Abstract
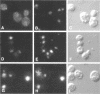
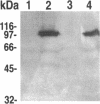
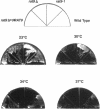
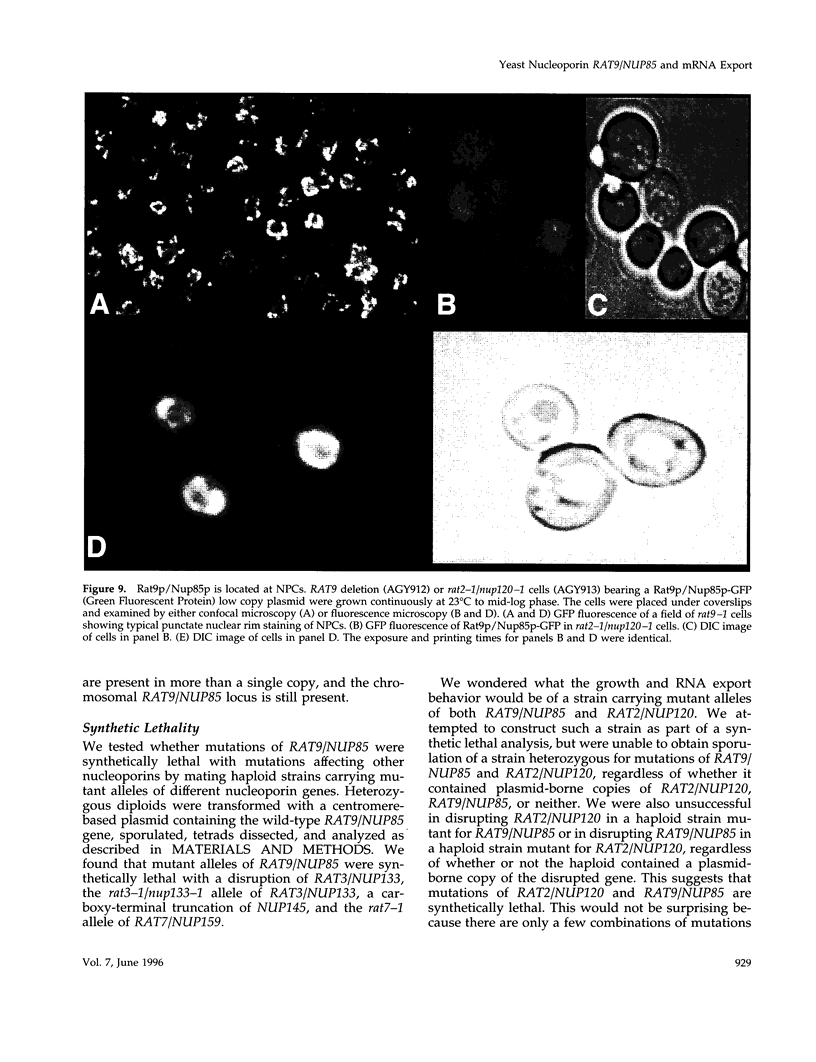
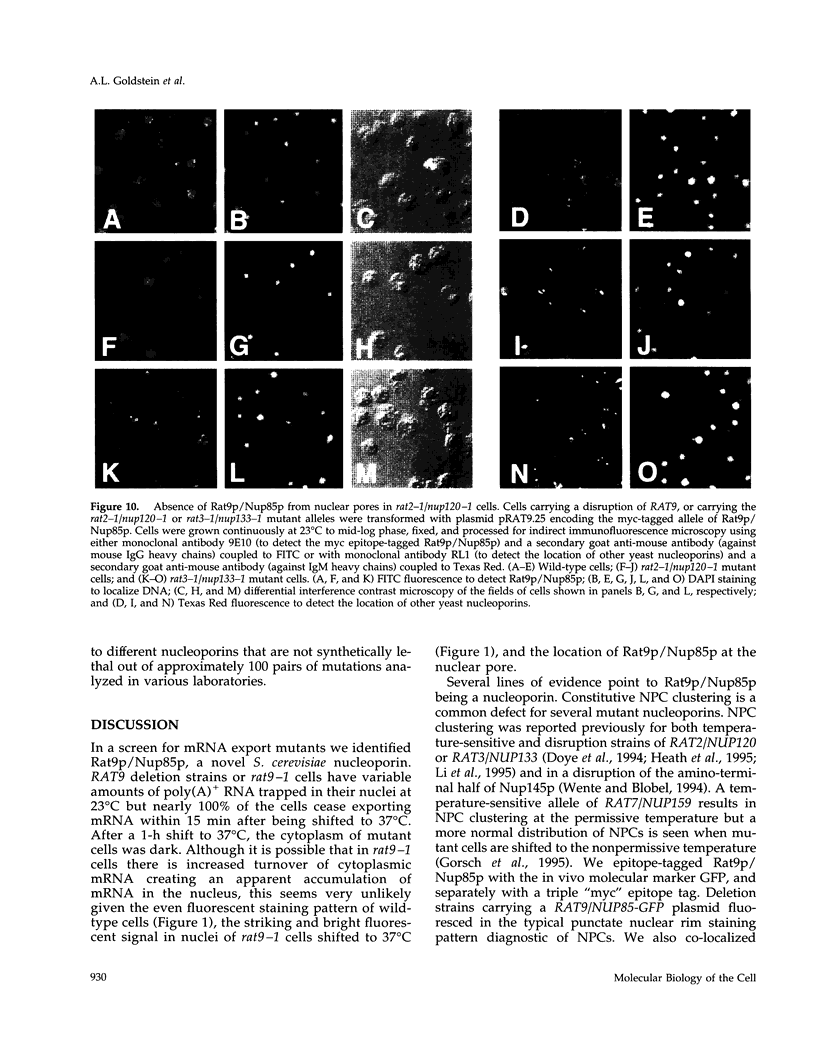
In a screen for mutants defective in nucleocytoplasmic export of mRNA, we have identified a new component of the Saccharomyces cerevisiae nuclear pore complex (NPC). The RAT9/NUP85 (ribonucleic acid trafficking) gene encodes an 84.9-kDa protein that we have localized to NPCs by tagging the RAT9/NUP85 gene with the in vivo molecular marker Green Fluorescent Protein. In cells containing either the rat9-1 allele or a complete deletion of the RAT9/NUP85 gene, poly(A)+ RNA accumulates rapidly in nuclei after a shift from 23 degrees C to 37 degrees C. Under these same conditions, rapid fragmentation of the nucleolus was also observed. At the permissive growth temperature in rat9-1 or RAT9 deletion strains, the nuclear envelope (NE) becomes detached from the main body of the nucleus, forming long thin double sheets of NE. NPCs within these sheets are clustered and some appear to be locked together between opposing sheets of NE such that their nucleoplasmic faces are in contact. The Rat9/Nup85 protein could not be detected in cells carrying a mutation of RAT2/NUP120, suggesting that Rat9p/Nup85p cannot be assembled into NPCs in the absence of Rat2p/Nup120p. In contrast,Rat9/ Nup85 protein was readily incorporated into NPCs in strains carrying mutant alleles of other nucleoporin genes. The possible role of Rat9p/Nup85p in NE integrity and the loss of nucleoporins when another nucleoporin is mutant or absent are discussed.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aitchison J. D., Blobel G., Rout M. P. Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J Cell Biol. 1995 Dec;131(6 Pt 2):1659–1675. doi: 10.1083/jcb.131.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amberg D. C., Goldstein A. L., Cole C. N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992 Jul;6(7):1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Aris J. P., Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol. 1988 Jul;107(1):17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993 Jul 11;21(14):3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Bogerd A. M., Hoffman J. A., Amberg D. C., Fink G. R., Davis L. I. nup1 mutants exhibit pleiotropic defects in nuclear pore complex function. J Cell Biol. 1994 Oct;127(2):319–332. doi: 10.1083/jcb.127.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975 Oct;124(1):511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland C. S., Snyder M. Nuclear pore complex antigens delineate nuclear envelope dynamics in vegetative and conjugating Saccharomyces cerevisiae. Yeast. 1993 Mar;9(3):235–249. doi: 10.1002/yea.320090304. [DOI] [PubMed] [Google Scholar]
