Abstract
Dietary restriction (DR) extends lifespan in multiple species. To examine the mechanisms of lifespan extension upon DR, we assayed genome-wide translational changes in Drosophila. A number of nuclear encoded mitochondrial genes, including those in Complex I and IV of the electron transport chain, showed increased ribosomal loading and enhanced overall activity upon DR. We found that various mitochondrial genes possessed shorter and less structured 5′UTRs, which were important for their enhanced mRNA translation. The translational repressor 4E-BP, the eukaryotic translation initiation factor 4E binding protein, was upregulated upon DR and mediated DR dependent changes in mitochondrial activity and lifespan extension. Inhibition of individual mitochondrial subunits from Complex I and IV diminished the lifespan extension obtained upon DR, reflecting the importance of enhanced mitochondrial function during DR. Our results implicate translational regulation of mitochondrial gene expression by 4E-BP which plays an important role in lifespan extension upon DR.
Introduction
Dietary restriction (DR), the reduction of nutrient intake without malnutrition, is a method of lifespan extension conserved from yeast to mammals. DR also slows the progression of many age-related diseases including cancer. It has been suggested that DR extends lifespan by inducing a shift from growth and reproduction towards somatic maintenance. The well established paradigms to study DR in flies, the conservation of signaling pathways and their short lifespan make Drosophila melanogaster an excellent model to understand the mechanisms by which DR slows aging and age-related diseases. It has become apparent in recent years that nutrient sensing growth pathways are important regulators of lifespan. The target of rapamycin (TOR) pathway, a key nutrient sensing pathway conserved from yeast to humans, integrates nutrient and environmental signals to mediate growth and metabolism.
Mutants in the nutrient sensing TOR pathway display pleiotropic phenotypes. In Drosophila melanogaster, larvae lacking TOR show similarities to amino acid-deprived animals, such as reduced nucleolar size, developmental arrest and lipid vesicle aggregation in the larval fat body (Zhang et al., 2000). We have previously shown that modulation of various genes that encode components of the TOR signaling pathway, including the products of the tuberous sclerosis complex genes (Tsc1 and Tsc2), TOR, and S6K, extend lifespan in Drosophila melanogaster (Kapahi et al., 2004). Lifespan extension by inhibition of the TOR pathway is nutrient dependent in both Drosophila melanogaster (Kapahi et al., 2004) and Saccharomyces cerevisiae (Kaeberlein et al., 2005), establishing the role of TOR in mediating the effects of DR. In Caenorhabditis elegans, inhibition of let-363, the worm ortholog of TOR or daf-15, the worm ortholog of the mammalian protein raptor (regulatory associated protein of mammalian TOR) extends lifespan (Jia et al., 2004; Vellai et al., 2003). Lifespan extension by inhibition of S6 kinase is also observed in Caenorhabditis elegans and is independent of the ILS pathway (Hansen et al., 2007; Pan et al., 2007). Interestingly, neither the developmental arrest nor the fat accumulation of homozygous daf-15 mutants is suppressible by daf-16 mutations, suggesting its effects on lifespan extension are independent of the ILS pathway in worms (Jia et al., 2004).
Regulation of downstream processes like mRNA translation, autophagy, stress responsive signaling pathways and amino acid transport by TOR are potential mediators of its effects on lifespan (Kaeberlein et al., 2007; Kapahi and Zid, 2004). 4E-BP (eukaryotic initiation factor eIF4E binding protein), a protein that is phosphorylated by TOR regulates mRNA translation and growth in flies and mammals (Harris and Lawrence, 2003; Shamji et al., 2003). When the TOR pathway is inhibited, the hypophosphorylated form of 4E-BP acts as a translational repressor by binding the protein synthesis initiation factor eIF4E blocking the activity of the eIF4F complex. The eukaryotic initiation factor 4F (eIF4F) complex mediates growth-dependent protein synthesis. This is accomplished by regulating the association of the mRNA cap binding protein eIF4E to the scaffold protein eIF4G, both components of the eIF4F complex. eIF4G helps assemble the eIF4F complex by bridging the poly(A) binding proteins (PABPs) with eIF4E (Sonenberg, 2000). This leads to the circularization of mRNAs, which has a synergistic effect on the rate of translation (Richter and Sonenberg, 2005). Transcripts with extensive secondary structure in their 5′ untranslated regions (UTRs) are particularly sensitive to the activity of the eIF4F cap binding complex (Koromilas et al., 1992; Shantz et al., 1996). Overexpression of eIF4E in cultured mammalian cells causes transformation of fibroblasts and increased cell size, which can be reversed by increasing the abundance of 4E-BP (Lazaris-Karatzas et al., 1990). eIF4E is also critical for growth in Drosophila (Lachance et al., 2002) and upregulation of eIF4E has been observed in a broad spectrum of cancers (Petroulakis et al., 2006). Overexpression of eIF4E has been shown to increase cellular senescence in mammalian cells (Ruggero et al., 2004). It has been shown in Caenorhabditis elegans that downregulation of various components of the eIF4F cap binding complex extends lifespan (Hansen et al., 2007; Henderson et al., 2006; Pan et al., 2007; Syntichaki et al., 2007). Inhibition of ribosomal genes has been shown to extend lifespan in both Saccharomyces cerevisiae (Steffen et al., 2008) and Caenorhabditis elegans (Hansen et al., 2007). Recently, two independent screens for genes that extend lifespan upon inhibition during adulthood but compromise development if reduced during development have identified a number of translation factors to be important for this process (Chen et al., 2007; Curran and Ruvkun, 2007). Together these findings suggest an important role of mRNA translation in modulating the aging process.
Genome-wide transcriptional profiling has led to major advances in various biological fields including aging. However, as regulation of gene expression lies not only in transcriptional changes, but also translational control, studies on mRNA translational state may provide new insights into the aging process. Currently established genome-wide approaches to study mRNA translation state (Arava et al., 2003; Zong et al., 1999) were used to identify novel processes that are modulated upon DR in Drosophila melanogaster. This method assesses the mRNA translation state based on the separation of mRNAs bound to varying number of polysomes via density gradient centrifugation. Using this technology we have made a novel observation that under DR some mRNAs are differentially loaded onto ribosomes compared to rich nutrient conditions. We found that mitochondrial electron transport components were one class of genes that are translationally upregulated upon DR. Inhibition of mitochondrial ETC subunits diminished the DR dependent lifespan extension, suggesting a key role for the enhancement of mitochondrial function upon DR. The translational repressor d4E-BP was upregulated under DR and played a key role in modulating the changes in mRNA translation upon DR. The modulation of mitochondrial function was dependent on d4E-BP and was associated with the structural properties of the 5′UTR. Furthermore, the DR dependent extension in lifespan required d4E-BP which was also found to be sufficient to extend lifespan.
