Abstract
In health, mucosal inflammation is prevented by tightly regulated responses via Toll-like receptors (TLR) that interact with specific microbe associated molecular patterns. Currently, 13 TLRs have been identified. Based on the specificity of ligand recognition, TLR-2 and TLR-4 can recognize most oral commensal microorganisms. Recent identification of some soluble TLRs (sTLRs) suggests additional regulatory roles for these receptors. We report here the presence of sTLR-4 polypeptides in adult human saliva. Functionally, the salivary sTLR-4 suppressed cytokine secretion by activated macrophages. The sTLR-4 levels were elevated significantly in oral lichen planus (OLP), a chronic inflammatory condition of the oral mucosa characterized by clinical persistence. In contrast, the epithelial cells in the saliva of OLP subjects expressed significantly reduced TLR-2 and TLR-4 mRNA that correlated with fewer bacteria/salivary epithelial cells. Investigating the soluble and cellular components of saliva is useful in identifying potential biomarkers for oral mucosal lesions.
Keywords: epithelial cells, OLP, saliva, TLR
Introduction
Lichen planus is a chronic inflammatory condition that affects the skin, oral and genital mucous membranes [1]. Oral lichen planus (OLP) occurs as the primary manifestation in 15–35% of patients [2]. The mucosal lesions are extremely uncomfortable, painful and persistent [3]. Pathologically, OLP is characterized by active epithelium and subepithelial accumulation of lymphocytes. A putative epithelium-specific autoantigen or signals from activated epithelial cells could initiate the inflammatory process [4–6].
In general, epithelial cells may be activated by chemical/microbial stimuli via Toll-like receptors (TLRs). TLRs recognize conserved pathogen-associated molecular patterns (PAMPs) shared by a large group of microorganisms [7]. Currently, 13 mammalian TLRs and many of their ligands are known. TLR-2 and TLR-4 recognize the peptidoglycan and the lipopolysaccharide (LPS) of Gram+ and Gram− bacteria respectively [8,9]. TLR-2 also acts as a receptor for the LPS of Porphyromonas gingivalis, a common Gram− oral pathogen [10]. Both TLR-2 and TLR-4 recognize PAMPs presented by Candida, a common fungal pathogen [11]. Thus, collectively, TLR-2 and TLR-4 can interact with most oral commensal and pathogen. CD14 is a co-receptor that facilitates PAMP recognition by both TLR-2 and TLR-4 [12]. In addition to PAMPs, TLRs also bind endogenous molecules such as the heat shock proteins (HSP) [13]. HSP are stress-related molecules that play an important role in immune and inflammatory reactions. The epithelial cells in OLP express elevated levels of HSP-60 and HSP-70 [14]. HSP-60 is a ligand for TLR-2 [15].
Stimulation of epithelial TLRs by pathogen or endogenous ligands mediates chemokine secretion, upregulates intercellular adhesion molecule-1 expression and promotes lymphocyte recruitment leading to cytokine secretion [5,6]. It has been observed that while proinflammatory cytokines [interleukin (IL)-1β/interferon (IFN)-γ/tumour necrosis factor (TNF)-α] promote TLR-4 expression, anti-inflammatory (IL-4, IL-13) cytokines down-regulate TLR-4 significantly in epithelial cells [16,17]. Although most TLRs are transmembrane receptors, recently soluble forms of some TLRs (sTLR) have been identified in various body fluids and cellular secretions [18–21]. It has been suggested that the sTLRs may protect the host by sequestrating PAMPs [7,22].
Because OLP occurs in constantly renewing epithelia continuously exposed to commensal microorganisms, we hypothesized that altered microbiota–epithelial interactions may play a critical role in the persistence and/or re-occurrence of the mucosal lesions. In recent years, considerable focus has been directed towards identifying molecular markers in saliva for monitoring disease progression [23]. In addition to the soluble components, saliva is a rich source of oral epithelial cells [24]. In this study we investigated the presence and diagnostic potential of salivary TLR-2 and TLR-4 in OLP.
Material and methods
Study population
The study group consisted of 30 subjects reporting to the Indiana University School of Dentistry (IUSD) presenting characteristic clinical lesions of OLP [4], and 40 age- and sex-matched control subjects with no known medical illness or clinically identifiable oral lesions. Informed consent was obtained from all subjects in accordance with the Indiana University's Institutional Review Board (IRB).
Collection and processing of saliva
Unstimulated whole saliva (UWS) was collected into a chilled centrifuge tube [20]. Each sample was divided into two parts. One part was clarified by centrifuging at 4000 g at 4°C for 10 min and stored in Complete™ Protease Inhibitor Cocktail (Roche, Mannheim, Germany) at −80°C. Epithelial cells were isolated from the second part of UWS [25]. The samples diluted 1:10 in isotonic diluent (Hematronix, Inc., Benicia, CA, USA) and two drops of Zap-o-Globin lytic reagent (Beckman Coulter, Fullerton, CA, USA) were centrifuged at 220–300 g for 10 min. The cell pellet resuspended in sterile phpsphate-buffered saline was passed over a 20 µm sterile nylon membrane (Small Parts Inc., Miami Lakes, FL, USA). The membrane-adherent epithelial cell-enriched population was collected and stored at −80°C.
The UWS sample preparation
The UWS samples were depleted of amylase and immunoglobulins by incubating serially with anti-human amylase monoclonal antibody (mAb) (1:2500, catalogue no. ab8944; Abcam, Cambridge, MA, USA) and protein G beads (Miltenyi Biotec, Inc., Auburn, CA, USA) at 4°C. No significant difference was observed in the amount of total protein in the UWS between the two groups, as determined by spectrophotometry (data not shown). For detection of sTLR-4, the precleaned UWS samples were depleted further of sCD14 and sTLR-2 by incubating sequentially with anti-human CD14 mAb (clone 134603; R&D Systems, Minneapolis, MN, USA) and anti-human TLR-2 mAb (clone: 1030A5.138; Imgenex Corp., San Diego, CA, USA) respectively. For detection of sTLR-2 and sCD14, the UWS samples were depleted similarly of sCD14 and sTLR-2/sTLR-4 using anti-human TLR-4 mAb (clone 76B357; Imgenex Corp.). For cytokine analysis, specific BD OptEIA™ enzyme-linked immunosorbent assay (ELISA) kits (BD Biosciences, San Jose, CA, USA) were used.
