties) | Unk | Unk | Sural nerve biopsy: loss of thinly myelinated fibers, partial caudacompression syndrome age 34 yrs. traumatic cataract right eye, hearing loss due to ear drum perforation | Novel family, clinical description previously reported (Kruyt et al., 2007) |
| PN-1192.03 | NTRK1 Gln626GlnfsX7 | – | – | – | – | + | + | Normal | Normal | Skin biopsy: absent innervation of capillaries and sweat glands, loss of myelinated and unmyelinated axons | (Verpoorten et al., 2006b) |
| CMT-886.01 | NTRK1 c.354_359+ 3 delTCGCCTGAA | – | Delayed motor milestones | – | – | + | + | Normal | Normal | Recurrent infections respiratory and gastrointestinal tract due to hypogammaglobulinemia, recurrent hip dislocation left | (Kilic et al., 2009) |
AA = amino acid; LL = lower limb; UL = upper limb; + = present; − = absent; ↓ = decreased; ↑ = increased; unk = unknown; R = right; L = left; Ach = Achilles tendon.
Mutations in SPTLC1
In Patient CMT-791.01, we detected a heterozygous missense mutation (c.992C>T; p.Ser331Phe), which was absent in both healthy parents (Fig. 1A). Paternity was confirmed in this family, pointing to a de novo mutation. In contrast to previously reported HSAN type I patients, the patient displayed a severe phenotype characterized by congenital onset with severe growth and mental retardation, hypotonia and vocal cord paralysis. In Patient CMT-186.05 a missense mutation (c.1055C>T; p.Ala352Val) was identified (Fig. 1B). A third heterozygous missense mutation (c.1160G>C; p.Gly387Ala) was found in Patient CMT-155.01 and her twin sister CMT-155.02 (Verhoeven et al., 2004). This mutation was also found in Patient CMT-820.01. However, the healthy mother of this index patient has the same variant in the homozygous state. This finding suggests that the Gly387Ala variation is not pathogenic, but a rare polymorphism. This has recently been confirmed by the analysis of serine palmitoyl transferase (SPT) activity by measuring the incorporation of [U-13C]-L-serine in protein extracts from stably transfected HEK293 cells and complementation testing using an SPTLC1-deficient CHO cell line (Hornemann et al., 2009).
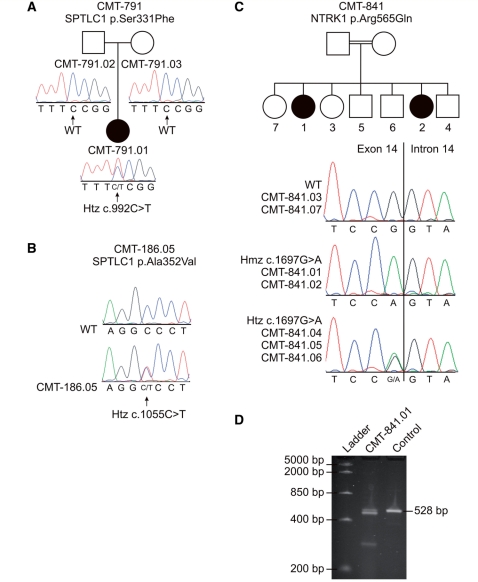
Figure 1.
Segregation of p.Ser331Phe and p.Ala352Val missense mutations in SPTLC1 (A and B) and segregation and cDNA analysis of the p.Arg565Gln missense mutation in NTRK1 (C and D). Segregation analysis of the p.Ser331Phe missense mutation in SPTLC1 reveals that this mutation occurred de novo (A). Panel B shows the sequence trace file of the p.Ala352Val missense mutation found in SPTLC1 in an isolated Patient CMT-186.05. Segregation of the p.Arg565Gln mutation in NTRK1 is shown in panel C. Two CIPA patients in family CMT-841 (CMT-841.01 and CMT-841.02) had a homozygous Arg565Gln mutation in NTRK1. The healthy siblings of these patients had either the wild-type allele (CMT-841.03 and CMT-841.07) or carried the Arg565Gln mutation in the heterozygous state (CMT-841.04, CMT-841.05 and CMT-841.06). The parents of the patients were first cousins. The mutated nucleotide (c.1697G > A) was the last nucleotide of exon 14, which could affect proper splicing of this exon. cDNA analysis of CMT-841.01 showed the absence of the expected band (528 bp), which was present in the control and confirmed by direct DNA sequencing (D). We could not determine the sequence of the three lower bands present in the patient. square = male, circle = female, black filled symbol = affected, empty symbol = unaffected.
