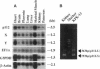Abstract
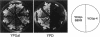
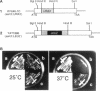
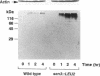
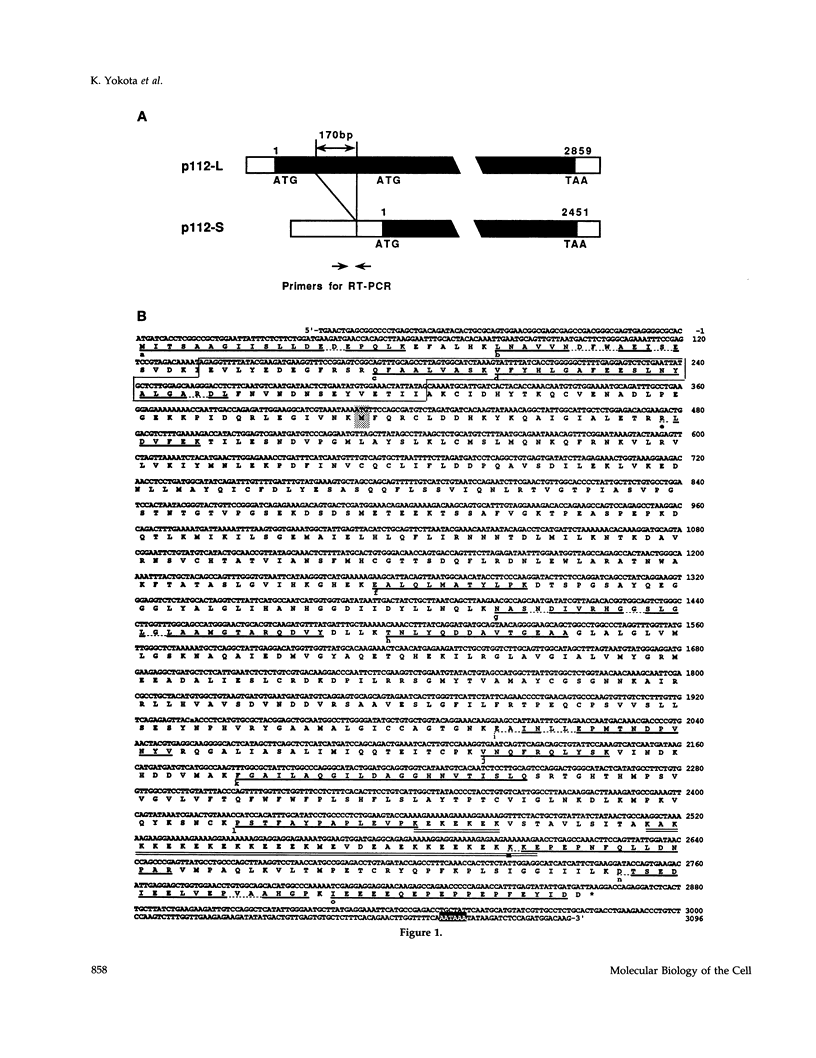
The 26S proteasome is a large multisubunit protease complex, the largest regulatory subunit of which is a component named p112. Molecular cloning of cDNA encoding human p112 revealed a polypeptide predicted to have 953 amino acid residues and a molecular mass of 105,865. The human p112 gene was mapped to the q37.1-q37.2 region of chromosome 2. Computer analysis showed that p112 has strong similarity to the Saccharomyces cerevisiae Sen3p, which has been listed in a gene bank as a factor affecting tRNA splicing endonuclease. The SEN3 also was identified in a synthetic lethal screen with the nin1-1 mutant, a temperature-sensitive mutant of NIN1. NIN1 encodes p31, another regulatory subunit of the 26S proteasome, which is necessary for activation of Cdc28p kinase. Disruption of the SEN3 did not affect cell viability, but led to temperature-sensitive growth. The human p112 cDNA suppressed the growth defect at high temperature in a SEN3 disruptant, indicating that p112 is a functional homologue of the yeast Sen3p. Maintenance of SEN3 disruptant cells at the restrictive temperature resulted in a variety of cellular dysfunctions, including defects in proteolysis mediated by the ubiquitin pathway, in the N-end rule system, in the stress response upon cadmium exposure, and in nuclear protein transportation. The functional abnormality induced by SEN3 disruption differs considerably from various phenotypes shown by the nin1-1 mutation, suggesting that these two regulatory subunits of the 26S proteasome play distinct roles in the various processes mediated by the 26S proteasome.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahn J. Y., Tanahashi N., Akiyama K., Hisamatsu H., Noda C., Tanaka K., Chung C. H., Shibmara N., Willy P. J., Mott J. D. Primary structures of two homologous subunits of PA28, a gamma-interferon-inducible protein activator of the 20S proteasome. FEBS Lett. 1995 Jun 5;366(1):37–42. doi: 10.1016/0014-5793(95)00492-r. [DOI] [PubMed] [Google Scholar]
- Akioka H., Forsberg N. E., Ishida N., Okumura K., Nogami M., Taguchi H., Noda C., Tanaka K. Isolation and characterization of the HC8 subunit gene of the human proteasome. Biochem Biophys Res Commun. 1995 Feb 6;207(1):318–323. doi: 10.1006/bbrc.1995.1190. [DOI] [PubMed] [Google Scholar]