Immunoblot for sCD14, sTLR-2 and sTLR-4
Western blot analysis of salivary protein was performed [18,19]. Ten µg of total protein in the precleaned UWS samples were separated on 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane. After blocking and washing the membrane was incubated with anti-human CD14 mAb (1:1000), anti-human TLR-2 mAb (1:500), anti-human TLR-4 mAb (1:1000; catalogue no. AF1478; R&D Systems) or anti-human TLR-4 mAb (1:500) overnight at 4°C. Binding was detected using horseradish peroxidase (HRP)-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) and visualized with an enhanced chemiluminescence system (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK). The images were obtained using GelLogic systems (Eastman Kodak Company, Rochester, NY, USA). Specificity of detection was investigated by performing competition immunoblot assays with 5× molar excess of protein (CD14Fc or TLR-4Fc) or peptides (TLR-2). The salivary sTLR-4 was eluted from a parallel gel using a protein elution kit. Recombinant human CD14, TLR-2 and TLR-4 fusion proteins (R&D Systems) were used as positive controls.
Cellular assay
Human macrophage-like Thp-1 cells (gift from Dr Klemz, Department of Microbiology and Immunology, Indiana University School of Medicine) (1 × 104 cells/well) were cultured in the presence or absence of Escherichia coli LPS (1 µg/ml) (Sigma, St Louis, MO, USA) and either 10 µg of gel eluted sTLR-4 or recombinant TLR-4Fc or a combination of sTLR-4/TLR-4Fc and 10 µg of recombinant sCD14 and sMD2. Supernatants were collected at 4 h and stored at −80°C. In a separate experiment, the Thp-1 cells were prestimulated with 1 µg/ml of LPS, rested for 8 h and restimulated with LPS (0·1 µg/ml) in the presence of precleaned UWS only, precleaned UWS preincubated with 10 µg of anti-human CD14/anti-human TLR-2/anti-human TLR-4 or both anti-human CD14 and anti-human TLR-4. The two-step stimulation up-regulated TLR-4 expression in Thp-1 cells (data not shown) facilitating their use for the in-vitro functional studies [21]. Unmanipulated and LPS (0·1 µg/ml) only treated cultures were included as controls. Supernatants collected at 2 h were stored at −80°C.
The ELISA for sCD14, sTLR-2 and sTLR-4
One µg of protein from each precleaned UWS sample was assessed for the presence of sCD14/sTLR-2/sTLR-4 by ELISA. For sCD14 a sandwich ELISA kit was used [20,22]. The sTLR-2 and sTLR-4 were detected using anti-human TLR-2 mAb and anti-human TLR-4 mAb (R&D Systems) respectively. Bound antibodies were detected using HRP-conjugated anti-mouse immunoglobulin (Ig)G followed by TMB (3,3′,5,5′-tetramethylbenzidine) substrate (Pharmingen, San Diego, CA, USA). Purified recombinant human CD14Fc, TLR-2Fc and TLR-4Fc (R&D Systems) were used to develop a standard curve. Absorbance at 450 nm was read in a microplate reader (model 680; Biorad Laboratories, Hercules, CA, USA). The amount of TNF-α in the culture supernatants was measured using the BD OptEIA™ ELISA kit.
Determination of bacterial count/salivary epithelial cell
A monolayer of salivary epithelial cells smeared on a histological slide was stained by the basic Gram method [26]. The slides were visualized under a light microscope (Microphot-Fxa; Nikon, Instruments Inc, Melville, NY, USA). The images captured with a three-colour charge-coupled device (red–green–blue, DXM1200; Nikon) were analysed using the National Institutes of Health image version 1·62 (http://rsbweb.nih.gov/ij/). The red, blue and purple stains were separated by colour deconvolution and the intensities of purple stain were used to determine the characteristics of bacterial staining [27]. The histograms of 15 epithelial cells in each UWS sample were used to calculate the average staining intensity/epithelial cell, referred to as the Gram staining intensity (GSI) [28]. The GSI represents an objective measurement of bacterial count/epithelial cell in each UWS sample.
Multiplex polymerase chain reaction for bacterial analysis of salivary epithelial cells
Polymerase chain reaction (PCR) identification of bacteria in oral epithelial cells was performed as described previously [29]. Briefly, DNA from epithelial cells was extracted using DNA extraction kits (Oragene™; DNA Genotek, Inc., Ottawa, Ontario, Canada). 16S rRNA was amplified from equal amounts of DNA with conserved (universal) 16S rRNA gene-specific primers (forward primer 5′-AGAGTTTGATCMTGGCTCAG-3′ and reverse primer 5′-TACGGYTACCTTGTTACGACTT-3′) in a PTC-100 programmable Thermocycler (MJ Research Inc., Watertown, MA, USA). The amplified products were processed for multiplex PCR, as described previously, using the previously published primer sets Bacteroides forsythus (Bf), Actinobacillus actinomycetemcomitans (Aa), Porphyromonas gingivalis (Pg) and conserved reverse primer C11R. Streptococcus mutans was amplified in a separate PCR reaction [30]. The PCR products were visualized by gel electrophoresis and the images acquired using GelLogic systems. The molecular sizes of the amplicons were determined by comparison with a commercial DNA molecular size marker (1 Kb DNA ladder; Invitrogen, Carlsbad, CA, USA).