Mutations in RAB7
In our cohort, two different missense mutations in RAB7 have been identified in seven anamnestically unrelated index patients. The index Patients CMT-90.01 and CMT-195.01 of two multigenerational pedigrees as well as two additional Patients with a positive familial history (CMT-186.28 and PN-626.01) carried the same heterozygous transition c.484G>A resulting in a Val162Met missense mutation. The c.385C>T (p.Leu129Phe) missense mutation was detected in three Austrian index Patients: CMT-126.01, CMT-140.01 and CMT-186.26. Additional haplotype analysis revealed that the Val162Met mutation arose independently in the reported families/patients, but the Leu129Phe mutation resides on a common disease haplotype indicating a founder effect (Verhoeven et al., 2003). All index patients carrying a RAB7 mutation present with an adolescent or adult-onset HMSN II phenotype and are characterized by distal atrophy and weakness in the lower limbs with pronounced distal sensory loss complicated by ulcerations and amputations.
Mutations in WNK1/HSN2
A recent report showed that HSN2 is a nervous system-specific exon of the WNK1-gene. A compound heterozygous mutation in WNK1 and HSN2 was identified as the cause for HSAN type II (Shekarabi et al., 2008). In view of this recent finding, the entire coding region of WNK1 was screened in our patient cohort. No additional disease-related sequence variants were identified outside of the HSN2 exon.
We identified a total of four different WNK1/HSN2 mutations in three previously reported patients: one compound heterozygous mutation consisting of a 1bp-deletion resulting in a frameshift mutation with a premature stop codon (c.254delC; p.Pro85HisfsX14) and an insertion of a thymine (c.1089_1090insT) predicted to cause a frameshift mutation with premature stop codon (p.Gln364SerfsX16) in Patient CMT-451.01. One homozygous non-sense mutation (c.550C>T; p.Gln184X) and one homozygous 2-bp deletion (c.1064_1065delTC) predicted to cause a frameshift and premature stop codon (p.Ile355AsnfsX7) were detected in Patients CMT-260.01 and CMT-178.01, respectively (Coen et al., 2006).
Mutations in NTRK1
In our cohort, seven mutations in NTRK1 were found, of which one was a novel homozygous missense mutation (c.1697G>A; p.Arg565Gln) in Patient CMT-841.01 (Tables 1 and 2). An affected sibling CMT-841.02 carried the same homozygous mutation (Fig. 1C). This mutation targets the tyrosine kinase domain of the neurotrophin tyrosine kinase receptor (Fig. 2). The mutated nucleotide is the last base pair of exon 14, resulting in aberrant splicing (Fig. 1D). Furthermore, we identified a known 9 bp-deletion (c.354_359 + 3delTCGCCTGAA) (Tuysuz et al., 2008) in a recently reported Turkish Patient (CMT-886.01) (Kilic et al., 2009). This deletion spans the splice-donor site of exon 3. cDNA analysis revealed two splice variants; one with skipping of exon 3 and the second with skipping of exons 2 and 3. These exons contain a part of the leucine-rich motif of NTRK1, important for ligand binding and signal transduction. The patient displayed a CIPA phenotype complicated by recurrent infections secondary to hypogammaglobulinemia, a feature not previously known to be associated with CIPA.
Figure 2.
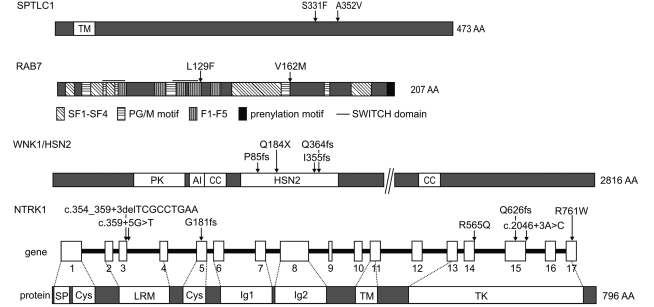
Schematic presentation of protein structures of SPTLC1, RAB7 (Pereira-Leal and Seabra, 2000), WNK1/HSN2 and NTRK1 (Indo, 2001) with mutations identified in this study causing HSAN.
Six additional known mutations were found in six index patients. Homozygous splice site mutations were found in Patients CMT-197.01 (c.359+5G>T) and CMT-826.01 (IVS15 +3A>C). Homozygous frameshift mutations were present in Patients PN-1192.03 (c.1877-1878insA; p.Gln626GlnfsX7) and CMT-179.01 (c.543delG; p.Gly181GlyfsX16). Finally, we identified a homozygous splice site mutation in Patient CMT-197.01 (c.359 + 5G>T) and a homozygous missense mutation (c.2281C>T; p.Arg761Trp) in Patient CMT-366.01 (Verpoorten et al., 2006b). Haplotype analysis suggested a common founder effect with previously described patients carrying this missense mutation (Indo et al., 2001).