Results
Analysis of DR Dependent Changes in mRNA Translation
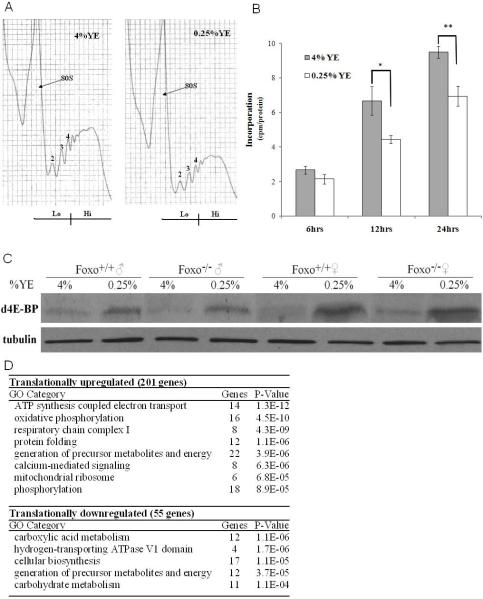
Reducing the concentration of yeast or yeast extract in the fly diet has been shown to be sufficient to extend lifespan (Chippindale et al., 1993; Kapahi et al., 2004; Mair et al., 2005; Nusbaum and Rose, 1999). We investigated the changes in mRNA translation under a paradigm of DR in which the yeast extract (YE) was varied while sucrose, the major carbohydrate source, was constant. The relative translation rate of an mRNA can be inferred from the number of ribosomes (polysomes) it recruits, as initiation is the rate limiting step for the translation of most mRNAs (Sonenberg et al., 2000). Investigation of the polysomal profiles showed that under DR, there was an overall reduction in the number of polysomes bound to mRNAs (Figure 1A). To investigate whether the changes in mRNA translation also influence protein synthesis rates, incorporation of 35S-methionine into proteins was measured under DR. Upon DR there is an approximately 30% decrease of labeled methionine incorporation into newly synthesized proteins after 24hrs (Figure 1B). As there is a decrease in translation initiation upon DR we examined the levels of the translational suppressor d4E-BP upon DR. Both male and female flies show an upregulation of the d4E-BP protein upon DR (Figure 1C). Previously it has been shown that induction of the d4E-BP transcript upon starvation is dependent on dFOXO (Teleman et al., 2005), a downstream component of the insulin-like signaling pathway. Our results suggest that upregulation of d4E-BP upon DR may involve different mechanisms from those under starvation, as unlike starvation this effect is independent of dFOXO (Figure 1C).
Figure 1. mRNA Translation Associated Changes Upon DR.
(A) Polysomal distribution of mRNAs of young adult male flies on 4% YE and 0.25% YE for 6 days showing the individual ribosomal subunits and the polysome peaks, normalized to body weight of flies. RNA was prepared from the low (Lo) and high (Hi) translation fractions for microarray analysis performed in triplicate.
(B) Levels of 35S-methionine incorporation in 7 day old control flies normalized to protein content on 4% YE or 0.25% YE. (±SEM) (N=3) (*p<0.05, ** p<0.01).
(C) 4E-BP protein is induced on 0.25% YE (DR) as measured by western blot using 30μg of protein probed with a polyclonal anti-d4E-BP (Miron et al., 2001). This induction is not dependent on dFOXO, as dFOXO null mutant flies show a normal response. β-tubulin levels were measured as a loading control. The blot is representative of three replicates.
(D) Functional grouping of differentially translated genes according to GO terminology using DAVID analysis software.
A number of studies have hinted at the lack of correlation between the transcriptome and proteome and the importance of regulation of mRNA translation especially under environmental stresses (Sonenberg, 2000). We examined the possibility that specific mRNAs are differentially translated upon DR in Drosophila melanogaster. To examine changes in the translation state of individual mRNAs at the genome-wide level under DR, translation state array analysis (TSAA) was performed (Arava et al., 2003; Zong et al., 1999). This method allows the identification of differentially translated genes based on the association of mRNAs with polysomes. Polysomes were separated over a sucrose gradient and divided into low (<5 ribosomes per mRNA) and high translated fractions, (5 or more ribosomes per mRNA). The two fractions were hybridized to microarray chips and the ratio of expression in the two fractions was used to determine the translation index (ratio of RNA in high to low translated fractions) of each mRNA. Therefore, comparing the translation index under two conditions using this method can detect mRNAs that show differential ribosomal loading and are likely to be under translational regulation.
Using a false discovery rate (FDR) of less than 5%, 55 genes were differentially translationally downregulated and 201 upregulated upon DR (Table S1). We used Gene Ontology (GO) classification to identify biological themes overrepresented in the differentially translated genes. Categories enriched among the downregulated genes were carboxylic acid, cellular biosynthesis and carbohydrate metabolism genes, while increased translation genes were enriched for mitochondrial ATP generation, oxidative phosphorylation and protein folding genes (Figure 1D). Components of the mitochondria overrepresented included nuclear encoded Complex I and IV subunits of the electron transport chain (ETC) and mitochondrial ribosomal proteins. The enhanced translation of various nuclear encoded ETC components and mitochondrial ribosomal proteins suggests an overall increase in mitochondrial proteins.