Semiquantitative measurement of reverse transcription–PCR for CD14, TLR-2 and TLR-4 mRNA
Because the cohort included in this study consisted of patients with a clinical diagnosis of OLP, we obtained 15 tissues each with histological diagnosis of OLP or fibroma from the IUSD repository following IRB approval. The epithelial cells were dissected selectively using a PixCell II Laser capture microdissection system (Arcturus Engineering, Mountain View, CA, USA) [31]. Total cellular RNA was isolated using a Qiagen RNA isolation kit (Invitrogen) and reverse-transcribed using an iScript cDNA synthesis kit (Biorad, Austin, TX, USA). An equal amount of cDNA was used to amplify CD14, TLR-2 and TLR-4 using Platinum PCR Supermix (Invitrogen). Primers were designed using Primer Express software as follows: CD14 forward: 5′ CGTGCGCGACAGGGCGTTCT-3′, reverse: 5′TAAAGGTGGGCAGGTT-3′; TLR-2 forward: 5′ ACCTGTGTGACTCTCCATCC-3′, reverse: 5′GCAGCATCATTGTTCTCTC-3′; TLR-4 forward: 5′TTCCTCTCCTGCGTGAGAC-3′, reverse: 5′TTCATAGGGTTCAGGGACAG-3′ and small proline-rich protein 2a (SPRR2a) forward: 5′AGT GCCAGCAGAAATATCCTCC-3′, reverse: 5′GAACGAGGTGAGCCAAATATC C-3′. The CD14, TLR-2 and TLR-4 mRNA in Thp-1 cells were amplified as positive controls. The PCR products were visualized and images acquired as before. The intensity of the amplified bands, determined using Kodak 1D image analysis software (Eastman Kodak Company, Rochester, MN, USA), were normalized with the corresponding SPRR2a signal [20].
Statistical analysis
The differences in the analysed soluble proteins in UWS of normal and OLP subjects were determined by one-way anova and Tukey's post hoc analysis. Student's t-test was used to determine the differences in the mRNA levels of CD14, TLR-2 and TLR-4 and GSI/epithelial cells between the groups. P values less than 0·05 were considered significant.
Results
Clinical features
The average age for OLP and healthy subjects was 61·7 ± 0·56 years and 57·4 ± 0·9 years respectively. OLP was common in women, with a female : male ratio of 3·4:1. Clinically, OLP patients presented papular, erosive or ulcerative mucosal lesions with characteristic Wickham's striae [3]. UWS was collected from all individuals and evaluated as follows.
The sTLR-4 polypeptides are found in adult human saliva
We first investigated the UWS for the presence of sTLR-4 by immunoblot analyses. The detection limit for TLR-4Fc was 0·25 µg with anti-human TLR-4 polyclonal antibody (Fig. 1a) and 1 µg with anti-human TLR-4 mAb (data not shown). Analysis of precleaned normal UWS samples using anti-human TLR-4 mAb showed four sTLR-4 polypeptides of 90, 78, 54 and 44 kDa (Fig. 1b; data not shown). The intensity of each band varied among donors. Preincubation of anti-human TLR-4 mAb with TLR-4Fc abrogated or reduced substantially the detection of most polypeptides (Fig. 1c). Only a weak signal of the 54 kDa peptide was detected, suggesting that part of this signal may be non-specific or may represent cross-reaction of anti-TLR-4 mAb with other polypeptide(s), such as the sCD14, migrating to the same position.
Fig. 1.

Detection of soluble Toll-like receptor (sTLR)-4 in human saliva: (a) decreasing concentration of recombinant TLR-4Fc was subjected to immunoblot analysis with goat anti-human TLR-4 polyclonal antibody. (b) Precleaned unstimulated whole saliva (UWS) samples (lanes 3–5) from normal individuals containing 10 µg of proteins were probed with the anti-human TLR-4 polyclonal antibody. Up to four sTLR-4 peptides were detected. Lanes 1 and 2 represent molecular weight (MW) and recombinant TLR-4-Fc respectively. (c) The specificity of the detection was confirmed by performing protein (TLR-4Fc) competition by immunoblotting.
The sTLR-4 in saliva is functional
Next we investigated the functional potential of the sTLR-4 in saliva. Stimulation of human macrophages with LPS has been shown to mediate TNF-α secretion [21]. We observed that the presence of either recombinant human TLR-4Fc or salivary sTLR-4 isolated by gel electrophoresis suppressed TNF-α secretion by LPS-stimulated Thp-1 cells (data not shown). This inhibition was overcome when cultures were added to recombinant human CD14-Fc and MD-2Fc. Previously, the CD14 and MD2 fusion proteins have been shown to bind TLR-4Fc with high avidity [32]. Hence, it is likely that the CD14-Fc and MD2Fc sequestered the sTLR-4 in UWS, allowing for unhindered cellular stimulation by LPS. Collectively, these observations support a functional potential for the 90 kDa sTLR-4 peptide isolated from saliva.
In order to determine the inhibitory potential of sTLR-4 polypeptides in the UWS, we stimulated Thp-1 cells with LPS in the presence of UWS depleted of sCD14/sTLR-4/sTLR-2/sCD14 and sTLR-4. Thp-1 cells stimulated with LPS in the presence of UWS preincubated with increasing concentrations (0–10 µg/ml) of anti-human TLR-4 mAb exhibited dose-dependent suppression of TNF-α secretion. A significant suppression of TNF-α secretion was also observed in the Thp-1 cell cultures stimulated with LPS in the presence of UWS-preincubated with anti-human CD14 mAb at 2·5 µg/ml (data not shown). It is speculated the sCD14 concentration in the UWS is the limiting factor. Significantly pronounced dose-dependent suppression of TNF-α secretion was observed in the Thp-1 cell cultures stimulated with LPS in the presence of UWS preincubated with increasing concentrations of both anti-human CD14 and anti-human TLR-4 antibody. The responsiveness of the Thp-1 cells to LPS was not inhibited in the presence of UWS preincubated with anti-human TLR-2 antibody. Taken together, these results suggest that sCD14 and sTLR-4 in the UWS either bind LPS and sequester PAMP or block LPS delivery to cell surface TLR-4.