- Akiyama K., Yokota K., Kagawa S., Shimbara N., Tamura T., Akioka H., Nothwang H. G., Noda C., Tanaka K., Ichihara A. cDNA cloning and interferon gamma down-regulation of proteasomal subunits X and Y. Science. 1994 Aug 26;265(5176):1231–1234. doi: 10.1126/science.8066462. [DOI] [PubMed] [Google Scholar]
- Bachmair A., Finley D., Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986 Oct 10;234(4773):179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Bartel B., Wünning I., Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990 Oct;9(10):3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Chu-Ping M., Vu J. H., Proske R. J., Slaughter C. A., DeMartino G. N. Identification, purification, and characterization of a high molecular weight, ATP-dependent activator (PA700) of the 20 S proteasome. J Biol Chem. 1994 Feb 4;269(5):3539–3547. [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994 Oct 7;79(1):13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- Confalonieri F., Duguet M. A 200-amino acid ATPase module in search of a basic function. Bioessays. 1995 Jul;17(7):639–650. doi: 10.1002/bies.950170710. [DOI] [PubMed] [Google Scholar]
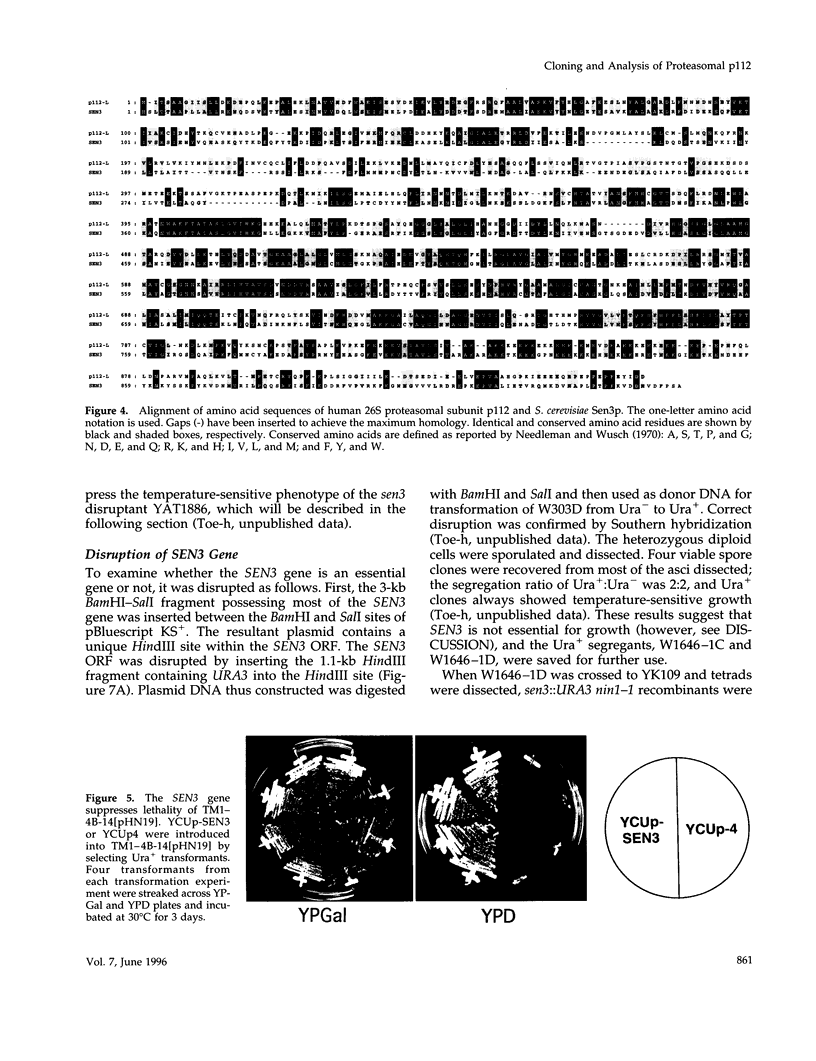
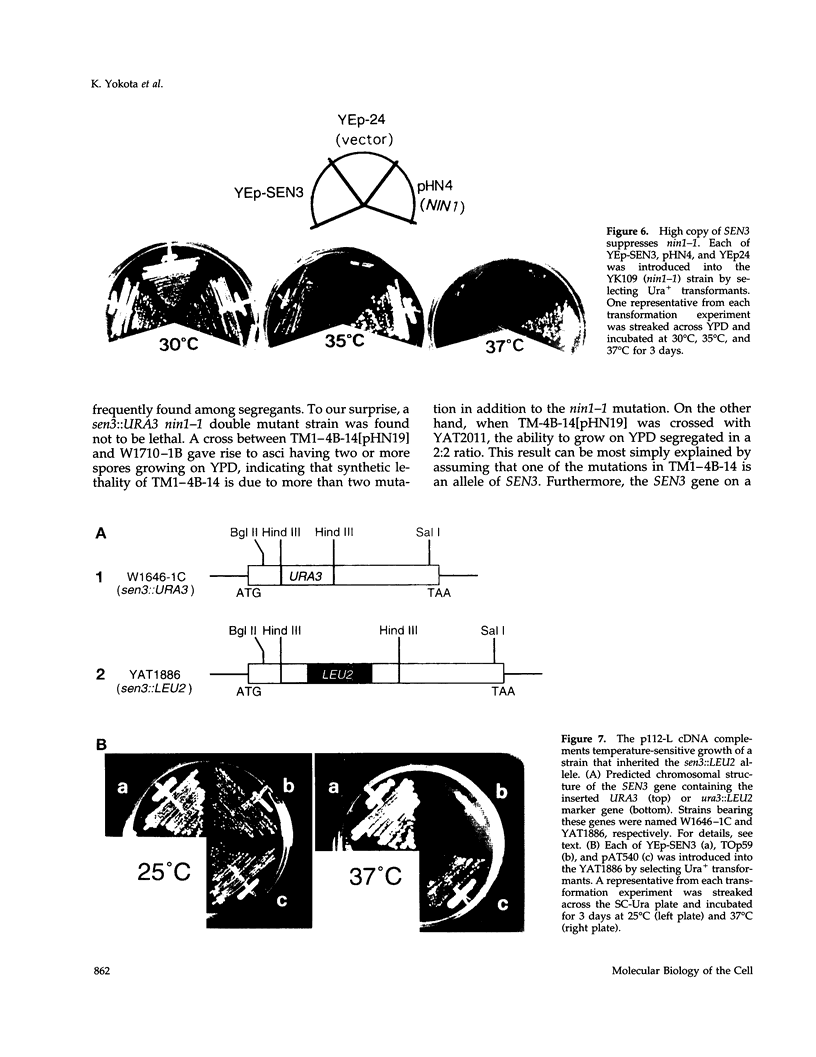
- DeMarini D. J., Papa F. R., Swaminathan S., Ursic D., Rasmussen T. P., Culbertson M. R., Hochstrasser M. The yeast SEN3 gene encodes a regulatory subunit of the 26S proteasome complex required for ubiquitin-dependent protein degradation in vivo. Mol Cell Biol. 1995 Nov;15(11):6311–6321. doi: 10.1128/mcb.15.11.6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMartino G. N., Moomaw C. R., Zagnitko O. P., Proske R. J., Chu-Ping M., Afendis S. J., Swaffield J. C., Slaughter C. A. PA700, an ATP-dependent activator of the 20 S proteasome, is an ATPase containing multiple members of a nucleotide-binding protein family. J Biol Chem. 1994 Aug 19;269(33):20878–20884. [PubMed] [Google Scholar]
- DeMartino G. N., Proske R. J., Moomaw C. R., Strong A. A., Song X., Hisamatsu H., Tanaka K., Slaughter C. A. Identification, purification, and characterization of a PA700-dependent activator of the proteasome. J Biol Chem. 1996 Feb 9;271(6):3112–3118. doi: 10.1074/jbc.271.6.3112. [DOI] [PubMed] [Google Scholar]
- Deveraux Q., Ustrell V., Pickart C., Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994 Mar 11;269(10):7059–7061. [PubMed] [Google Scholar]