Mitochondrial Processes are Upregulated upon DR and Play an Important Role in Mediating the Lifespan Extension Effects of DR
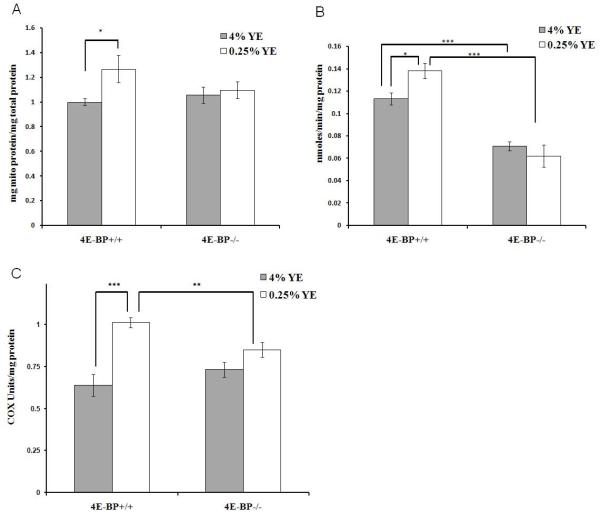
Next we examined whether the effects of DR on the translation state of nuclear encoded mitochondrial mRNAs also leads to changes in mitochondrial protein levels and function. Overall mitochondrial protein density was investigated by measuring protein levels in total homogenates and isolated mitochondria and normalizing to citrate synthase activity. Upon DR mitochondrial protein density increased 25% in control flies, while in d4E-BP null mutant flies there was no change (Figure 2A). Mitochondrial purity was confirmed by western blotting the mitochondrial fractions against antibodies to nuclear protein Lamin A and cytoplasmic dAKT (Figure S1). Next we investigated the activity of specific electron chain complexes. Complex I activity was investigated by measuring the activity of NADH: ubiquinone reductase in isolated mitochondria. A 20% upregulation in Complex I activity was seen in DR flies (Figure 2B), while d4E-BP null mutant flies had lower overall Complex I activity and no beneficial effect from DR (Figure 2B). Similarly, Cytochrome C oxidase (COX) activity increased 60% in control flies upon DR, when normalized to total protein, while COX activity was unchanged in d4E-BP null mutant flies upon DR (Figure 2C). Microarray analysis of total RNA from control flies upon DR showed minimal transcriptional changes in mitochondrial ETC components, which do not explain the large changes in mitochondrial protein density and mitochondrial activities observed (Table S2). These data demonstrate a post-transcriptional upregulation of components of the mitochondria under DR that is d4E-BP dependent and support our TSAA results that mitochondrial mRNAs are translationally upregulated upon DR.
Figure 2. 4E-BP Dependent Induction of Mitochondrial Processes upon DR.
(A) Mitochondrial protein density was calculated by measuring the protein concentration from crude homogenates and from isolated mitochondrial fractions and normalized to citrate synthase activity in control (4E-BP +/+) and d4E-BP null mutant (4E-BP-/-) flies. (±SEM) (n≥7).
(B) Complex I activity measured on isolated mitochondria from whole male flies normalized to mitochondrial protein content in control and d4E-BP null mutant flies. (±SEM) (Control n=3, Null n=4).
(C) Cytochrome C oxidase (COX) activity measured on crude homogenates from whole male control and d4E-BP null mutant flies upon DR, normalized to total protein content (±SEM) (n=7).
(*p<0.05, ** p<0.01, *** p<0.001).
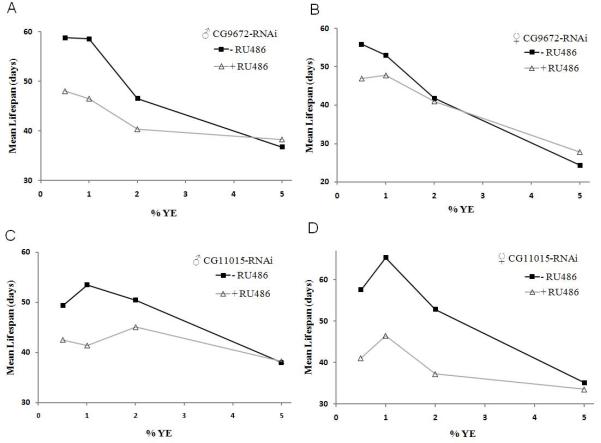
Next we investigated whether the increase in mitochondrial ETC activity was important for the lifespan extension upon DR. This was investigated by reduction of individual mitochondrial ETC Complex I and Complex IV subunits, CG9762 and CG11015 respectively using RNAi. Lifespan was measured under different dietary conditions using the RU486 inducible Act5C (actin5C) P [Switch] GAL4 driver which has been shown to have ubiquitous expression (Ford et al., 2007). Under rich nutritional conditions the lifespan of control flies (-RU486) were indistinguishable from CG9762 and CG11015 knockdown flies (+RU486) in both males and females (Figures 3A-3D, Table S3). However, upon DR, RNAi knockdown of CG9762 and CG11015 diminished the lifespan extension in both male and female flies compared to their respective controls (Figures 3A-3D, Table S3). RU486 had no impact on the lifespan of control flies upon DR (Figures S2A and S2B, Table S3) The RNAi knockdown was sufficient to lower mRNA levels under different yeast concentrations (data not shown). Together these data demonstrate the importance of the mitochondrial ETC function in the lifespan extension upon DR in Drosophila.
Figure 3. Reduction of Mitochondrial ETC Subunits Shortens Lifespan upon DR.
(A-B) Mean lifespan for Complex I (CG9762) knockdown using UAS-RNAi and Act5C-Gal4 P [Switch] (+RU486) compared to genetically identical control flies (-RU486) on various yeast YE concentrations.
(C-D) Mean lifespan for Complex IV (CG11015) knockdown using UAS-RNAi and Act5C-Gal4 P [Switch] (+RU486) compared to genetically identical control flies (-RU486) on various YE concentrations. (Statistical analyses are shown in Table S3).
The Role of 5′UTR Structural Properties in mediating changes in mRNA translation upon DR
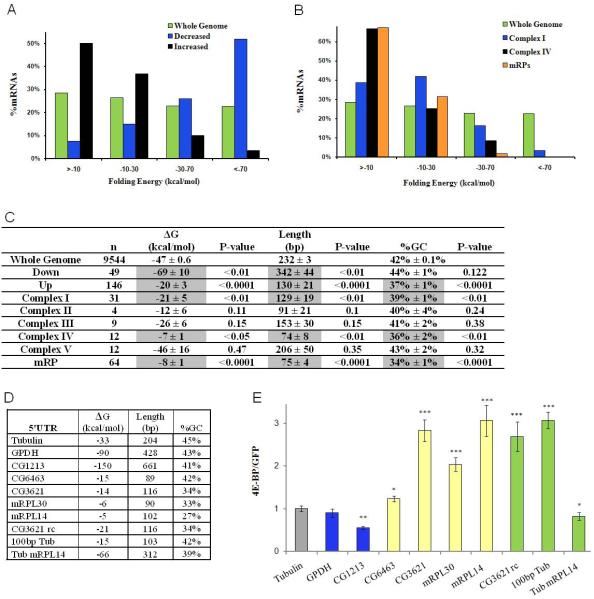
As transcripts with extensive 5′UTR secondary structure are sensitive to the activity of eIF4E (Koromilas et al., 1992), the structural properties of 5′UTRs from differentially translated mRNAs were assessed. Theoretical free folding energies (ΔG), 5′UTR length and the GC content were calculated using DAMBE Software (Xia and Xie, 2001). Differentially translated genes were selected using a FDR of less than 1% to reduce the number of false positives. The mean ΔG for the 5′UTRs of all mRNAs in the Drosophila genome is -47 kcal/mol. Under DR, translationally downregulated mRNAs have highly structured 5′UTRs with an average ΔG of -69 kcal/mol, whereas upregulated mRNAs have significantly less secondary structure, averaging -20 kcal/mol (Figures 4A and 4C). The 5′UTRs of translationally downregulated genes were also significantly longer than the average Drosophila 5′UTR, while translationally upregulated transcripts have shorter 5′UTRs with lower GC content (Figure 4C).