The sTLR-2 and sTLR-4 are elevated in the UWS of OLP patients
Previously, we and others have reported the presence of sCD14 in the UWS [20,22]. Here we observed equivalent amounts of sCD14 in the UWS of both normal and OLP patients (Fig. 2a and d). Immunoblot analysis showed four sTLR-2 polypeptides of 54, 40, 34 and 30 kDa in precleaned normal UWS (Fig. 2b, lanes 1–4). Preincubation of anti-TLR-2 polyclonal antibody with blocking peptides abrogated the 54, 40 and 30 kDa polypeptides (Fig. 2b, lanes 1a–2a). Interestingly, the 34 kDa and 40 kDa sTLR-2 peptides was absent in the UWS of OLP patients (Fig. 2b, lanes 5–8). Four polypeptides sTLR-4 of 90, 78, 54 and 44 kDa were detected in precleaned normal adult UWS (Fig. 2c, lanes 8–13). Only the higher molecular weight TLR-4 polypeptide (90 kDa and78 kDa) was detected in the UWS of OLP patients (Fig. 3, lanes 1–7). Quantitatively, the sTLR-2 and sTLR-4 levels were elevated significantly in the UWS of OLP patients compared with those of control subjects (Fig. 2d).
Fig. 2.
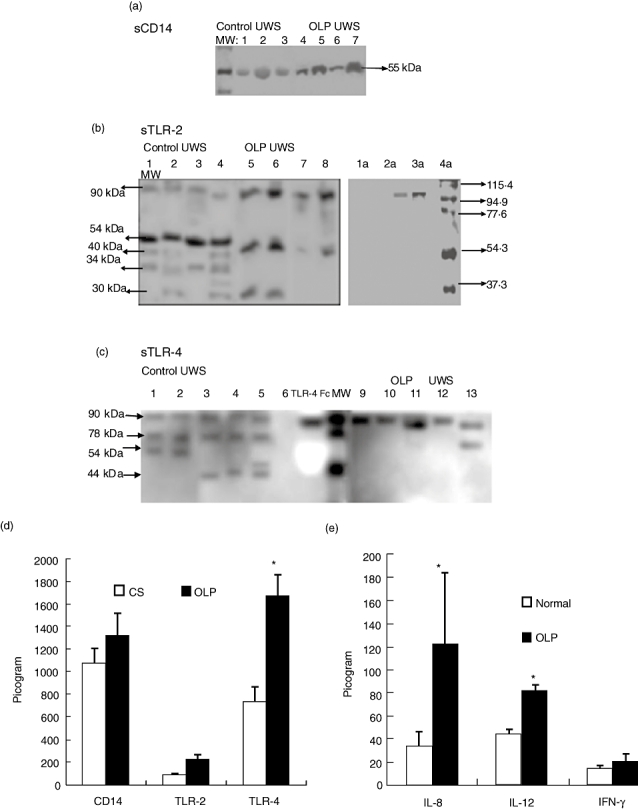
Soluble Toll-like receptor (sTLR)-2 and sTLR-4 are increased in the unstimulated whole saliva (UWS) of oral lichen planus (OLP) patients: sCD14, sTLR-2 and sTLR-4 were investigated by immunoblot (a, b and c respectively) and measured by enzyme-linked immunosorbent assay (ELISA) (d). The sCD14 was detected as a 55 kDa protein (lane 10). Lanes 1–3 and 4–7 represent UWS samples from control and oral lichen planus (OLP) subjects respectively. (b) sTLR-2 polypeptides present in precleaned UWS (10 µg total protein) by Western blotting. The specificity of the detection was confirmed by performing peptide competition by immunoblotting. The anti-TLR-2 monoclonal antibody (mAb) was preincubated (+) (lanes 1a, 2a) or not (–) (lanes 3a, 4a) with excess of the peptide/TLR-2Fc. Lanes 1–4 and 5–8 represent UWS samples from control and OLP subjects respectively. (Sup) Culture supernatants of human peripharal blood mononuclear cell (PBMC) cultures stimulated with concanavalin A (ConA) (lane 11) or recombinant human TLR-4Fc (lane 12) were loaded as controls. (c) Precleaned UWS samples containing 10 µg of proteins were subjected to immunoblot analysis with anti-human TLR-4 mAb. Recombinant human TLR-4Fc was included as control (lane 14). Lanes 1–7 and 8–13 represent UWS samples from OLP and control subjects respectively. MW, molecular weight. (d) The amount of sCD14, sTLR-2 and sTLR-4/µg of total proteins in the precleaned UWS of normal and OLP patients were determined by ELISA. (e) Quantitative estimation of the cytokines interleukin (IL-8), IL-12 and interferon-γ/µg of total proteins in the precleaned UWS of normal and OLP patients as determined by ELISA. = P < 0·05 and represents statistical significance compared with control saliva.
Fig. 3.

CD14, Toll-like receptor (sTLR)-2 and TLR-4 are expressed in epithelial cells in unstimulated whole saliva (UWS). (a) Epithelial cells from unstimulated whole saliva (UWS) of patients with oral lichen planus (OLP) (lanes 6–10) or control subjects (lanes 1–5) were assessed for the expression of CD14 [758 base pairs (bp)], TLR-2 (498 bp), TLR-4 (398 bp) mRNA and small proline-rich protein 2a (SPRR2a) (110 bp) mRNA by reverse transcription–polymerase chain reaction using specific primers. (b) Semiquantitative measurement of CD14, TLR-2 and TLR-4 mRNA normalized to the SPRR2a levels. *P < 0·05.
Consistent with previous reports, we observed significantly higher levels of IL-8, IL-12-p40 and IFN-γ in the UWS of patients with OLP compared with control subjects (Fig. 2e) [33].
Exfoliated epithelial cells in saliva and tissue epithelium express reduced TLR-2 mRNA in OLP
The cellular components of saliva include leucocytes and functional epithelial cells [25]. We determined the amount of mRNA of CD14, TLR-2 and TLR-4 in salivary epithelial cells by reverse transcription (RT)–PCR. The amount of each mRNA was normalized to that of SPRR2a, an epithelial differentiation-specific gene (Fig. 3) [34]. Previously, bacterial infection has been associated with elevated SPRR2a mRNA in the intestinal mucosa in mice [35]. However, we did not observe significant difference in the SPRR2a mRNA expression in oral epithelial cells derived from the UWS of control individuals (475 ± 219·6 arbitrary units) and OLP patients (485 ± 197 arbitrary units). The discrepancy may be attributed to the variations in the degree of differentiation between the oral and the intestinal epithelial cells [36]. Alternatively, differences in the inherent properties of the keratinocytes in different species and/or at different sites may contribute to the variation [37]. The level of TLR-2 mRNA in oral epithelial cells was significantly lower in patients with OLP compared with those of controls (Fig. 3a and b). The TLR-4 mRNA was also decreased in the salivary epithelial cells of OLP patients, although the reduction was less pronounced than that of TLR-2 mRNA. The amount of CD14 mRNA did not differ significantly between the groups (Fig. 3b).
