Abstract
Diamond-Blackfan anemia (DBA) is a severe congenital anemia characterized by a specific decrease of erythroid precursors. The disease is also associated with growth retardation, congenital malformations, a predisposition for malignant disease and heterozygous mutations in either of the ribosomal protein (RP) genes RPS7, RPS17, RPS19, RPS24, RPL5, RPL11 and RPL35a. We show herein that primary fibroblast from DBA patients with truncating mutations in RPS19 or in RPS24 have a marked reduction in proliferative capacity. Mutant fibroblasts are associated with extended cell cycles and normal levels of p53 when compared to w.t. cells. RPS19 mutant fibroblasts accumulate in the G1 phase, whereas the RPS24 mutant cells show an altered progression in the S phase resulting in reduced levels in the G2/M phase. RPS19 deficient cells exhibit reduced levels of Cyclin-E, CDK2 and retinoblastoma (Rb) protein supporting a cell cycle arrest in the G1 phase. In contrast, RPS24 deficient cells show increased levels of the cell cycle inhibitor p21 and a seemingly opposing increase in Cyclin-E, CDK4 and CDK6. In combination, our results show that RPS19 and RPS24 insufficient fibroblasts have an impaired growth caused by distinct blockages in the cell cycle. We suggest this proliferative constraint to be an important contributing mechanism for the complex extra-hematological features observed in DBA.
Keywords: RPS19, RPS24, Diamond-Blackfan Anemia, Proliferation, Cell cycle, Cell cycle regulation
1. Introduction
Diamond-Blackfan Anemia (DBA) is a rare bone marrow failure syndrome characterized by absent or decreased number of erythroid precursors whereas other lineages remain unaffected [1]. At least 50% of patients display variable developmental defects [2–5] and/or short stature [6]. The disease is also associated with an increased risk of developing malignancies [7–9]. Approximately 35% of patients with DBA are heterozygous for mutations in one of the ribosomal protein (RP) genes RPS7, RPS17, RPS19, RPS24, RPL5, RPL11 and RPL35a which suggest that impaired ribosomal function causes the disease [5, 10–12]. The mutations result in perturbed rRNA processing and an impaired ribosomal subunit formation [13, 14]. Still, the mechanisms whereby disruption of ribosome biogenesis results in anemia, marrow failure, congenital malformations, short stature and malignant transformation remain to be defined. Several explanations have been suggested including downstream effects on protein translation [15] or impaired extra-ribosomal functions related to individual ribosomal proteins [16–18]. Another possibility is a direct effect induced by impaired ribosome biogenesis on the cellular stress response or through cellular toxicity [19, 20].
It has been widely accepted that DBA results from a cellular defect which makes erythroid precursor cells sensitive to apoptosis [19, 21]. Hematopoietic cells expressing RPS19 siRNA show an increase in apoptosis [22], reduced proliferation [23] and cell cycle arrest [24]. A proliferative defect in hematopoietic progenitors is also supported from studies of primary CD34+ CD38- cells in patients with RPS19 mutations [23].
We hypothesized that the phenotypic complexity seen in DBA patients is associated with a cellular growth defect in non-hematopoietic tissues. We addressed the questions by the investigation of primary fibroblasts from two DBA patients with truncating and heterozygous mutations in RPS19 and RPS24, respectively. We report here a marked reduction in proliferative capacity of RPS19 and RPS24 mutant fibroblasts when compared to control fibroblasts. The reduced growth is mainly attributed to a prolonged cell cycle with distinct cell cycle profiles for RPS19 and RPS24 mutations, respectively. The cell cycle arrest in RPS19 mutant is associated with reduced Cyclin-E/CDK2 levels, hypophosphorylation of Rb and a G1 arrest. The RPS24 mutant cells show increased p21 levels and increased levels of Cyclin-E, CDK 4 and CDK6. These results have implications for our understanding of the mechanisms mediated by ribosomal protein insufficiency in DBA.
2. Materials and Methods
2.1. Patients, mutant cell lines, culture conditions
Primary fibroblasts were obtained by skin punch biopsies from two DBA patients with a RPS19 acceptor splice site mutation (c. 72–2A>C) and a RPS24 start codon mutation (c. 1A>G), respectively, and healthy control individuals. The patient with an RPS19 mutation is a 5 years old girl treated with corticosteroids and she has a left sided thumb malformation (loss of thenar muscles) and reduced growth (−3 SD). The patient with RPS24 mutation is a 26 years old transfusion dependent female. She has a short stature (−4SD) and a dysmorphic facial appearance. Only two fibroblast cell lines were available from DBA patients with defined mutations. Cells were cultured in 50 % Ham’s F-10 (GIBCO) plus 50 % RPMI 1640 (GIBCO) supplemented with 10 % fetal bovine serum (GIBCO), 20 IU/ml PEST (GIBCO), 2 mM L-glutamine (GIBCO) and 3 % Ultroser G (PALL Life Sciences). This study and the use of human samples were approved by the regional ethical committee of Uppsala and after informed consent from the patients and/or their parents.