- Deveraux Q., van Nocker S., Mahaffey D., Vierstra R., Rechsteiner M. Inhibition of ubiquitin-mediated proteolysis by the Arabidopsis 26 S protease subunit S5a. J Biol Chem. 1995 Dec 15;270(50):29660–29663. doi: 10.1074/jbc.270.50.29660. [DOI] [PubMed] [Google Scholar]
- Dubiel W., Ferrell K., Dumdey R., Standera S., Prehn S., Rechsteiner M. Molecular cloning and expression of subunit 12: a non-MCP and non-ATPase subunit of the 26 S protease. FEBS Lett. 1995 Apr 17;363(1-2):97–100. doi: 10.1016/0014-5793(95)00288-k. [DOI] [PubMed] [Google Scholar]
- Dubiel W., Ferrell K., Rechsteiner M. Subunits of the regulatory complex of the 26S protease. Mol Biol Rep. 1995;21(1):27–34. doi: 10.1007/BF00990967. [DOI] [PubMed] [Google Scholar]
- Eytan E., Armon T., Heller H., Beck S., Hershko A. Ubiquitin C-terminal hydrolase activity associated with the 26 S protease complex. J Biol Chem. 1993 Mar 5;268(7):4668–4674. [PubMed] [Google Scholar]
- Fujimuro M., Sawada H., Yokosawa H. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 1994 Aug 1;349(2):173–180. doi: 10.1016/0014-5793(94)00647-4. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L. The mechanism and functions of ATP-dependent proteases in bacterial and animal cells. Eur J Biochem. 1992 Jan 15;203(1-2):9–23. doi: 10.1111/j.1432-1033.1992.tb19822.x. [DOI] [PubMed] [Google Scholar]
- Gridley T., Gray D. A., Orr-Weaver T., Soriano P., Barton D. E., Francke U., Jaenisch R. Molecular analysis of the Mov 34 mutation: transcript disrupted by proviral integration in mice is conserved in Drosophila. Development. 1990 May;109(1):235–242. doi: 10.1242/dev.109.1.235. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Udvardy A. Cloning and sequencing a non-ATPase subunit of the regulatory complex of the Drosophila 26S protease. Eur J Biochem. 1995 Aug 1;231(3):720–725. doi: 10.1111/j.1432-1033.1995.tb20753.x. [DOI] [PubMed] [Google Scholar]
- Heemels M. T., Ploegh H. Generation, translocation, and presentation of MHC class I-restricted peptides. Annu Rev Biochem. 1995;64:463–491. doi: 10.1146/annurev.bi.64.070195.002335. [DOI] [PubMed] [Google Scholar]