Discussion
In this study, we investigated a cohort of 100 HSAN patients and determined the relative contribution of mutations in six genes known to be involved in various forms of HSAN (SPTLC1, RAB7, WNK1/HSN2, NTRK1, NGFB and CCT5). In addition, we studied the functional candidate gene NGFR. The known subforms of HSAN and their most important clinical characteristics have been summarized in Table 3. In four genes (SPTLC1, RAB7, WNK1/HSN2, NTRK1), we identified disease-causing mutations in 19 index patients representing a mutation frequency of 19%. Only nine of these patients had a clear familial history suggestive of HSAN with the remaining 10 being isolated patients. This results in a relative mutation frequency of 31% (9/29) for familial patients and 14% (10/71) for isolated patients. In the group of dominantly inherited HSAN (RAB7 and SPTLC1), the mutation frequency is 33% (7/21) and for recessive HSAN (WNK1/HSN2 and NTRK1), the frequency is 25% (2/8). Conversely, 20 familial patients in our screening cohort (14 dominant and 6 recessive) remain unsolved. These findings clearly indicate that additional genes must be involved in the pathogenesis of HSAN. RAB7 and NTRK1 were the most frequently mutated genes in our cohort (both 7%), followed by WNK1/HSN2 with 3% and SPTLC1 with 2%.
Table 3.
Overview of HSAN types with corresponding gene/locus, inheritance pattern, cardinal phenotypic features and references to the Online Mendelian Inheritance in Man (OMIM) database and literature
| Type | Gene | Locus | Inh | Clinical features | AAO | OMIM | Reference |
|---|---|---|---|---|---|---|---|
| HSAN I | SPTLC1 | 9q22.2 | AD | Predominant loss of pain and temperature sensation, preservation of vibration sense, lancinating pain, variable distal motor involvementa | Adulta | 162400 | Bejaoui et al., 2001; Dawkins et al., 2001 |
| HSAN IB | unknown | 3p24-p22 | AD | Predominant sensory neuropathy with cough and gastroesophageal reflux, rarely foot ulcerations | Adult | 608088 | Kok et al., 2003 |
| CMT2B | RAB7 | 3q21.3 | AD | Prominent distal motor involvement, sensory loss of all qualities, acro-mutilating complications | Adult | 600882 | Verhoeven et al., 2003 |
| HSAN II | WNK1/HSN2 | 12p13.3 | AR | Prominent sensory loss and mutilations in hands and feet, acropathy | Childhood | 201300 | Lafreniere et al., 2004 |
| HSAN III (Riley–Day syndrome) | IKBKAP | 9q31 | AR | Familial dysautonomia, prominent autonomic disturbances and complications, absence of fungiform papillae of the tongue, alacrimia, excessive sweating | Congenital | 223900 | Slaugenhaupt et al., 2001 |
| HSAN IV (CIPA) | NTRK1 | 1q21-22 | AR | No or reduced response to painful stimuli, anhidrosis, episodic fever, mild mental retardation, skin and cornea lesions, joint deformities, hypogammaglobulinemia in one patient (this study) | Congenital | 256800 | Indo et al., 1996 |
| HSAN V | NGFB (NTRK1 in rare cases) | 1p13.1 (1q21-22) | AR | Congenital insensitivity to pain, severe loss of deep pain perception, painless fractures, joint deformities, normal intelligence | Congenital | 608654 | Einarsdottir et al., 2004 (Houlden et al., 2001) |
| HSAN with spastic paraplegia | CCT5 | 5p15-p14 | AR | Prominent sensory neuropathy with sensory loss of all qualities, mutilating acropathy, spastic paraplegia. | Early childhood | 256840 | Bouhouche et al., 2006a |
The three mutations previously described in SPTLC1 (Cys133Trp, Cys133Tyr and Val144Asp) are associated with an ulcero-mutilating sensory neuropathy with a spectrum of clinical and electrophysiological features that is variable within and between families (Bejaoui et al., 2001; Dawkins et al., 2001; Auer-Grumbach, 2004; Houlden et al., 2006). A fourth SPTLC1 mutation (Gly387Ala) was recently shown not to be disease causing (Verhoeven et al., 2004; Hornemann et al., 2009).
In the present study, we describe a novel SPTLC1 mutation (Ser331Phe) in a patient who presented with a severe congenital phenotype. This mutation occurred de novo, was absent from 600 control chromosomes and affected a highly conserved amino acid. So far, all known missense mutations are located within a 12-amino acid segment encoded by exons 5 and 6 of SPTLC1. The Ser331Phe mutation is located downstream of this segment (Fig. 2). It is possible that the Ser331Phe mutation, due to its different location in the protein, exerts a different effect on SPT activity leading to a more severe phenotype. These results suggest a broadening of the phenotype associated with HSAN type I. We identified a second sequence variant (Ala352Val) in SPTLC1, in an isolated patient with a sensory neuropathy. However, the pathogenicity of this sequence variant could not be verified because DNA of the family members was not available for segregation analysis. Furthermore, the amino acid targeted is not well-conserved in evolution and the change from alanine to valine is mild considering their similar chemical properties.
We found mutations in SPTLC1 in only 2% of our patients confirming the rare occurrence of SPTLC1 mutations in isolated and familial HSAN, as found in previous studies (Klein et al., 2005; Houlden et al., 2006).