Figure 4. The role of 5′UTR Structural Properties of Differentially Translated mRNAs.
(A-C) 5′UTR secondary structure analysis of differentially translated mRNAs upon DR and mitochondrial genes was calculated using DAMBE.
(A) Distribution of theoretical 5′UTR folding free energies, ΔG, for mRNAs which showed higher or lower translation ratios upon DR and the whole genome.
(B) Distribution of theoretical 5′UTR folding free energies of genes in Complex I, Complex IV and ribosomal proteins of the mitochondria.
(C) Mean ΔG, length and GC content (±SEM) are given the whole genome, translationally up and downregulated genes and all Complexes of the mitochondrial ETC. P-values were calculated by generating sampling distributions for each experimental distribution, as described in the methods.
(D, E) Analysis of 5′UTR dependent translational control. (D) Structural properties of 5′UTRs tested.
(E) 5′UTRs were cloned into a mono-cistronic firefly luciferase plasmid, downstream of the Act5C promoter and upstream of FLuc. DNA transfection of various constructs in S2 cells, with data presented as the ratio of FLuc activity in the presence of activated d4E-BP, (d4E-BPLLAA) versus the presence of GFP, a nonspecific control, with all data normalized to a RLuc transfection control plasmid (±SEM) (n≥6) (*p<0.05, ** p<0.01, *** p<0.001)
Since many genes from Complexes I and IV, and mitochondrial ribosomal proteins, were translationally upregulated, we examined the structural properties for the 5′UTRs of all subunits of these complexes and observed that they are shorter, with lower GC content and weak secondary structure (Figures 4B and 4C). The structural properties of Complexes II, III and V were also investigated and while the overall trend for Complexes II and III showed similar structural properties to Complex I and IV, they were not significantly different from the whole genome (Figure 4C). Complex V had mean secondary structure, length and GC content similar to the whole genome control, but this was skewed by three subunits having long, highly structured (<-100kcal/mol) 5′UTRs (Figure 4C and Table S2). This may be of biological importance, with specific subunits being differentially regulated under various conditions as has recently been shown under hypoxia (Fukuda et al., 2007).
To investigate the sufficiency of various 5′UTRs in 4E-BP dependent translational control, 5′UTRs were cloned into a firefly luciferase (FLuc) reporter vector under the control of the Act5C promoter. The constructs were then transfected into S2 cells along with a transfection control Renilla luciferase (RLuc) vector in the presence or absence of a constitutively active form of Act-d4E-BPLLAA or an Act-GFP vector control. All values were normalized to the αTub84B 5′UTR, which had a theoretical free folding energy (-33 kcal/mol) and length (204 bp) similar to the average 5′UTR of the Drosophila genome and was used as a control. GPDH and CG1213, a putative glucose transporter, were translationally downregulated upon DR and possess extensive 5′UTR secondary structure (Figure 4D). While the 5′UTR for GPDH was not sufficient to confer translational changes upon activated d4E-BP expression, the 5′UTR of CG1213 caused an almost 2 fold decrease in the expression of the FLuc reporter upon d4E-BPLLAA expression (Figure 4E). The sufficiency of translational control by mitochondrial 5′UTRs was investigated by analyzing two Complex I subunit 5′UTRs (CG3621, CG6463) and two mitochondrial ribosomal protein 5′UTRs (mRPL30, mRPL14) (Figure 4D). These short 5′UTRs with weak secondary structure were sufficient to confer translational upregulation to the reporter construct upon activated d4E-BP expression (Figure 4E). To further elucidate whether there may be a specific sequence necessary for this translational control or if it is a general property of short length and minimal secondary structure the CG3621 5′UTR was cloned in a reverse orientation (Figure 4D). This 5′UTR responded similarly and caused preferential translation upon activated d4E-BP expression. The truncated form of the Tubulin 5′UTR, that included only the 103bp upstream of the start codon, enhanced translation 3-fold higher than the full length Tubulin 5′UTR (Figure 4E). Finally we inserted the full length Tubulin 5′UTR, upstream of the mRPL14 5′UTR. This hybrid 5′UTR had length and secondary structure comparable to an average translationally downregulated DR gene from the TSAA. This caused an almost 20% reduction in translation compared to the Tubulin 5′UTR (Figure 4E). No significant transcriptional changes in FLuc mRNA levels were observed upon d4E-BPLLAA expression using qRT-PCR (data not shown). These data imply that upon DR, d4E-BP levels increase and lead to a preferential translation of weak secondary structure mRNAs, including mitochondrial ETC components.
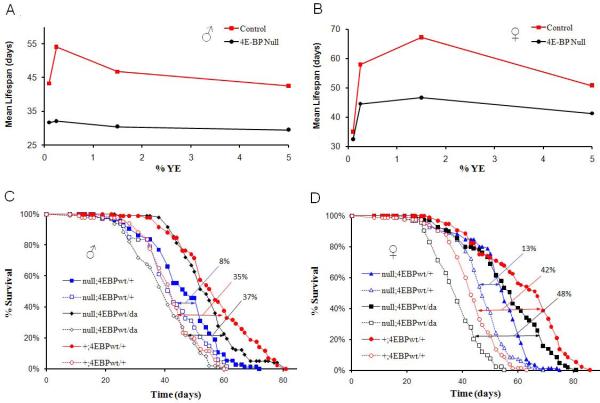
Maximal Lifespan Extension by DR is Dependent on d4E-BP
As d4E-BP appears to be important for the translational shift towards increased ribosomal loading of mitochondrial ETC components, we asked whether d4E-BP is necessary for lifespan extension upon DR. Control flies showed lifespan extension of 27% in males and 32% in females upon DR (Figures 5A and 5B). In contrast, d4E-BP null mutant flies, (Bernal and Kimbrell, 2000), showed a diminished change in lifespan when the nutrient conditions were varied (Figures 5A and 5B and Table S4). To confirm that lack of d4E-BP was causal for the diminished DR response, wild-type d4E-BP was ubiquitously expressed with daughterless-Gal4, using the Gal4-UAS system (Brand and Perrimon, 1993) in the d4E-BP null mutant background. This rescued the DR response from 8% in 4E-BP null mutant males to 37% and from 13% in d4E-BP null mutant females to 48%. This was similar to the 35% and 42% lifespan extension upon DR seen in control males and females respectively (Figures 5C and 5D and Table S5).As many labs use different yeast sources in fly media, we also investigated the lifespan effect due to DR using Brewer’s Yeast (BY). Male and female control flies had a 24% and 15% lifespan increase upon DR with BY, while neither male nor female d4E-BP null mutants showed a significant change in lifespan upon DR (Figures S3A and S3B and Table S6). While an inverse correlation between reproduction and lifespan extension has been noted (Partridge et al., 2005), this link can be decoupled (Mair et al., 2004). We found that both control and d4E-BP null mutant flies showed similar reductions in egg production upon YE restriction despite their differences in lifespan (Figure S4). These observations suggest that d4E-BP is necessary for the full lifespan extension upon DR, but not for reproductive changes.