- Heinemeyer W., Gruhler A., Möhrle V., Mahé Y., Wolf D. H. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J Biol Chem. 1993 Mar 5;268(7):5115–5120. [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Hilt W., Wolf D. H. Proteasomes of the yeast S. cerevisiae: genes, structure and functions. Mol Biol Rep. 1995;21(1):3–10. doi: 10.1007/BF00990964. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995 Apr;7(2):215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- Hoffman L., Pratt G., Rechsteiner M. Multiple forms of the 20 S multicatalytic and the 26 S ubiquitin/ATP-dependent proteases from rabbit reticulocyte lysate. J Biol Chem. 1992 Nov 5;267(31):22362–22368. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jariel-Encontre I., Pariat M., Martin F., Carillo S., Salvat C., Piechaczyk M. Ubiquitinylation is not an absolute requirement for degradation of c-Jun protein by the 26 S proteasome. J Biol Chem. 1995 May 12;270(19):11623–11627. doi: 10.1074/jbc.270.19.11623. [DOI] [PubMed] [Google Scholar]
- Johnson E. S., Ma P. C., Ota I. M., Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995 Jul 21;270(29):17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- Jungmann J., Reins H. A., Schobert C., Jentsch S. Resistance to cadmium mediated by ubiquitin-dependent proteolysis. Nature. 1993 Jan 28;361(6410):369–371. doi: 10.1038/361369a0. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Richardson W. D., Markham A. F., Smith A. E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984 Sep 6;311(5981):33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Kilmartin J. V., Adams A. E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984 Mar;98(3):922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami K., DeMartino G. N., Moomaw C. R., Slaughter C. A., Shimbara N., Fujimuro M., Yokosawa H., Hisamatsu H., Tanahashi N., Shimizu Y. Nin1p, a regulatory subunit of the 26S proteasome, is necessary for activation of Cdc28p kinase of Saccharomyces cerevisiae. EMBO J. 1995 Jul 3;14(13):3105–3115. doi: 10.1002/j.1460-2075.1995.tb07313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami K., Toh-e A. Characterization of the function of the NIN1 gene product of Saccharomyces cerevisiae. Exp Cell Res. 1994 Apr;211(2):203–211. doi: 10.1006/excr.1994.1079. [DOI] [PubMed] [Google Scholar]
- Koshland D., Kent J. C., Hartwell L. H. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 1985 Feb;40(2):393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- Lupas A., Koster A. J., Baumeister W. Structural features of 26S and 20S proteasomes. Enzyme Protein. 1993;47(4-6):252–273. doi: 10.1159/000468684. [DOI] [PubMed] [Google Scholar]