- Doye V., Wepf R., Hurt E. C. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J. 1994 Dec 15;13(24):6062–6075. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworetzky S. I., Feldherr C. M. Translocation of RNA-coated gold particles through the nuclear pores of oocytes. J Cell Biol. 1988 Mar;106(3):575–584. doi: 10.1083/jcb.106.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E., Boelens W. C., Wimmer C., Mattaj I. W., Hurt E. C. Nup145p is required for nuclear export of mRNA and binds homopolymeric RNA in vitro via a novel conserved motif. Cell. 1994 Jul 29;78(2):275–289. doi: 10.1016/0092-8674(94)90297-6. [DOI] [PubMed] [Google Scholar]
- Feldherr C. M., Kallenbach E., Schultz N. Movement of a karyophilic protein through the nuclear pores of oocytes. J Cell Biol. 1984 Dec;99(6):2216–2222. doi: 10.1083/jcb.99.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes D. J. Structure and function of the nuclear pore complex. Annu Rev Cell Biol. 1992;8:495–527. doi: 10.1146/annurev.cb.08.110192.002431. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988 Dec 30;74(2):527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gorsch L. C., Dockendorff T. C., Cole C. N. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J Cell Biol. 1995 May;129(4):939–955. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Emig S., Weise C., Hucho F., Pohl T., Hurt E. C. A novel nuclear pore protein Nup82p which specifically binds to a fraction of Nsp1p. J Cell Biol. 1995 Sep;130(6):1263–1273. doi: 10.1083/jcb.130.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath C. V., Copeland C. S., Amberg D. C., Del Priore V., Snyder M., Cole C. N. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J Cell Biol. 1995 Dec;131(6 Pt 2):1677–1697. doi: 10.1083/jcb.131.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R., Cubitt A. B., Tsien R. Y. Improved green fluorescence. Nature. 1995 Feb 23;373(6516):663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- Hurwitz M. E., Blobel G. NUP82 is an essential yeast nucleoporin required for poly(A)+ RNA export. J Cell Biol. 1995 Sep;130(6):1275–1281. doi: 10.1083/jcb.130.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T., Chen S., Hitomi M., Jacobs E., Kumagai C., Liang S., Schneiter R., Singleton D., Wisniewska J., Tartakoff A. M. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J Cell Biol. 1994 Aug;126(3):649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T., Hitomi M., Chen S., Tartakoff A. M. Nuclear mRNA accumulation causes nucleolar fragmentation in yeast mtr2 mutant. Mol Biol Cell. 1994 Nov;5(11):1253–1263. doi: 10.1091/mbc.5.11.