To date, four missense mutations (Leu129Phe, Lys157Asn, Asn161Thr and Val162Met) have been reported in RAB7 (Verhoeven et al., 2003; Houlden et al., 2004b; Meggouh et al., 2006). In our cohort, we found the Leu129Phe mutation in two families (CMT-126 and CMT-140) and one isolated Patient (CMT-186.26), and the Val162Met mutation in families CMT-90 and CMT-195 and in two additional patients with a positive family history (CMT-186.28 and PN626.01) (Verhoeven et al., 2003). The patients carrying the Leu129Phe mutation were all of Austrian descent and shared a common disease haplotype, indicating a founder effect. However, the patients with the Val162Met mutation did not share a common haplotype, suggesting independently arising mutations in the same residue, possibly pointing towards a mutational hotspot for RAB7. This mutation is located in a highly conserved domain, important for the formation of the nucleotide-binding site of RAB7 (Fig. 2). The functional importance of this domain in peripheral neuron integrity is further underlined by the identification of mutations in adjacent amino acids (Lys157Asn and Asn161Thr) in patients with CMT2B (Houlden et al., 2004b; Meggouh et al., 2006).
In our cohort, RAB7 mutations were exclusively found in patients diagnosed with HSMN II, with ulcero-mutilations also known as CMT2B. Apart from the rare exception of a patient with a typical HSAN type I phenotype carrying the Asn161Thr RAB7 mutation (Houlden et al., 2004b), the phenotype associated with RAB7 mutations seems to be largely confined to HMSN II (CMT2B). Because of the marked motor involvement, RAB7-neuropathy was originally classified as hereditary motor and sensory neuropathy type 2B (CMT2B) (Kwon et al., 1995). Due to the prominent presence of ulcerations, however, CMT2B should be considered part of the spectrum of HSAN (Vance et al., 1996).
The mutation frequency in the CMT2B-subgroup of our study cohort was very high (7 out of 13 CMT2B patients). The high mutation frequency of RAB7 found in the present study contrasts with a previous report (Klein et al., 2005), where the known RAB7 mutations were shown to be absent from a group of 25 families with adult-onset HSAN I or HMSN II with prominent sensory involvement and from an additional 92 idiopathic patients. The frequency of the RAB7 mutation in our cohort remained high, even when the founder effect of the Leu129Phe mutation was taken into account.
The phenotype associated with mutations in WNK1/HSN2 is a severe AR ulcero-mutilating sensory neuropathy with mild autonomic disturbances beginning in early childhood (Axelrod and Gold-von Simson, 2007). So far, 11 different non-sense and frameshift mutations in WNK1/HSN2 have been reported in the literature, all resulting in a complete loss of protein. Recently, it was shown that HSN2 is not a separate gene residing in intron 8 of WNK1 but in fact is a neuron-specific exon of WNK1 itself, with high expression in dorsal root ganglia (DRG) and sciatic nerves (Shekarabi et al., 2008). In our cohort, we identified four loss-of-function mutations in WNK1/HSN2, in three patients, all of which resided in the HSN2 exon (Fig. 2) (Coen et al., 2006). From the first large-scale screening of WNK1 in HSAN patients performed in this study, we can conclude that mutations outside of the HSN2 exon are likely to be rare.
The most frequently mutated gene in this study, together with RAB7, was NTRK1. The NTRK1 protein is a receptor tyrosine kinase, which is phosphorylated in response to nerve growth factor (NGF), supporting survival of sympathetic ganglion neurons and nociceptive sensory neurons in DRG (Levi-Montalcini, 1987). To date, more than 40 different missense, non-sense, frameshift and splice site mutations in NTRK1 have been described in families from various ethnic origins (http://www.molgen.ua.ac.be/CMTMutations/). The corresponding syndrome, CIPA, consists of characteristic features: recurrent episodic fevers due to anhidrosis, absence of reaction to painful stimuli, self-mutilating behaviour and mental retardation (Axelrod and Gold-von Simson, 2007).
In our cohort of 100 HSAN patients, seven different mutations were identified in NTRK1 (Fig. 2), of which six were previously reported (Verpoorten et al., 2006b; Kilic et al., 2009). We identified a previously unreported splice site mutation (Arg565Gln), which comprised the last nucleotide of exon 14 and further broadens the genetic spectrum of NTRK1 mutations. This mutation resides in the tyrosine kinase domain of the NTRK1 receptor, which regulates autophosphorylation of NTRK1 in response to NGF. Both the index patient (CMT-841.01) and her affected sister (CMT-841.02) were diagnosed with CIPA and carried the same homozygous mutation. However, Patient CMT-841.01 had a normal intelligence whereas the affected sib (CMT-841.02) had learning difficulties. The phenotype of these patients has been described previously (Kruyt et al., 2007).