- Löwe J., Stock D., Jap B., Zwickl P., Baumeister W., Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995 Apr 28;268(5210):533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- Michalek M. T., Grant E. P., Gramm C., Goldberg A. L., Rock K. L. A role for the ubiquitin-dependent proteolytic pathway in MHC class I-restricted antigen presentation. Nature. 1993 Jun 10;363(6429):552–554. doi: 10.1038/363552a0. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T., Tanaka K., Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992 Dec 10;360(6404):597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Nelson M. K., Kurihara T., Silver P. A. Extragenic suppressors of mutations in the cytoplasmic C terminus of SEC63 define five genes in Saccharomyces cerevisiae. Genetics. 1993 May;134(1):159–173. doi: 10.1093/genetics/134.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni R., Tomita Y., Tokunaga F., Liang T. J., Noda C., Ichihara A., Tanaka K. Molecular cloning of two types of cDNA encoding subunit RC6-I of rat proteasomes. Biochim Biophys Acta. 1995 Oct 17;1264(1):45–52. doi: 10.1016/0167-4781(95)00113-u. [DOI] [PubMed] [Google Scholar]
- Nisogi H., Kominami K., Tanaka K., Toh-e A. A new essential gene of Saccharomyces cerevisiae, a defect in it may result in instability of nucleus. Exp Cell Res. 1992 May;200(1):48–57. doi: 10.1016/s0014-4827(05)80070-9. [DOI] [PubMed] [Google Scholar]
- Papa F. R., Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993 Nov 25;366(6453):313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- Peters J. M., Franke W. W., Kleinschmidt J. A. Distinct 19 S and 20 S subcomplexes of the 26 S proteasome and their distribution in the nucleus and the cytoplasm. J Biol Chem. 1994 Mar 11;269(10):7709–7718. [PubMed] [Google Scholar]
- Peters J. M. Proteasomes: protein degradation machines of the cell. Trends Biochem Sci. 1994 Sep;19(9):377–382. doi: 10.1016/0968-0004(94)90115-5. [DOI] [PubMed] [Google Scholar]
- Realini C., Dubiel W., Pratt G., Ferrell K., Rechsteiner M. Molecular cloning and expression of a gamma-interferon-inducible activator of the multicatalytic protease. J Biol Chem. 1994 Aug 12;269(32):20727–20732. [PubMed] [Google Scholar]
- Realini C., Rogers S. W., Rechsteiner M. KEKE motifs. Proposed roles in protein-protein association and presentation of peptides by MHC class I receptors. FEBS Lett. 1994 Jul 11;348(2):109–113. doi: 10.1016/0014-5793(94)00569-9. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M., Hoffman L., Dubiel W. The multicatalytic and 26 S proteases. J Biol Chem. 1993 Mar 25;268(9):6065–6068. [PubMed] [Google Scholar]