Microscopically, OLP tissues exhibited epithelial atrophy, basement membrane zone degeneration and presence of a dense band of subepithelial inflammatory cell infiltration (data not shown). RT–PCR analysis showed reduced TLR-2 mRNA expression significantly in the epithelium of OLP tissues compared with that of normal mucosa. The CD14 and TLR-4 mRNA expressions did not vary between the groups (Fig. 4a and b).
Fig. 4.

Pure populations of epithelial cells were obtained from histologically normal buccal mucosa (lanes 6–10) or tissues that exhibit features of oral lichen planus (OLP) (lanes 1–5) by laser capture microdissection. (a) Expression of CD14 mRNA [758 base pairs (bp)], Toll-like receptor (TLR)-2 mRNA (498 bp) and TLR-4 mRNA (398 bp). The first lane (indicated) in the top three panels represent respective mRNA expression in Thp-1 cells. Expression of SPRR2a (110 bp) abundant in oral epithelium was amplified as control. (b) Quantitation of the band intensity was performed by densitometry. Data are presented as average intensity ± standard error. *P = 0·05.
The GSI/oral epithelial cell is decreased in OLP
Shedding oral epithelial cells have been shown to act as reservoirs for commensal oral bacteria [38]. In order to investigate the relationship between TLR expression and microbial association/colonization, a portion of the epithelial cells from each UWS sample was stained by the Gram method [26]. Very few, if any, Gram− bacteria were identified in the epithelial cells. Image analysis of stained individual epithelial cells after colour deconvolution suggested fewer bacteria in the salivary epithelial cells of OLP patients compared with those of controls. The density of bacteria associated with the epithelial cells measured in terms of GSI/epithelial cells was significantly lower in the OLP cohort than that of control subjects (data not shown).
Amplification of common oral commensals was performed to confirm the bacterial presence in salivary epithelial cells. Multiplex PCR analyses suggested the presence of both Gram− (B. forsythus and P. gingivalis) and Gram+ bacteria (S. mutans and A. actimomycetemcomitans) in these cells. Although the PCR method does not distinguish between intracellular and cell surface bacteria, it corroborates their association with the epithelial cells (data not shown). Others have used a similar strategy for determining the bacterial loads in oral epithelial cells by incorporating fluorescent reporter molecules into the PCR products [38].
Discussion
The presence of juxta-epithelial band of lymphocytes is a pathognomonic feature of OLP. Chemokines secreted by activated epithelial cells facilitate recruitment of lymphocytes to the mucosa [5,6]. Oral epithelial cells are activated by exogenous/endogenous stimuli via TLRs [9]. Based on the specificity of ligand recognition, TLR-2, TLR-4 and CD14 constitute a set of receptors that can recognize most oral microorganisms [9,10,39]. We report here the presence of sTLR-4 in the UWS and discuss salivary TLRs as potential biomarkers for OLP.
We detected four sTLR-4 polypeptides in the normal UWS. The extracellular domain (ECD) of human TLR-4 has been estimated to be ∼70 kDa, that increases to ∼95 kDa with glycosylation [21,40]. Hence, it is speculated that the 90 kDa sTLR-4 polypeptide in the UWS may correspond to the full ECD and the other sTLR-4 polypeptides may represent its splice variants. An alternative-spliced TLR-4 mRNA encoding a 20 kDa soluble form of TLR-4 has been reported in mice [41]. Alternatively, the lower molecular weight sTLR-4 polypeptides may originate from proteolytic processing of TLR-4. Previously, the sTLR-2 polypeptides in human plasma, breast milk and saliva have been suggested to represent processed fragments of the TLR-2 protein [18,19].
As we analysed whole saliva for its presence, the source of sTLR-4 in the UWS can be only speculated. Possible sources include secretions of salivary glands, mucosal epithelial cells and salivary leucocytes. Interestingly, the endocervical glands in the female genital mucosa have been suggested to secrete a granular form of sTLR-4 in the human cervix [42,43]. sTLRs are thought to function as decoy receptors to sequestrate circulating PAMPs [7,22]. Recombinant TLRs attenuate TLR-mediated cellular activation [19,41,44]. Here we showed that the response of human macrophage-like cells to LPS is abrogated in the presence of the 90 kDa sTLR-4 polypeptide/UWS. Depletion of salivary sCD14 and sTLR-4, the PRRs that recognize bacterial LPS abrogated this suppression.
Polymorphisms in TLR-2 and TLR-4 are associated variably with chronic mucosal inflammatory conditions such as asthma, atopy and colitis [45,46]. The observed absence of select TLR-2 and TLR-4 polypeptides in the UWS of OLP patients may be suggestive of associated TLR-2 and/or TLR-4 gene polymorphism. Clearly, careful and extensive studies are needed to investigate the possible correlation. Quantitative estimation showed significantly higher sTLR-4 in the UWS of patients with OLP. Interestingly, the inflammatory cytokines TNF-α and IFN-γ have been shown to up-regulate TLR-4 expression in human monocytes [47]. Pathologically, OLP tissues exhibit subepithelial accumulation of mononuclear cells and increased proinflammatory cytokines [48]. It is tempting to speculate that the oral epithelial cells in this local milieu may exhibit up-regulation of TLR-4 and either active secretion or proteolytic cleavage may account for the elevated sTLR-4 in the UWS of OLP patients. It has been suggested that the intestinal epithelial cells secrete sCD14 and modulate mucosal immune responses in colitis [49].