Interestingly, a 9-bp deletion (c.354_359 + 3delTCGCCTGAA), resulting in skipping of exon 3 and of exons 2 and 3, was found in a CIPA patient of Turkish origin (CMT-860.01). This patient presented with a multisystem involvement including recurrent infections due to immunological abnormalities as hypogammaglobulinemia (Kilic et al., 2009). The same 9-bp deletion was recently reported in an unrelated Turkish HSAN type IV patient (Tuysuz et al., 2008). Therefore, this mutation is likely to be a founder mutation in the Turkish population. The phenotype in our patient broadens the clinical spectrum of CIPA.
It has previously been suggested (Indo et al., 2001) that the current literature reveals little to no genetic and clinical heterogeneity in HSAN IV. Although some degree of phenotypic variability was observed in our CIPA-patients with proven NTRK1 mutations, overall the clinical presentation seems to correspond to a readily recognizable syndrome that is indeed genetically homogenous.
The phenotype caused by NGFB mutations (HSAN type V) is similar to the CIPA phenotype (HSAN type IV). So far, only one homozygous missense mutation has been reported, in a recessive Swedish family (Einarsdottir et al., 2004; Minde et al., 2004). Detailed clinical, neurophysiological and genetic analysis of this family revealed that the heterozygous carriers presented with a variable but mild phenotype (Minde et al., 2009). The main difference with CIPA was the absence of obvious mental retardation and less-pronounced anhidrosis. However, one family with the HSAN type V phenotype was described with pathogenic mutations in NTRK1 indicating the overlap between HSAN types IV and V (Houlden et al., 2001, 2004a). These findings underscore the relevance of genetic screenings outside of the known phenotypes and modes of inheritance. In our study cohort, no heterozygous or homozygous sequence variations were found in NGFB confirming the rare occurrence of NGFB mutations in HSAN patients.
Recessive mutations in CCT5 were identified in a consanguineous Moroccan family presenting with HSAN with spastic paraplegia (Bouhouche et al., 2006a). In our cohort, only two HSAN patients presented with an associated spastic paraplegia. However, we did not identify mutations in CCT5. Additional screening of 25 unrelated index patients with hereditary spastic paraplegia with sensory involvement did not reveal any mutations in CCT5 (data not shown). Our results suggest that mutations in CCT5 are a rare cause for HSAN and make it unlikely to find any mutations outside of the known phenotype.
Because of the phenotypical resemblance among Ntrk1−/−, Ngfb−/− and Ngfr−/− knockout mice, the patient cohort was screened for NGFR (p75/NTR) (Lee et al., 1992). No mutations were found in this gene making its contribution to the pathogenesis of HSAN uncertain.
In summary, we examined the distribution of mutations in genes associated with AD and AR forms of HSAN in a large group of familial and sporadic patients. The genotype–phenotype correlations in this study revealed little variability when compared with previous reports, with the sole exception of a de novo SPTLC1 mutation in a severe phenotype with congenital onset. Taken together, these results show that the relevant clinical phenotypes are recognizable and should be used to orient molecular diagnosis. Screening of NTRK1 should be confined to patients presenting with a CIPA phenotype (both AR and isolated patients) as no NTRK1 mutations were found in other HSAN phenotypes. RAB7 screening is mandatory in patients presenting with an AD axonal sensory-motor neuropathy with ulcerations (CMT2B) as we found a high mutation frequency in this subgroup (54% or 7/13). Mutations in WNK1/HSN2 and SPTLC1 seem to be rare in HSAN. We found no mutations in NGFB and CCT5 indicating that these genes are only rarely involved in HSAN. No pathogenic sequence variations were identified in the functional candidate gene NGFR, making its contribution to the pathogenesis of HSAN uncertain. The overall mutation rate was relatively low (19%) suggesting that other genes must be involved in the pathogenesis of HSAN.
At the present time, the precise nature of the mechanism underlying the pathogenesis of the various HSAN forms remains unclear. Although it is particularly challenging to link the different genes, some preliminary disease pathways may already take form. Of special interest are disturbances of vesicular transport. Both RAB7 and SPTLC1 have a function in endocytotic membrane trafficking. In addition, the NGFB/NTRK1 signalling complex, which is critically important in the development and function of nociceptive neurons, is also dependent upon retrograde transport through signalling endosomes (Verhoeven et al., 2006). Future research is needed to improve our still very incomplete understanding of these mechanisms.
Additional descriptions of HSAN families and patients with known or novel genetic defects are needed to further refine the existing classification and to get a better insight into the molecular basis of these disorders.
Funding
University of Antwerp; the Fund for Scientific Research (FWO-Flanders); the Medical Foundation Queen Elisabeth (GSKE); the ‘Association Belge contre les Maladies Neuromusculaires’ (ABMM); the Interuniversity Attraction Poles P6/43 program of the Belgian Federal Science Policy Office (BELSPO); the IZKF RWTH Aachen (TV M3); the German Research Council (DFG; WE1406/13-1); Austrian Science Fond (FWF; P19455-B05); PhD fellowships of the Institute for Science and Technology (IWT; to A.R.) and FWO-Flanders (J.B.).