- Richter-Ruoff B., Heinemeyer W., Wolf D. H. The proteasome/multicatalytic-multifunctional proteinase. In vivo function in the ubiquitin-dependent N-end rule pathway of protein degradation in eukaryotes. FEBS Lett. 1992 May 11;302(2):192–196. doi: 10.1016/0014-5793(92)80438-m. [DOI] [PubMed] [Google Scholar]
- Rock K. L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A. L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994 Sep 9;78(5):761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Rubin D. M., Finley D. Proteolysis. The proteasome: a protein-degrading organelle? Curr Biol. 1995 Aug 1;5(8):854–858. doi: 10.1016/s0960-9822(95)00172-2. [DOI] [PubMed] [Google Scholar]
- Seufert W., Jentsch S. In vivo function of the proteasome in the ubiquitin pathway. EMBO J. 1992 Aug;11(8):3077–3080. doi: 10.1002/j.1460-2075.1992.tb05379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Silva Pereira I., Bey F., Coux O., Scherrer K. Two mRNAs exist for the Hs PROS-30 gene encoding a component of human prosomes. Gene. 1992 Oct 21;120(2):235–242. doi: 10.1016/0378-1119(92)90098-a. [DOI] [PubMed] [Google Scholar]
- Sutton A., Immanuel D., Arndt K. T. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol Cell Biol. 1991 Apr;11(4):2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
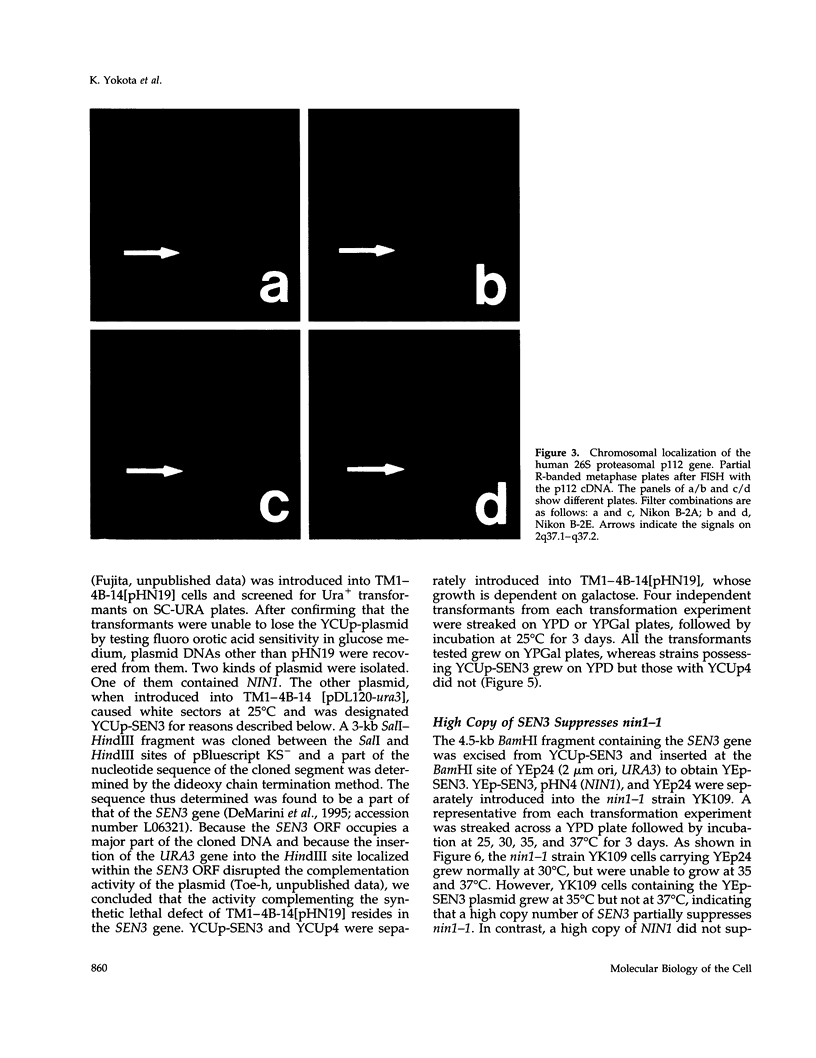
- Takahashi E., Hori T., O'Connell P., Leppert M., White R. Mapping of the MYC gene to band 8q24.12----q24.13 by R-banding and distal to fra(8)(q24.11), FRA8E, by fluorescence in situ hybridization. Cytogenet Cell Genet. 1991;57(2-3):109–111. doi: 10.1159/000133124. [DOI] [PubMed] [Google Scholar]
- Takahashi E., Yamauchi M., Tsuji H., Hitomi A., Meuth M., Hori T. Chromosome mapping of the human cytidine-5'-triphosphate synthetase (CTPS) gene to band 1p34.1-p34.3 by fluorescence in situ hybridization. Hum Genet. 1991 Nov;88(1):119–121. doi: 10.1007/BF00204942. [DOI] [PubMed] [Google Scholar]
- Tanahashi N., Tsurumi C., Tamura T., Tanaka K. Molecular structure of 20S and 26S proteasomes. Enzyme Protein. 1993;47(4-6):241–251. doi: 10.1159/000468683. [DOI] [PubMed] [Google Scholar]