Figure 5. 4E-BP is Required for Maximal Lifespan Extension upon DR in Drosophila.
(A, B) Lifespan of male and female revertant and d4E-BP null mutant flies on various YE concentrations.
(A) Male control and d4E-BP null mutant flies (B) Female control and d4E-BP null mutant flies. (C, D) Rescue of DR effect by ubiquitously expressing d4E-BP using daughterless (da)-Gal4 in the 4E-BP null mutant background. Solid symbols DR (0.25% YE), empty symbols control (5% YE). (C) Male 4E-BP null mutant flies have an 8% lifespan extension upon DR. Control flies have a lifespan extension of 35% and putting 4E-BP back in the null mutant line gives a lifespan extension of 37%.
(D) Female 4E-BP null mutant flies have a 13% lifespan extension upon DR. Control flies have a lifespan extension of 42% and putting 4E-BP back in the null mutant background gives a lifespan extension of 48%. (Statistical analyses with replicates are shown in Tables S4 and S5)
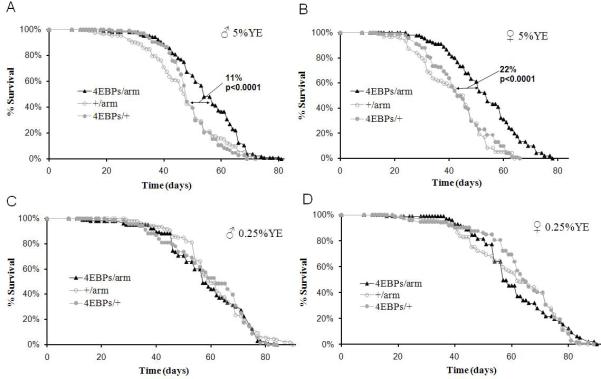
To ascertain whether elevated levels of 4E-BP are sufficient to extend lifespan, a wild-type d4E-BP (d4E-BPwt), as well as two activated alleles of d4E-BP (Miron et al., 2001) were overexpressed using the ubiquitously expressed driver, armadillo-Gal4. d4E-BPLL, has both its Met 59 and Lys 60 mutated to Leu, which increases its binding to deIF4E more than threefold (Miron et al., 2001). Two activated alleles were previously classified as weak (d4E-BPw) and strong (d4E-BPs) based on their growth inhibition properties. Overexpression of d4E-BPwt caused no change in lifespan on rich food in males or females, while overexpression of the weak activated allele extended mean lifespan of females 14% on rich food but no significant lifespan extension was observed in male flies (Figures S5A and S5B and Table S7). Induction of the strong allele extended male lifespan 11% and female lifespan 22% on rich food (Figures 6A and 6B and Table S7). In contrast, under DR (0.25% YE), there was no lifespan extension, beyond the effect of DR alone, in all of the d4E-BP alleles tested (Figures 6C, 6D, S5C, S5D and Table S7). These observations are consistent with the hypothesis that lifespan extension during DR is mediated by an increase in 4E-BP activity.
Figure 6. Overexpression of Activated d4E-BP Extends Lifespan in a Nutrient Dependent Manner in Drosophila.
(A-D) Male and female flies overexpressing d4E-BPLLs (4E-BPs) using armadillo (arm) GAL4 strain extends lifespan on high nutrition (5%YE) but not under DR (0.25% YE).
(A) Survival of male flies on high nutrition.
(B) Female flies on high nutrition.
(C) Male flies under DR.
(D) Female flies under DR. ‘+’ is the Benzer Lab w1118 strain, into which each of these lines was outcrossed 6 times. P values were obtained by comparing the survival curves with GraphPad Prism Software using the longest lived control. (Statistical analyses with replicates are shown in Table S7)
Discussion
A number of studies have hinted at the lack of correlation between the transcriptome and proteome and the importance of regulation of mRNA translation especially under environmental stresses (Sonenberg et al., 2000). A previous study shows that RAS and Akt contribute to tumor formation only modestly by transcriptional changes but largely by altered recruitment of mRNAs to large polysomes (Rajasekhar et al., 2003). Genes that regulate growth, transcriptional regulation, cell to cell interactions and morphology were most altered in their mRNA translation state. Selective increase in translation of particular mRNAs under conditions that result in most mRNAs being translationally repressed has been described in other systems. Inhibition of eIF4E in HeLA cells has been shown to decrease cell growth and protein synthesis but increase the protein levels of HSP 90, HSP 70, HSP 65 and HSP 27. This increase in protein was found to be due to an increased mRNA ribosome loading (Joshi-Barve et al., 1992). Measurement of genome wide changes in mRNA translation state under nutrient limiting conditions is therefore important to understand how DR modulates lifespan.
The translational regulator d4E-BP has been shown to be regulated upon starvation via dFOXO (Teleman et al., 2005). We found that dFOXO is not necessary for the induction of the d4E-BP protein under DR conditions where YE is limited but sucrose is kept constant. It has also been shown that under starvation, mitochondrial ribosomal proteins are transcriptionally downregulated in a dFOXO dependent manner (Teleman et al., 2008), yet we do not see a transcriptional downregulation of mitochondrial ribosomal proteins upon DR (Table S2). Furthermore, it has also been shown that dFOXO null mutant flies appear to respond normally to DR (Giannakou et al., 2008; Min et al., 2008). These results emphasize that there are differences between the acute nutritional deprivation under complete starvation and the nutritional manipulation of DR which extends lifespan.