Acknowledgements
We are grateful to the patients and their families for their willingness to co-operate in our research project. We also wish to thank the Genetic Service Facility (VIB) for the sequencing support (http://www.vibgeneticservicefacility.be/).
Glossary
Abbreviations
- AD
autosomal dominant
- AR
autosomal recessive
- CIPA
congenital insensitivity to pain and anhidrosis
- CMT
Charcot-Marie-Tooth disease
- HMSN
hereditary motor and sensory neuropathy
- HSAN
hereditary sensory and autonomic neuropathy
- NCV
nerve conduction velocity
- STRs
short tandem repeats
- TM
transmembrane domain
- SF
Rab subfamily domain
- PG/M
conserved domain implicated in binding of phosphate/Mg2+ and guanine binding
- F
Ras family domain
- PK
protein kinase domain
- AI
autoinhibitory domain
- CC
coiled coil domain
- SP
signal peptide
- Cys
cysteine cluster
- LRM
leucine rich motif
- Ig
immunoglobulin-like domain
References
- Auer-Grumbach M. Hereditary sensory neuropathies. Drugs Today. 2004;40:385–94. doi: 10.1358/dot.2004.40.5.850487. [DOI] [PubMed] [Google Scholar]
- Auer-Grumbach M, De Jonghe P, Verhoeven K, Timmerman V, Wagner K, Hartung HP, et al. Autosomal dominant inherited neuropathies with prominent sensory loss and mutilations: a review. Arch Neurol. 2003;60:329–34. doi: 10.1001/archneur.60.3.329. [DOI] [PubMed] [Google Scholar]
- Auer-Grumbach M, Mauko B, Auer-Grumbach P, Pieber TR. Molecular genetics of hereditary sensory neuropathies. Neuromolecular Med. 2006;8:147–58. doi: 10.1385/nmm:8:1-2:147. [DOI] [PubMed] [Google Scholar]
- Axelrod FB, Gold-von Simson G. Hereditary sensory and autonomic neuropathies: types II, III, and IV. Orphanet J Rare Dis. 2007;2:39. doi: 10.1186/1750-1172-2-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejaoui K, Wu C, Scheffler MD, Haan G, Ashby P, Wu L, et al. SPTLC1 is mutated in hereditary sensory neuropathy, type 1. Nat Genet. 2001;27:261–2. doi: 10.1038/85817. [DOI] [PubMed] [Google Scholar]
- Bouhouche A, Benomar A, Bouslam N, Chkili T, Yahyaoui M. Mutation in the epsilon subunit of the cytosolic chaperonin-containing t-complex peptide-1 (Cct5) gene causes autosomal recessive mutilating sensory neuropathy with spastic paraplegia. J Med Genet. 2006a;43:441–3. doi: 10.1136/jmg.2005.039230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhouche A, Benomar A, Bouslam N, Ouazzani R, Chkili T, Yahyaoui M. Autosomal recessive mutilating sensory neuropathy with spastic paraplegia maps to chromosome 5p15.31–14.1. Eur J Hum Genet. 2006b;14:249–52. doi: 10.1038/sj.ejhg.5201537. [DOI] [PubMed] [Google Scholar]
- Coen K, Pareyson D, Auer-Grumbach M, Buyse G, Goemans N, Claeys KG, et al. Novel mutations in the HSN2 gene causing hereditary sensory and autonomic neuropathy type II. Neurology. 2006;66:748–51. doi: 10.1212/01.wnl.0000201191.57519.47. [DOI] [PubMed] [Google Scholar]
- Dawkins JL, Hulme DJ, Brahmbhatt SB, Auer-Grumbach M, Nicholson GA. Mutations in SPTLC1, encoding serine palmitoyltransferase, long chain base subunit-1, cause hereditary sensory neuropathy type I. Nat Genet. 2001;27:309–12. doi: 10.1038/85879. [DOI] [PubMed] [Google Scholar]
- Dyck PJ. Neuronal atrophy and degeneration predominantly affecting peripheral sensory and autonomic neurons. In: Dyck PJ, Thomas PK, Griffin JW, Low PA, Poduslo JF, editors. Peripheral neuropathy. 3rd. Philadelphia: W.B. Saunders; 1993. pp. 1065–93. [Google Scholar]
- Einarsdottir E, Carlsson A, Minde J, Toolanen G, Svensson O, Solders G, et al. A mutation in the nerve growth factor beta gene (NGFB) causes loss of pain perception. Hum Mol Genet. 2004;13:799–805. doi: 10.1093/hmg/ddh096. [DOI] [PubMed] [Google Scholar]
- Hornemann T, Penno A, Richard S, Nicholson G, van Dijk FS, Rotthier A, et al. A systematic comparison of all mutations in hereditary sensory neuropathy type I (HSAN I) reveals that the G387A mutation is not disease associated. Neurogenetics. 2009;10:135–43. doi: 10.1007/s10048-008-0168-7. [DOI] [PubMed] [Google Scholar]