The oral epithelium exhibits a higher turnover rate in OLP [6,50]. This may precipitate exposure to hitherto unexposed/less-exposed microflora mediating altered TLR expressions and further epithelial activation. Analysis of salivary epithelial cells provides a non-invasive method of identifying biomarkers for mucosal lesions [23,25]. Molecular analysis showed dramatic reduction in the TLR-2 mRNA expression that correlated with the reduced GSI in oral epithelial cells from the UWS of OLP patients. The higher TLR-2 mRNA expression in oral epithelial cells of control individuals may be attributed to the continuous exposure to the predominantly Gram+ microflora in the healthy oral cavity [51]. Potential proliferation of Candida species, hepatitis C virus infection or a relative increase in the ratio of Gram− to Gram+ population in the local flora may alter the epithelial TLR homeostasis in OLP [4,6,52,53]. The reduced TLR-2 mRNA may represent one of the modulations.
Although the biopsy tissues evaluated for epithelial expressions of TLR-2 and TLR-4 were not from the same cohort as the UWS samples, it is interesting to observe that the TLR-2 mRNA was reduced significantly in the epithelium of the inflamed mucosa in OLP. Reduced TLR-2 expression in the epithelium in conditions of inflammation has been reported in chronic allergic rhinitis and acute anterior uveitis [54,55].
In conclusion, elevated sTLR-4 in the saliva and the increased ratio of the TLR-4 to TLR-2 expression in the tissue epithelium suggest altered microbial contacts in OLP. Perturbations in the expression and response of the epithelial TLRs to the local flora have been shown to modulate mucosal inflammation in colitis and psoriasis [42,56,57]. Future studies will elucidate the role of salivary TLRs in oral mucosal health and disease.
Acknowledgments
This study was supported partly by the Graduate Student Research Funds of the IUSD to Dr Largura Burton. The authors thank Krithika N. Kodumudi for assistance in the performance of experiments.
References
- 1.Lewis FM. Lichen sclerosus and autoimmune disease. J Reprod Med. 1998;43:1006. [PubMed] [Google Scholar]
- 2.Ingafou M, Leao JC, Porter SR, Scully C. Oral lichen planus: a retrospective study of 690 British patients. Oral Dis. 2006;12:463–8. doi: 10.1111/j.1601-0825.2005.01221.x. [DOI] [PubMed] [Google Scholar]
- 3.Al-Hashimi I, Schifter M, Lockhart PB, et al. Oral lichen planus and oral lichenoid lesions: diagnostic and therapeutic considerations. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2007;103(Suppl 25):e1–12. doi: 10.1016/j.tripleo.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Sugerman PB, Savage NW, Walsh LJ, et al. The pathogenesis of oral lichen planus. Crit Rev Oral Biol Med. 2002;13:350–65. doi: 10.1177/154411130201300405. [DOI] [PubMed] [Google Scholar]
- 5.Dutz JP. T-cell-mediated injury to keratinocytes: insights from animal models of the lichenoid tissue reaction. J Invest Dermatol. 2008 doi: 10.1038/jid.2008.242. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto T, Nakane T, Osaki T. The mechanism of mononuclear cell infiltration in oral lichen planus: the role of cytokines released from keratinocytes. J Clin Immunol. 2000;20:294–305. doi: 10.1023/a:1006671804110. [DOI] [PubMed] [Google Scholar]
- 7.Sugawara S, Uehara A, Tamai R, Takada H. Innate immune responses in oral mucosa. J Endotoxin Res. 2002;8:465–8. doi: 10.1179/096805102125001082. [DOI] [PubMed] [Google Scholar]
- 8.Ishii KJ, Coban C, Akira S. Manifold mechanisms of Toll-like receptor-ligand recognition. J Clin Immunol. 2005;25:511–21. doi: 10.1007/s10875-005-7829-1. [DOI] [PubMed] [Google Scholar]
- 9.Uehara A, Fujimoto Y, Fukase K, Takada H. Various human epithelial cells express functional Toll-like receptors, NOD1 and NOD2 to produce anti-microbial peptides, but not proinflammatory cytokines. Mol Immunol. 2007;44:3100–11. doi: 10.1016/j.molimm.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 10.Kikkert R, Laine ML, Aarden LA, van Winkelhoff AJ. Activation of Toll-like receptors 2 and 4 by Gram-negative periodontal bacteria. Oral Microbiol Immunol. 2007;22:145–51. doi: 10.1111/j.1399-302X.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 11.Netea MG, Van Der Graaf CA, Vonk AG, Verschueren I, Van Der Meer JW, Kullberg BJ. The role of Toll-like receptor (TLR) 2 and TLR4 in the host defense against disseminated candidiasis. J Infect Dis. 2002;185:1483–9. doi: 10.1086/340511. [DOI] [PubMed] [Google Scholar]
- 12.Vidal K, Donnet-Hughes A. CD14: a soluble pattern recognition receptor in milk. Adv Exp Med Biol. 2008;606:195–216. doi: 10.1007/978-0-387-74087-4_7. [DOI] [PubMed] [Google Scholar]
- 13.Asea A. Heat shock proteins and Toll-like receptors. Handb Exp Pharmacol. 2008:111–27. doi: 10.1007/978-3-540-72167-3_6. [DOI] [PubMed] [Google Scholar]
- 14.Chaiyarit P, Kafrawy AH, Miles DA, Zunt SL, Van Dis ML, Gregory RL. Oral lichen planus: an immunohistochemical study of heat shock proteins (HSPs) and cytokeratins (CKs) and a unifying hypothesis of pathogenesis. J Oral Pathol Med. 1999;28:210–15. doi: 10.1111/j.1600-0714.1999.tb02026.x. [DOI] [PubMed] [Google Scholar]
- 15.Kirschning CJ, Schumann RR. TLR2: cellular sensor for microbial and endogenous molecular patterns. Curr Top Microbiol Immunol. 2002;270:121–44. doi: 10.1007/978-3-642-59430-4_8. [DOI] [PubMed] [Google Scholar]