- Tanaka K. Molecular biology of proteasomes. Mol Biol Rep. 1995;21(1):21–26. doi: 10.1007/BF00990966. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Tamanoi F., Kaziro Y., Matsumoto K., Toh-e A. IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol Cell Biol. 1990 Aug;10(8):4303–4313. doi: 10.1128/mcb.10.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Yoshimura T., Kumatori A., Ichihara A., Ikai A., Nishigai M., Kameyama K., Takagi T. Proteasomes (multi-protease complexes) as 20 S ring-shaped particles in a variety of eukaryotic cells. J Biol Chem. 1988 Nov 5;263(31):16209–16217. [PubMed] [Google Scholar]
- Treier M., Staszewski L. M., Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994 Sep 9;78(5):787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- Tsurumi C., DeMartino G. N., Slaughter C. A., Shimbara N., Tanaka K. cDNA cloning of p40, a regulatory subunit of the human 26S proteasome, and a homolog of the Mov-34 gene product. Biochem Biophys Res Commun. 1995 May 16;210(2):600–608. doi: 10.1006/bbrc.1995.1701. [DOI] [PubMed] [Google Scholar]
- Udvardy A. Purification and characterization of a multiprotein component of the Drosophila 26 S (1500 kDa) proteolytic complex. J Biol Chem. 1993 Apr 25;268(12):9055–9062. [PubMed] [Google Scholar]
- Ugai S., Tamura T., Tanahashi N., Takai S., Komi N., Chung C. H., Tanaka K., Ichihara A. Purification and characterization of the 26S proteasome complex catalyzing ATP-dependent breakdown of ubiquitin-ligated proteins from rat liver. J Biochem. 1993 Jun;113(6):754–768. doi: 10.1093/oxfordjournals.jbchem.a124116. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule. Cell. 1992 May 29;69(5):725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]
- Viegas-Péquignot E., Li Z. L., Dutrillaux B., Apiou F., Paulin D. Assignment of human desmin gene to band 2q35 by nonradioactive in situ hybridization. Hum Genet. 1989 Aug;83(1):33–36. doi: 10.1007/BF00274143. [DOI] [PubMed] [Google Scholar]
- Yoshimura T., Kameyama K., Takagi T., Ikai A., Tokunaga F., Koide T., Tanahashi N., Tamura T., Cejka Z., Baumeister W. Molecular characterization of the "26S" proteasome complex from rat liver. J Struct Biol. 1993 Nov-Dec;111(3):200–211. doi: 10.1006/jsbi.1993.1050. [DOI] [PubMed] [Google Scholar]
- van Nocker S., Deveraux Q., Rechsteiner M., Vierstra R. D. Arabidopsis MBP1 gene encodes a conserved ubiquitin recognition component of the 26S proteasome. Proc Natl Acad Sci U S A. 1996 Jan 23;93(2):856–860. doi: 10.1073/pnas.93.2.856. [DOI] [PMC free article] [PubMed] [Google Scholar]