- Houlden H, Blake J, Reilly MM. Hereditary sensory neuropathies. Curr Opin Neurol. 2004a;17:569–77. doi: 10.1097/00019052-200410000-00007. [DOI] [PubMed] [Google Scholar]
- Houlden H, King R, Blake J, Groves M, Love S, Woodward C, et al. Clinical, pathological and genetic characterization of hereditary sensory and autonomic neuropathy type 1 (HSAN I) Brain. 2006;129:411–25. doi: 10.1093/brain/awh712. [DOI] [PubMed] [Google Scholar]
- Houlden H, King RH, Hashemi-Nejad A, Wood NW, Mathias CJ, Reilly M, et al. A novel TRK A (NTRK1) mutation associated with hereditary sensory and autonomic neuropathy type V. Ann Neurol. 2001;49:521–5. [PubMed] [Google Scholar]
- Houlden H, King RH, Muddle JR, Warner TT, Reilly MM, Orrell RW, et al. A novel RAB7 mutation associated with ulcero-mutilating neuropathy. Ann Neurol. 2004b;56:586–90. doi: 10.1002/ana.20281. [DOI] [PubMed] [Google Scholar]
- Indo Y. Molecular basis of congenital insensitivity to pain with anhidrosis (CIPA): mutations and polymorphisms in TRKA (NTRK1) gene encoding the receptor tyrosine kinase for nerve growth factor. Hum Mutat. 2001;18:462–71. doi: 10.1002/humu.1224. [DOI] [PubMed] [Google Scholar]
- Indo Y, Tsuruta M, Hayashida Y, Karim MA, Ohta K, Kawano T, et al. Mutations in the TRKA/NGF receptor gene in patients with congenital insensitivity to pain with anhidrosis. Nat Genet. 1996;13:485–8. doi: 10.1038/ng0896-485. [DOI] [PubMed] [Google Scholar]
- Indo Y, Mardy S, Miura Y, Moosa A, Ismail EA, Toscano E, et al. Congenital insensitivity to pain with anhidrosis (CIPA): novel mutations of the TRKA (NTRK1) gene, a putative uniparental disomy, and a linkage of the mutant TRKA and PKLR genes in a family with CIPA and pyruvate kinase deficiency. Hum Mutat. 2001;18:308–18. doi: 10.1002/humu.1192. [DOI] [PubMed] [Google Scholar]
- Kilic SS, Ozturk R, Sarisozen B, Rotthier A, Baets J, Timmerman V. Humoral immunodeficiency in congenital insensitivity to pain with anhidrosis. Neurogenetics. 2009;10:161–5. doi: 10.1007/s10048-008-0165-x. [DOI] [PubMed] [Google Scholar]
- Klein CJ, Wu Y, Kruckeberg KE, Hebbring SJ, Anderson SA, Cunningham JM, et al. SPTLC1 and RAB7 mutation analysis in dominantly inherited and idiopathic sensory neuropathies. J Neurol Neurosurg Psychiatry. 2005;76:1022–4. doi: 10.1136/jnnp.2004.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok C, Kennerson ML, Spring PJ, Ing AJ, Pollard JD, Nicholson GA. A locus for hereditary sensory neuropathy with cough and gastroesophageal reflux on chromosome 3p22-p24. Am J Hum Genet. 2003;73:632–7. doi: 10.1086/377591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruyt MC, Kruyt ND, Oner FC, Hanlo PW, Verbout AJ. [Congenital pain-insensitivity syndrome; a rare indication of the benefit of pain] Ned Tijdschr Geneeskd. 2007;151:1527–32. [PubMed] [Google Scholar]
- Kwon JM, Elliott JL, Yee WC, Ivanovich J, Scavarda NJ, Moolsintong PJ, et al. Assignment of a second Charcot-Marie-Tooth type II locus to chromosome 3q. Am J Hum Genet. 1995;57:853–8. [PMC free article] [PubMed] [Google Scholar]
- Lafreniere RG, MacDonald ML, Dube MP, MacFarlane J, O’Driscoll M, Brais B, et al. Identification of a novel gene (HSN2) causing hereditary sensory and autonomic neuropathy type II through the Study of Canadian Genetic Isolates. Am J Hum Genet. 2004;74:1064–73. doi: 10.1086/420795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KF, Li E, Huber LJ, Landis SC, Sharpe AH, Chao MV, et al. Targeted mutation of the gene encoding the low affinity NGF receptor p75 leads to deficits in the peripheral sensory nervous system. Cell. 1992;69:737–49. doi: 10.1016/0092-8674(92)90286-l. [DOI] [PubMed] [Google Scholar]
- Levi-Montalcini R. The nerve growth factor: thirty-five years later. EMBO J. 1987;6:1145–54. doi: 10.1002/j.1460-2075.1987.tb02347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meggouh F, Bienfait HM, Weterman MA, de Visser M, Baas F. Charcot-Marie-Tooth disease due to a de novo mutation of the RAB7 gene. Neurology. 2006;67:1476–8. doi: 10.1212/01.wnl.0000240068.21499.f5. [DOI] [PubMed] [Google Scholar]