- 16.Mueller T, Terada T, Rosenberg IM, Shibolet O, Podolsky DK. Th2 cytokines down-regulate TLR expression and function in human intestinal epithelial cells. J Immunol. 2006;176:5805–14. doi: 10.4049/jimmunol.176.10.5805. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki M, Hisamatsu T, Podolsky DK. Gamma interferon augments the intracellular pathway for lipopolysaccharide (LPS) recognition in human intestinal epithelial cells through coordinated up-regulation of LPS uptake and expression of the intracellular Toll-like receptor 4-MD-2 complex. Infect Immun. 2003;71:3503–11. doi: 10.1128/IAI.71.6.3503-3511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroishi T, Tanaka Y, Sakai A, Sugawara Y, Komine K, Sugawara S. Human parotid saliva contains soluble Toll-like receptor (TLR) 2 and modulates TLR2-mediated interleukin-8 production by monocytic cells. Mol Immunol. 2007;44:1969–76. doi: 10.1016/j.molimm.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 19.LeBouder E, Rey-Nores JE, Rushmere NK, et al. Soluble forms of Toll-like receptor (TLR)2 capable of modulating TLR2 signaling are present in human plasma and breast milk. J Immunol. 2003;171:6680–9. doi: 10.4049/jimmunol.171.12.6680. [DOI] [PubMed] [Google Scholar]
- 20.Srinivasan M, Kodumudi KN, Zunt SL. Soluble CD14 and Toll-like receptor-2 are potential salivary biomarkers for oral lichen planus and burning mouth syndrome. Clin Immunol. 2008;126:31–7. doi: 10.1016/j.clim.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 21.Jaresova I, Rozkova D, Spisek R, Janda A, Brazova J, Sediva A. Kinetics of Toll-like receptor-4 splice variants expression in lipopolysaccharide-stimulated antigen presenting cells of healthy donors and patients with cystic fibrosis. Microbes Infect. 2007;9:1359–67. doi: 10.1016/j.micinf.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 22.Uehara A, Sugawara S, Watanabe K, et al. Constitutive expression of a bacterial pattern recognition receptor, CD14, in human salivary glands and secretion as a soluble form in saliva. Clin Diagn Lab Immunol. 2003;10:286–92. doi: 10.1128/CDLI.10.2.286-292.2003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Xie H, Onsongo G, Popko J, et al. Proteomics analysis of cells in whole saliva from oral cancer patients via value-added three-dimensional peptide fractionation and tandem mass spectrometry. Mol Cell Proteomics. 2008;7:486–98. doi: 10.1074/mcp.M700146-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Amerongen AV, Veerman EC. Saliva – the defender of the oral cavity. Oral Dis. 2002;8:12–22. doi: 10.1034/j.1601-0825.2002.1o816.x. [DOI] [PubMed] [Google Scholar]
- 25.Lilly EA, Shetty KV, Leigh JE, Cheeks C, Fidel PL., Jr Oral epithelial cell antifungal activity: approaches to evaluate a broad range of clinical conditions. Med Mycol. 2005;43:517–23. doi: 10.1080/13693780500050655. [DOI] [PubMed] [Google Scholar]
- 26.Vaahtoniemi LH, Raisanen S, Stenfors LE. Attachment of bacteria to oral epithelial cells in vivo: a possible correlation to gingival health status. J Periodont Res. 1993;28:308–11. doi: 10.1111/j.1600-0765.1993.tb02098.x. [DOI] [PubMed] [Google Scholar]
- 27.Rasband W, Image J. ImageJ, U. S. http://rsb.info.nih.gov/ij/, In B. National Institutes of Health, Maryland, USA (Ed.
- 28.Saida H, Maekawa T, Satake T, Higashi Y, Seki H. Gram stain index of a natural bacterial community at a nutrient gradient in the freshwater environment. Environ Pollut. 2000;109:293–301. doi: 10.1016/s0269-7491(99)00257-2. [DOI] [PubMed] [Google Scholar]
- 29.Tran SD, Rudney JD. Multiplex PCR using conserved and species-specific 16S rRNA gene primers for simultaneous detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. J Clin Microbiol. 1996;34:2674–8. doi: 10.1128/jcm.34.11.2674-2678.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoshino T, Kawaguchi M, Shimizu N, Hoshino N, Ooshima T, Fujiwara T. PCR detection and identification of oral streptococci in saliva samples using gtf genes. Diagn Microbiol Infect Dis. 2004;48:195–9. doi: 10.1016/j.diagmicrobio.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Jewell SD, Srinivasan M, McCart LM, et al. Analysis of the molecular quality of human tissues: an experience from the Cooperative Human Tissue Network. Am J Clin Pathol. 2002;118:733–41. doi: 10.1309/VPQL-RT21-X7YH-XDXK. [DOI] [PubMed] [Google Scholar]
- 32.Shin HJ, Lee H, Park JD, et al. Kinetics of binding of LPS to recombinant CD14, TLR4, and MD-2 proteins. Mol Cells. 2007;24:119–24. [PubMed] [Google Scholar]
- 33.Rhodus NL, Cheng B, Myers S, Miller L, Ho V, Ondrey F. The feasibility of monitoring NF-kappaB associated cytokines: TNF-alpha, IL-1alpha, IL-6, and IL-8 in whole saliva for the malignant transformation of oral lichen planus. Mol Carcinog. 2005;44:77–82. doi: 10.1002/mc.20113. [DOI] [PubMed] [Google Scholar]
- 34.Hohl D, de Viragh PA, Amiguet-Barras F, Gibbs S, Backendorf C, Huber M. The small proline-rich proteins constitute a multigene family of differentially regulated cornified cell envelope precursor proteins. J Invest Dermatol. 1995;104:902–9. doi: 10.1111/1523-1747.ep12606176. [DOI] [PubMed] [Google Scholar]
- 35.Sun FJ, Kaur S, Ziemer D, Banerjee S, Samuelson LC, De Lisle RC. Decreased gastric bacterial killing and up-regulation of protective genes in small intestine in gastrin-deficient mouse. Dig Dis Sci. 2003;48:976–85. doi: 10.1023/a:1023068116934. [DOI] [PubMed] [Google Scholar]