Our data suggests that the translation repressor, d4E-BP, is required for maximal lifespan extension upon DR and that upregulation of activated d4E-BP is sufficient to extend the lifespan of the fly on a rich diet. The fact that overexpression of wildtype d4E-BP was not sufficient to extend lifespan on high nutrition suggests the importance of post-translational regulation of 4E-BP by the TOR pathway to determine lifespan. Thus, DR is likely to have two functions, firstly to increase levels of 4E-BP and secondly to reduce 4E-BP phosphorylation. Our experiments show that both males and females show significant differences upon overexpression of 4E-BP but not to the same extent. In general females show a greater extension of lifespan upon DR and it is possible that there is sexual dimorphism in responsive to d4E-BP effects. Sexual dimorphism with regards to lifespan extension has also been observed previously. Mutation in chico, an insulin receptor substrate, extends lifespan to a greater degree in females than males (Clancy et al., 2001). Given the important role for 4E-BP in modulating mRNA translation our results imply that lifespan extension by DR is likely to be due to changes in mRNA translation state of various mRNAs. The effects of DR have also been linked with alterations in mRNA translation in Saccharomyces cerevisiae. Recently, it was shown that specific reduction of 60S ribosomal subunit levels slows aging in yeast in a manner overlapping with the effects of DR and TOR. Genetic epistasis analyses suggest that DR reduced 60S subunit abundance, which mediates the translational regulation of GCN4. GCN4 was found to partially mediate the lifespan extension of Saccharomyces cerevisiae by DR (Steffen et al., 2008). Mitochondrial gene expression has also been shown to be translationally upregulated by inhibition of TOR in Saccharomyces cerevisiae and has been shown to confer increased resistance to starvation (Bonawitz et al., 2007). Along with our results this supports the idea that regulation of mRNA translation may play a key role in mediating lifespan extension across species.
Investigation of the genome wide translation changes upon DR identified a correlation between mRNA translation and 5′UTR structural properties. Though global translation is decreased upon DR, mitochondrial genes are one class which is translationally upregulated. Mitochondrial ETC and mitochondrial ribosomal protein 5′UTRs are significantly shorter and have weaker secondary structure compared to the whole Drosophila genome. We demonstrate that mitochondrial 5′UTRs were sufficient to confer differential translational upregulation upon activated d4E-BP expression. This upregulation did not seem to depend on specific sequences as the reverse complement of a Complex I 5′UTR and the 100 bp 5′ of the ATG translational start site of the Tubulin 5′UTR were also sufficient to upregulate translation upon activated d4E-BP expression. While the mechanism for this upregulation is unknown, it appears to be independent of an internal ribosome entry site (IRES) as the full length Tubulin 5′UTR upstream of the mRPL14 5′UTR was sufficient to suppress the preferential translation upon activated d4E-BP expression. Control of mRNA translation by gross 5′UTR secondary structure may represent a novel means of regulating gene expression under nutrient limitation. Such a regulatory mechanism would have the advantage of being faster than transcriptional regulation, yet more energy-efficient than posttranslational control. Along with our mitochondrial measurements, these data support the possibility of translational upregulation of mitochondrial components upon DR in Drosophila.
Mitochondrial biogenesis and efficiency have been suggested to increase under DR in mammals (Lopez-Lluch et al., 2006; Nisoli et al., 2005). Our data showing increased translation of mitochondrial genes upon DR, and the DR specific reduction in lifespan upon inhibition of specific mitochondrial ETC subunits support a causal link between enhanced mitochondrial function and protective effects of DR. Increased reliance on ATP generation by mitochondrial respiration during times of nutrient limitation may be advantageous. While glycolysis is much faster in ATP generation, there is a trade-off of lower efficiency compared to mitochondrial respiration (Pfeiffer et al., 2001). This phenomenon is similar to the Warburg effect in cancer, where cancer cells which have high nutrient uptake, and rely much more on glycolysis than oxidative phosphorylation for ATP generation, even in the presence of O2 (DeBerardinis et al., 2008; Warburg, 1956). Recently 4E-BP1 has been postulated as a funnel factor for cancer, as its phosphorylation status is associated with malignant progression in a large variety of cancers regardless of the upstream oncogenic alterations (Armengol et al., 2007). Many genes, including various growth factors and regulatory proteins, with high 5′UTR secondary structure are enhanced translationally in vivo upon increased eIF4E expression (Kozak, 1991; Mamane et al., 2004). Consistent with this we observe that upon DR mRNAs are differentially associated with ribosome depending upon its 5′UTR structure, which maybe important for both the anti oncogenic and lifespan extension effects of DR. Recently it has been suggested that an age-related decline in metabolic function as evidenced by reduced expression of genes in the electron transport chain may be a common feature between species as diverse as flies, worms and mammals (McCarroll et al., 2004; Zahn et al., 2006). It is therefore significant that we observe that DR which is known to slow aging in various species increases the translation of mitochondrial electron transport chain genes via the downstream TOR effector 4E-BP. This could have a protective effect by maintaining the function of the electron transport chain and hence ATP production with age. Our results suggest that d4E-BP modulates mRNA translation to induce a metabolic shift towards increased mitochondrial capacity which may prolong lifespan during times of nutrient limitation.
Supplementary Material
Acknowledgements
We thank David Nicholls, Martin Brand, Judith Campisi, Gordon Lithgow, Nagendra Yadav, Paul Sternberg, Eric Schwarz, Eimear Kenny and members of the Kapahi and Benzer Labs, for helpful discussions. We would also like to thank the Kimbrell, Lasko, Sonenberg, Tijan and Hafen labs for donation of strains and antibodies. This work was funded by grants from the National Institutes of Health (SB), a T32 NIH training grant fellowship (AR), grants from the Ellison Medical Foundation, American Foundation for aging research, Hillblom foundation, a Nathan Shock Startup award, a gift from the Harold J. and Reta Haynes Family Foundation and the NIH (RL1AAG032113, 1R21AG028241-01) (PK). The microarray work was supported by the Millard and Muriel Jacobs Genetics and Genomics Laboratory at California Institute of Technology.
Appendix
Experimental Procedures
Polysome profiling
Profiles were generated via optimization of previously used methods (Dinkova et al., 2005). In brief, 100 1-2 day old male flies kept on either 4% YE and 0.25% YE for 6 days were homogenized on ice in 350 ml of solublization buffer (300 mM NaCl, 50 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 1 mM EGTA, 200 mg heparin/ml, 400 U RNAsin/ml, 1.0 mM phenylmethylsulfonyl fluoride, 0.2 mg cycloheximide/ml, 1% Triton X100, 0.1% Sodium Deoxycholate) by 60 strokes with a Teflon homogenizer. 850 ml additional solublization buffer was added, vortexed for no longer than 1 second, and placed back on ice for 10 minutes. Samples were spun at 20,000g for 15 minutes at 4°C and 0.9 ml of the supernatant was applied to the top of a 10-50% sucrose gradient in high salt resolving buffer (140 mM NaCl, 25 mM Tris-HCl [pH 8.0], 10 mM MgCl2). Polysomes and ribosomal subunits were separated via centrifugation in a Beckman SW41Ti rotor at 38,000 rpm for 90 min at 4°C. Gradients were fractionated using a Teledyne density gradient fractionator with continuous monitoring of absorbance at 252 nm. Fractions containing low (1 to 4) or high (5 or more) numbers of ribosomes per transcript were isolated for RNA isolation. This was repeated on three biological replicates. Further details on RNA isolation can be found in the supplementary experimental procedures.