- Minde J, Toolanen G, Andersson T, Nennesmo I, Remahl IN, Svensson O, et al. Familial insensitivity to pain (HSAN V) and a mutation in the NGFB gene. A neurophysiological and pathological study. Muscle Nerve. 2004;30:752–60. doi: 10.1002/mus.20172. [DOI] [PubMed] [Google Scholar]
- Minde J, Andersson T, Fulford M, Aguirre M, Nennesmo I, Remahl IN, et al. A novel NGFB point mutation: a phenotype study of heterozygous patients. J Neurol Neurosurg Psychiatry. 2009;80:188–95. doi: 10.1136/jnnp.2007.136051. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, Seabra MC. The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J Mol Biol. 2000;301:1077–87. doi: 10.1006/jmbi.2000.4010. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Meth Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Shatzky S, Moses S, Levy J, Pinsk V, Hershkovitz E, Herzog L, et al. Congenital insensitivity to pain with anhidrosis (CIPA) in Israeli-Bedouins: genetic heterogeneity, novel mutations in the TRKA/NGF receptor gene, clinical findings, and results of nerve conduction studies. Am J Med Genet. 2000;92:353–60. doi: 10.1002/1096-8628(20000619)92:5<353::aid-ajmg12>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Shekarabi M, Girard N, Riviere JB, Dion P, Houle M, Toulouse A, et al. Mutations in the nervous system—specific HSN2 exon of WNK1 cause hereditary sensory neuropathy type II. J Clin Invest. 2008;118:2496–505. doi: 10.1172/JCI34088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaugenhaupt SA, Blumenfeld A, Gill SP, Leyne M, Mull J, Cuajungco MP, et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am J Hum Genet. 2001;68:598–605. doi: 10.1086/318810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuysuz B, Bayrakli F, Diluna ML, Bilguvar K, Bayri Y, Yalcinkaya C, et al. Novel NTRK1 mutations cause hereditary sensory and autonomic neuropathy type IV: demonstration of a founder mutation in the Turkish population. Neurogenetics. 2008;9:119–25. doi: 10.1007/s10048-008-0121-9. [DOI] [PubMed] [Google Scholar]
- Vance JM, Speer MC, Stajich JM, West S, Wolpert C, Gaskell P, et al. Misclassification and linkage of hereditary sensory and autonomic neuropathy type 1 as Charcot-Marie-Tooth disease, type 2B. Am J Hum Genet. 1996;59:258–62. [PMC free article] [PubMed] [Google Scholar]
- Verhoeven K, Coen K, De Vriendt E, Jacobs A, Van Gerwen V, Smouts I, et al. SPTLC1 mutation in twin sisters with hereditary sensory neuropathy type I. Neurology. 2004;62:1001–2. doi: 10.1212/01.wnl.0000115388.10828.5c. [DOI] [PubMed] [Google Scholar]
- Verhoeven K, De Jonghe P, Coen K, Verpoorten N, Auer-Grumbach M, Kwon JM, et al. Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot-Marie-Tooth type 2B neuropathy. Am J Hum Genet. 2003;72:722–7. doi: 10.1086/367847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpoorten N, De Jonghe P, Timmerman V. Disease mechanisms in hereditary sensory and autonomic neuropathies. Neurobiol Dis. 2006a;21:247–55. doi: 10.1016/j.nbd.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Verhoeven K, Timmerman V, Mauko B, Pieber TR, De Jonghe P, Auer-Grumbach M. Recent advances in hereditary sensory and autonomic neuropathies. Curr Opin Neurol. 2006;19:474–80. doi: 10.1097/01.wco.0000245370.82317.f6. [DOI] [PubMed] [Google Scholar]
- Verpoorten N, Claeys KG, Deprez L, Jacobs A, Van Gerwen V, Lagae L, et al. Novel frameshift and splice site mutations in the neurotrophic tyrosine kinase receptor type 1 gene (NTRK1) associated with hereditary sensory neuropathy type IV. Neuromuscul Disord. 2006b;16:19–25. doi: 10.1016/j.nmd.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Weckx S, De Rijk P, Van Broeckhoven C, Del-Favero J. SNPbox: web-based high-throughput primer design from gene to genome. Nucleic Acids Res. 2004;32:W170–2. doi: 10.1093/nar/gkh369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckx S, Del-Favero J, Rademakers R, Claes L, Cruts M, De Jonghe P, et al. novoSNP, a novel computational tool for sequence variation discovery. Genome Res. 2005;15:436–42. doi: 10.1101/gr.2754005. [DOI] [PMC free article] [PubMed] [Google Scholar]