- 36.Gibbs S, Ponec M. Intrinsic regulation of differentiation markers in human epidermis, hard palate and buccal mucosa. Arch Oral Biol. 2000;45:149–58. doi: 10.1016/s0003-9969(99)00116-8. [DOI] [PubMed] [Google Scholar]
- 37.Fischer DF, Gibbs S, van De Putte P, Backendorf C. Interdependent transcription control elements regulate the expression of the SPRR2A gene during keratinocyte terminal differentiation. Mol Cell Biol. 1996;16:5365–74. doi: 10.1128/mcb.16.10.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudney JD, Chen R, Sedgewick GJ. Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Tannerella forsythensis are components of a polymicrobial intracellular flora within human buccal cells. J Dent Res. 2005;84:59–63. doi: 10.1177/154405910508400110. [DOI] [PubMed] [Google Scholar]
- 39.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 40.Smirnova I, Poltorak A, Chan EK, McBride C, Beutler B. Phylogenetic variation and polymorphism at the Toll-like receptor 4 locus (TLR4) Genome Biol. 2000;1:RESEARCH002. doi: 10.1186/gb-2000-1-1-research002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwami KI, Matsuguchi T, Masuda A, Kikuchi T, Musikacharoen T, Yoshikai Y. Cutting edge: naturally occurring soluble form of mouse Toll-like receptor 4 inhibits lipopolysaccharide signaling. J Immunol. 2000;165:6682–6. doi: 10.4049/jimmunol.165.12.6682. [DOI] [PubMed] [Google Scholar]
- 42.Fazeli A, Bruce C, Anumba DO. Characterization of Toll-like receptors in the female reproductive tract in humans. Hum Reprod. 2005;20:1372–8. doi: 10.1093/humrep/deh775. [DOI] [PubMed] [Google Scholar]
- 43.Pioli PA, Amiel E, Schaefer TM, Connolly JE, Wira CR, Guyre PM. Differential expression of Toll-like receptors 2 and 4 in tissues of the human female reproductive tract. Infect Immun. 2004;72:5799–806. doi: 10.1128/IAI.72.10.5799-5806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hyakushima N, Mitsuzawa H, Nishitani C, et al. Interaction of soluble form of recombinant extracellular TLR4 domain with MD-2 enables lipopolysaccharide binding and attenuates TLR4-mediated signaling. J Immunol. 2004;173:6949–54. doi: 10.4049/jimmunol.173.11.6949. [DOI] [PubMed] [Google Scholar]
- 45.Leung TF, Tang NL, Sung YM, et al. The C-159T polymorphism in the CD14 promoter is associated with serum total IgE concentration in atopic Chinese children. Pediatr Allergy Immunol. 2003;14:255–60. doi: 10.1034/j.1399-3038.2003.00048.x. [DOI] [PubMed] [Google Scholar]
- 46.Paulus SC, Hirschfeld AF, Victor RE, Brunstein J, Thomas E, Turvey SE. Common human Toll-like receptor 4 polymorphisms – role in susceptibility to respiratory syncytial virus infection and functional immunological relevance. Clin Immunol. 2007;123:252–7. doi: 10.1016/j.clim.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 47.Farina C, Theil D, Semlinger B, Hohlfeld R, Meinl E. Distinct responses of monocytes to Toll-like receptor ligands and inflammatory cytokines. Int Immunol. 2004;16:799–809. doi: 10.1093/intimm/dxh083. [DOI] [PubMed] [Google Scholar]
- 48.Tao XA, Li CY, Rhodus NL, Xia J, Yang XP, Cheng B. Simultaneous detection of IFN-gamma and IL-4 in lesional tissues and whole unstimulated saliva from patients with oral lichen planus. J Oral Pathol Med. 2008;37:83–7. doi: 10.1111/j.1600-0714.2007.00593.x. [DOI] [PubMed] [Google Scholar]
- 49.Funda DP, Tuckova L, Farre MA, Iwase T, Moro I, Tlaskalova-Hogenova H. CD14 is expressed and released as soluble CD14 by human intestinal epithelial cells in vitro: lipopolysaccharide activation of epithelial cells revisited. Infect Immun. 2001;69:3772–81. doi: 10.1128/IAI.69.6.3772-3781.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonzalez-Moles MA, Bascones-Ilundain C, Gil Montoya JA, Ruiz-Avila I, Delgado-Rodriguez M, Bascones-Martinez A. Cell cycle regulating mechanisms in oral lichen planus: molecular bases in epithelium predisposed to malignant transformation. Arch Oral Biol. 2006;51:1093–103. doi: 10.1016/j.archoralbio.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 51.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Backman K, Jontell M. Microbial-associated oral lichenoid reactions. Oral Dis. 2007;13:402–6. doi: 10.1111/j.1601-0825.2006.01312.x. [DOI] [PubMed] [Google Scholar]
- 53.Madara J. Building an intestine – architectural contributions of commensal bacteria. N Engl J Med. 2004;351:1685–6. doi: 10.1056/NEJMcibr042621. [DOI] [PubMed] [Google Scholar]
- 54.Chang JH, Hampartzoumian T, Everett B, et al. Toll-like receptor (TLR)-2 and TLR4 expression and function but not polymorphisms are associated with acute anterior uveitis. Invest Ophthalmol Vis Sci. 2007;48:1711–17. doi: 10.1167/iovs.06-0807. [DOI] [PubMed] [Google Scholar]
- 55.Vanhinsbergh LJ, Powe DG, Jones NS. Reduction of TLR2 gene expression in allergic and nonallergic rhinitis. Ann Allergy Asthma Immunol. 2007;99:509–16. doi: 10.1016/S1081-1206(10)60379-1. [DOI] [PubMed] [Google Scholar]
- 56.Furrie E. Is Bifidobacterium a more effective probiotic therapy than Lactobacillus for patients with irritable bowel syndrome? Nat Clin Pract Gastroenterol Hepatol. 2005;2:304–5. doi: 10.1038/ncpgasthep0219. [DOI] [PubMed] [Google Scholar]
- 57.Kollisch G, Kalali BN, Voelcker V, et al. Various members of the Toll-like receptor family contribute to the innate immune response of human epidermal keratinocytes. Immunology. 2005;114:531–41. doi: 10.1111/j.1365-2567.2005.02122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