35S-Methionine Incorporation
Details can be found in the supplementary experimental procedures.
Microarray Analysis
Data was normalized and analyzed using the Rosetta Resolver gene expression data analysis system. Translation ratios were calculated by dividing the high translation intensity by the low translation intensity. The translation changes under DR were calculated by dividing the translation ratios from 0.25% YE by the translation ratio for 4% YE. From the translation ratios, P-values were assigned using the Benjamini-Hochberg (FDR) test for multiple correction. Setting a FDR of <5% significantly regulated genes were examined using the DAVID bioinformatics resource (niaid.abcc.ncifcrf.gov) to identify overrepresented categories by GO classification. Data available for download at NCBI Gene Expression Omnibus (accession GSE16738).
Strains
Details on strains can be found in the supplementary experimental procedures.
Western Blot
Further details can be found in the supplementary experimental procedures.
Mitochondrial Density
Citrate synthase activity was measured in whole male fly homogenates and isolated mitochondrial fractions and normalized to their respective protein content. The specific citrate synthase activity in whole-fly homogenates divided by the specific activity in isolated mitochondria gives a ratio of mitochondrial protein per whole-fly protein or the mitochondrial protein density. Citrate synthase activity was measured as the increase in absorbance due to reduction of DTNB [(5,5′-dithiobis-(2-nitrobenzoic acid)] at 412 nm, coupled to the reduction of CoA by citrate synthase in the presence of oxaloacetate (Magwere et al., 2006).
Complex I activity
The activity of NADH: ubiquinone reductase was measured as the decrease in absorbance due to the reduction of 2,6-dichloroindophenol (DCIP) at 600 nm (e = 21,000 M-1 cm-1), coupled to the reduction of decylubiquinone. Mitochondria were isolated from whole male flies (25 flies/preparation) at 4°C according to Katewa and Ballard (Katewa and Ballard, 2007). Further details can be found in the supplementary experimental procedures.
Cytochrome C Oxidase Activity
10 whole male flies were ground in 400μL PBST, and centrifuged at 8000g 5min 4°C, and supernatant isolated. Cytochrome C 0.22mM was made fresh and reduced with 0.1M dTT. Assay Buffer contained 0.01M TrisCl, 2.5mM MgCl and 10μM Cyto C. 10μL of extract was added to 165uL Assay buffer and monitored at 550nm over 10s intervals for 1min. COX activity was normalized to total protein quantified using Pierce BCA Protein Assay.
5′UTR analysis
5′UTRs were downloaded from Flybase, annotation 5.1. 5′UTRs which were shorter then 10bp were excluded from our analysis. For any gene that had more than one annotated 5′UTR, the longest 5′UTR was analyzed and any gene without an annotated 5′UTR was omitted from further analysis. The sequences were analyzed with DAMBE (data analysis in molecular biology and evolution) software(Xia and Xie, 2001), with settings of 37°C, with no lone pairs and no GU pairs at the end of helices. DAMBE uses the Vienna RNA folding package, which gives similar folding energies to mFold (www.bioinfo.rpi.edu/applications/hybrid/quikfold.php), yet unlike mFold, it returns only non-positive folding energies. Only nuclear encoded mitochondrial ETC genes were analyzed and they were classified based on Mitodrome annotation (www2.ba.itb.cnr.it/MitoDrome/), mitochondrial ribosomal proteins were classified by Affymetrix annotation. To determine the probability that the 5′UTR folding energies of our experimentally derived subgroups (downregulated n=49, upregulated n=146, Complex I n=31, Complex IV n=12, mRPs n=64, Complex II n=4, Complex III n=9, Complex V n=12) would be expected by chance, we compared the mean of each subgroup to the mean of an appropriate sampling distribution. Sampling distributions were generated by randomly sampling 5′UTR folding energies from the pool of all 5′UTR’s for each sample size. This was repeated 10,000 times and a sampling distribution was constructed from the sample means. The mean of each subgroup was then compared to the mean and standard deviation of their respective sampling distribution and p-values obtained.
UTR cloning and Cell Culture
Details on cloning and the cell culture experiments can be found in the supplementary experimental procedures.
Lifespan Analysis
Flies were developed on Standard Caltech food (8.6% Cornmeal, 1.6% Yeast, 5% Sucrose 10% Dextrose, 0.46% Agar, 1% Acid mix) (Lewis, 1960), and adults transferred within 24 hr of eclosion to YE food (8.6% Cornmeal 5% Sucrose, 0.46% Agar, 1% Acid mix, and yeast extract (#212750 Bacto™ Yeast Extract, B.D. Diagnostic Systems, Sparks,MD) or Brewer’s Yeast (#903312 MP Biomedicals.LLC, Solon, OH). For the induction of RNAi 200μM RU486 (Mifepristone, Sigma) in 100% ethanol was added to the food (+RU486) while the controls received the vehicle (-RU486). Further details on lifespan and husbandry can be found in the supplementary experimental procedures
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengol G, Rojo F, Castellvi J, Iglesias C, Cuatrecasas M, Pons B, Baselga J, Ramon y Cajal S. 4E-binding protein 1: a key molecular “funnel factor” in human cancer with clinical implications. Cancer research. 2007;67:7551–7555. doi: 10.1158/0008-5472.CAN-07-0881. [DOI] [PubMed] [Google Scholar]
- Bernal A, Kimbrell DA. Drosophila Thor participates in host immune defense and connects a translational regulator with innate immunity. Proc Natl Acad Sci U S A. 2000;97:6019–6024. doi: 10.1073/pnas.100391597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development (Cambridge, England) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Chen D, Pan KZ, Palter JE, Kapahi P. Longevity determined by developmental arrest genes in Caenorhabditis elegans. Aging Cell. 2007 doi: 10.1111/j.1474-9726.2007.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippindale AK, Leroi AM, Kim SB, Rose MR. Phenotypic plasticity and selection in Drosophila life-history evolution. I. Nutrition and the cost of reproduction. J Evol Biology. 1993;6:171–193. [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell metabolism. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Dinkova TD, Keiper BD, Korneeva NL, Aamodt EJ, Rhoads RE. Translation of a small subset of Caenorhabditis elegans mRNAs is dependent on a specific eukaryotic translation initiation factor 4E isoform. Molecular and cellular biology. 2005;25:100–113. doi: 10.1128/MCB.25.1.100-113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford D, Hoe N, Landis GN, Tozer K, Luu A, Bhole D, Badrinath A, Tower J. Alteration of Drosophila life span using conditional, tissue-specific expression of transgenes triggered by doxycyline or RU486/Mifepristone. Exp Gerontol. 2007;42:483–497. doi: 10.1016/j.exger.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Partridge L. Role of dFOXO in lifespan extension by dietary restriction in Drosophila melanogaster: not required, but its activity modulates the response. Aging cell. 2008;7:187–198. doi: 10.1111/j.1474-9726.2007.00362.x. [DOI] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Harris TE, Lawrence JC., Jr. TOR signaling. Sci STKE. 2003;2003:re15. doi: 10.1126/stke.2122003re15. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Bonafe M, Johnson TE. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J Gerontol A Biol Sci Med Sci. 2006;61:444–460. doi: 10.1093/gerona/61.5.444. [DOI] [PubMed] [Google Scholar]
- Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- Joshi-Barve S, De Benedetti A, Rhoads RE. Preferential translation of heat shock mRNAs in HeLa cells deficient in protein synthesis initiation factors eIF-4E and eIF-4 gamma. J Biol Chem. 1992;267:21038–21043. [PubMed] [Google Scholar]
- Kaeberlein M, Burtner CR, Kennedy BK. Recent developments in yeast aging. PLoS Genet. 2007;3:e84. doi: 10.1371/journal.pgen.0030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid B. TOR pathway: linking nutrient sensing to life span. Sci Aging Knowledge Environ. 2004;2004:PE34. doi: 10.1126/sageke.2004.36.pe34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katewa SD, Ballard JW. Sympatric Drosophila simulans flies with distinct mtDNA show difference in mitochondrial respiration and electron transport. Insect biochemistry and molecular biology. 2007;37:213–222. doi: 10.1016/j.ibmb.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Koromilas AE, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 1992;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. The Journal of cell biology. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance PE, Miron M, Raught B, Sonenberg N, Lasko P. Phosphorylation of eukaryotic translation initiation factor 4E is critical for growth. Mol Cell Biol. 2002;22:1656–1663. doi: 10.1128/MCB.22.6.1656-1663.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- Lewis EB. A standard new food medium. Drosoph Inf Serv. 1960;34:117–118. [Google Scholar]
- Lopez-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S, Cascajo MV, Allard J, Ingram DK, Navas P, et al. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci U S A. 2006;103:1768–1773. doi: 10.1073/pnas.0510452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwere T, Goodall S, Skepper J, Mair W, Brand MD, Partridge L. The effect of dietary restriction on mitochondrial protein density and flight muscle mitochondrial morphology in Drosophila. J Gerontol A Biol Sci Med Sci. 2006;61:36–47. doi: 10.1093/gerona/61.1.36. [DOI] [PubMed] [Google Scholar]
- Mair W, Piper MD, Partridge L. Calories do not explain extension of life span by dietary restriction in Drosophila. PLoS Biol. 2005;3:e223. doi: 10.1371/journal.pbio.0030223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Sgro CM, Johnson AP, Chapman T, Partridge L. Lifespan extension by dietary restriction in female Drosophila melanogaster is not caused by a reduction in vitellogenesis or ovarian activity. Experimental gerontology. 2004;39:1011–1019. doi: 10.1016/j.exger.2004.03.018. [DOI] [PubMed] [Google Scholar]
- Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N. eIF4E--from translation to transformation. Oncogene. 2004;23:3172–3179. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin CS, Jan YN, Kenyon C, Bargmann CI, Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nature genetics. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- Min KJ, Yamamoto R, Buch S, Pankratz M, Tatar M. Drosophila lifespan control by dietary restriction independent of insulin-like signaling. Aging cell. 2008;7:199–206. doi: 10.1111/j.1474-9726.2008.00373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron M, Verdu J, Lachance PE, Birnbaum MJ, Lasko PF, Sonenberg N. The translational inhibitor 4E-BP is an effector of PI(3)K/Akt signalling and cell growth in Drosophila. Nat Cell Biol. 2001;3:596–601. doi: 10.1038/35078571. [DOI] [PubMed] [Google Scholar]
- Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- Nusbaum TJ, Rose MR. The effects of nutritional manipulation and laboratory selection on lifespan in Drosophila melanogaster. J Gerontol A Biol Sci Med Sci. 1999;54:B192–198. doi: 10.1093/gerona/54.5.b192. [DOI] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Gems D, Withers DJ. Sex and death: what is the connection? Cell. 2005;120:461–472. doi: 10.1016/j.cell.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Petroulakis E, Mamane Y, Le Bacquer O, Shahbazian D, Sonenberg N. mTOR signaling: implications for cancer and anticancer therapy. Br J Cancer. 2006;94:195–199. doi: 10.1038/sj.bjc.6602902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 2001;292:504–507. doi: 10.1126/science.1058079. [DOI] [PubMed] [Google Scholar]
- Rajasekhar VK, Viale A, Socci ND, Wiedmann M, Hu X, Holland EC. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol Cell. 2003;12:889–901. doi: 10.1016/s1097-2765(03)00395-2. [DOI] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The Translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nature. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- Shamji AF, Nghiem P, Schreiber SL. Integration of growth factor and nutrient signaling: implications for cancer biology. Mol Cell. 2003;12:271–280. doi: 10.1016/j.molcel.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Shantz LM, Hu RH, Pegg AE. Regulation of ornithine decarboxylase in a transformed cell line that overexpresses translation initiation factor eIF-4E. Cancer Res. 1996;56:3265–3269. [PubMed] [Google Scholar]
- Sonenberg N, Hershey JWB, Mathews BM. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2000. [Google Scholar]
- Sonenberg N, Hershey JWB, Mathews M. Translational control of gene expression. 2nd edn Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syntichaki P, Troulinaki K, Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007 doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Chen YW, Cohen SM. 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev. 2005;19:1844–1848. doi: 10.1101/gad.341505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman AA, Hietakangas V, Sayadian AC, Cohen SM. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell metabolism. 2008;7:21–32. doi: 10.1016/j.cmet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. The Journal of heredity. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Zahn JM, Sonu R, Vogel H, Crane E, Mazan-Mamczarz K, Rabkin R, Davis RW, Becker KG, Owen AB, Kim SK. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS genetics. 2006;2:e115. doi: 10.1371/journal.pgen.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000;14:2712–2724. doi: 10.1101/gad.835000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Q, Schummer M, Hood L, Morris DR. Messenger RNA translation state: the second dimension of high-throughput expression screening. Proc Natl Acad Sci U S A. 1999;96:10632–10636. doi: 10.1073/pnas.96.19.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.