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T., Schneiter R., Hitomi M., Tartakoff A. M. Mutations in nucleolar proteins lead to nucleolar accumulation of polyA+ RNA in Saccharomyces cerevisiae. Mol Biol Cell. 1995 Sep;6(9):1103–1110. doi: 10.1091/mbc.6.9.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer D. M., Strambio-de-Castillia C., Blobel G., Rout M. P. The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J Biol Chem. 1995 Aug 11;270(32):19017–19021. doi: 10.1074/jbc.270.32.19017. [DOI] [PubMed] [Google Scholar]
- Li O., Heath C. V., Amberg D. C., Dockendorff T. C., Copeland C. S., Snyder M., Cole C. N. Mutation or deletion of the Saccharomyces cerevisiae RAT3/NUP133 gene causes temperature-dependent nuclear accumulation of poly(A)+ RNA and constitutive clustering of nuclear pore complexes. Mol Biol Cell. 1995 Apr;6(4):401–417. doi: 10.1091/mbc.6.4.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F., Paschal B., Evans J., Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993 Dec;123(6 Pt 2):1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzayan C., Copeland C. S., Snyder M. The NUF1 gene encodes an essential coiled-coil related protein that is a potential component of the yeast nucleoskeleton. J Cell Biol. 1992 Mar;116(6):1319–1332. doi: 10.1083/jcb.116.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. S., Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993 Oct 14;365(6447):661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- Moroianu J., Blobel G. Protein export from the nucleus requires the GTPase Ran and GTP hydrolysis. Proc Natl Acad Sci U S A. 1995 May 9;92(10):4318–4322. doi: 10.1073/pnas.92.10.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M., Nogi Y., Clark M. W., Nomura M. Structural alterations of the nucleolus in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Mol Cell Biol. 1993 Apr;13(4):2441–2455. doi: 10.1128/mcb.13.4.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A., Moore M. S., Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995 Apr 21;81(2):215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Reichelt R., Holzenburg A., Buhle E. L., Jr, Jarnik M., Engel A., Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J Cell Biol. 1990 Apr;110(4):883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riles L., Dutchik J. E., Baktha A., McCauley B. K., Thayer E. C., Leckie M. P., Braden V. V., Depke J. E., Olson M. V. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics. 1993 May;134(1):81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash M., Singer R. H. RNA travel: tracks from DNA to cytoplasm. Cell. 1993 Nov 5;75(3):399–401. doi: 10.1016/0092-8674(93)90373-x. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Rout M. P., Blobel G. Isolation of the yeast nuclear pore complex. J Cell Biol. 1993 Nov;123(4):771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M. P., Wente S. R. Pores for thought: nuclear pore complex proteins. Trends Cell Biol. 1994 Oct;4(10):357–365. doi: 10.1016/0962-8924(94)90085-x. [DOI] [PubMed] [Google Scholar]
- Schneiter R., Kadowaki T., Tartakoff A. M. mRNA transport in yeast: time to reinvestigate the functions of the nucleolus. Mol Biol Cell. 1995 Apr;6(4):357–370. doi: 10.1091/mbc.6.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Wimmer C., Rieger M., Doye V., Tekotte H., Weise C., Emig S., Segref A., Hurt E. C. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996 Jan 26;84(2):265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Snow C. M., Senior A., Gerace L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J Cell Biol. 1987 May;104(5):1143–1156. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathmann M., Hamilton B. A., Mayeda C. A., Simon M. I., Meyerowitz E. M., Palazzolo M. J. Transposon-facilitated DNA sequencing. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1247–1250. doi: 10.1073/pnas.88.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J. R. Labeling of RNA and phosphoproteins in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:423–428. doi: 10.1016/0076-6879(91)94033-9. [DOI] [PubMed] [Google Scholar]
- Wente S. R., Blobel G. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J Cell Biol. 1993 Oct;123(2):275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente S. R., Blobel G. NUP145 encodes a novel yeast glycine-leucine-phenylalanine-glycine (GLFG) nucleoporin required for nuclear envelope structure. J Cell Biol. 1994 Jun;125(5):955–969. doi: 10.1083/jcb.125.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente S. R., Rout M. P., Blobel G. A new family of yeast nuclear pore complex proteins. J Cell Biol. 1992 Nov;119(4):705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R., Rine J. Transmission electron microscopy and immunocytochemical studies of yeast: analysis of HMG-CoA reductase overproduction by electron microscopy. Methods Cell Biol. 1989;31:473–512. doi: 10.1016/s0091-679x(08)61624-6. [DOI] [PubMed] [Google Scholar]
- Wu J., Matunis M. J., Kraemer D., Blobel G., Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem. 1995 Jun 9;270(23):14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]