2.2. RNA isolation and Northern blotting
Fibroblasts were harvested before they reached 70% confluence. RNA was isolated using Trizol® (Invitrogen) and rRNA analysis was performed by northern blot as described [14].
2.3. Western blotting
Lysates from primary fibroblasts were separated on a 12% Bis-Tris SDS-PAGE, transferred to PVDF Immobilon-FL membranes (Millipore) and hybridized using primary antibodies against RPS19, RPS24 (Swedish Human Proteome Resource project), p53, p-Rb (Cell Signaling), Rb, CDK2, CDK4, CDK6, Cyclin-D1, Cyclin-E, p21 and p27 (Santa Cruz). Antibodies detecting β-actin (Abcam) were used as internal controls.
2.4. Cell cycle analysis and apoptosis assay
Primary fibroblasts were harvested by and fixed in 70% ethanol, incubated with RNase solution and stained with propidium iodide (PI). 20,000 cells were subjected to flow cytometry analysis using FACS flow (BD Bioscience) and BD FACSDiva. The software Modfit LT was used for cell cycle analysis. For apoptosis, the primary fibroblasts were harvested after 24 hours and 48 hours of exponential growth and stained using the TACS Annexin V-FITC assay kit (R&D systems) prior to flow cytometry using FACS flow (BD Bioscience). The data were analysed with BD FACSDiva program.
2.5. Calculation of cell growth, proliferation and generation time
We seeded 6,800 fibroblasts/cm2 in duplicates. Cells were counted after 48 and 72 hours. Generation times (tgen) for the distinct cell lines were calculated as follows: (Equation 1). Where tn= time (hours) and Nn = number of cells at time point n. The growth rates were estimated assuming exponential growth by incorporating the calculated generation time into an exponential growth equation N= Noeμt (Equation 2), describing the increase in cell numbers over time (t), as defined by their respective growth rate (μ). Growth rate is defined as μ= 0,693/tgen. N = number of cells at time t; No= initial number of cells at time to. Proliferation was re-estimated by introducing a distinct apoptosis rate correction factor (ka) into equation 2 to obtain an adjusted growth curve: N=Noekaμt (Equation 3).
3. Results and Discussion
3.1. Downregulation of RPS19 and RPS24 in primary fibroblast cells from DBA patients
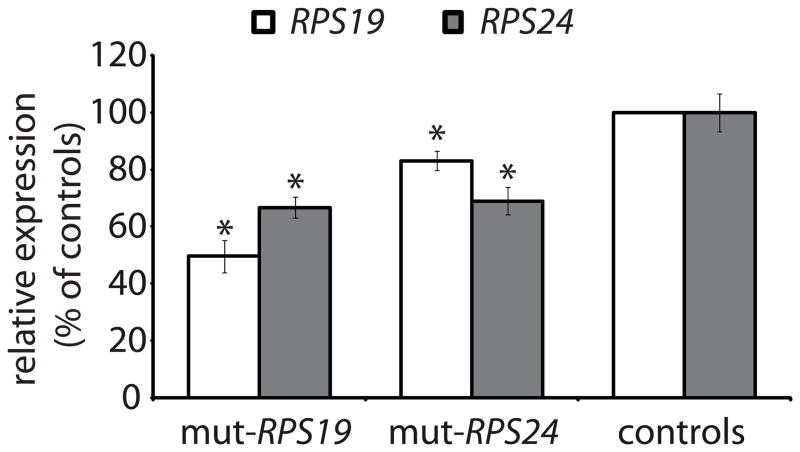
The molecular mechanism mediated by ribosomal protein deficiency in DBA is poorly understood [19]. The predominant clinical feature is restricted to the erythroid lineage but up to 50% of patients with DBA show developmental abnormalities and short stature (>−2SD). Similar to other bone marrow failure syndromes a predisposition for acute myeloid leukemia and osteosarcoma is recorded [9]. The ribosomes have critical functions in all cells and the relative tissue specificity observed in DBA has led to the suggestion of extraribosomal functions for the mutant proteins in erythropoiesis [17, 18]. Yet another explanation, based on the recent identification of perturbed ribosome biogenesis in ribosomal protein-deficient hematopoietic cells, may be that erythroblasts, because of a high proliferation rate, are particularly sensitive to defects in ribosome biogenesis [25]. This may increase their sensitivity to cellular stress and an impaired response to proliferation and differentiation. In order to clarify the possible effect of ribosomal protein gene mutations in non-hematopoietic tissues we analyzed primary fibroblasts from DBA patients with truncating RPS19 and RPS24 mutations, respectively. Western blot analysis on fibroblasts derived from the two DBA patients and three healthy controls were performed using polyclonal rabbit antisera against RPS19 and RPS24, respectively. The level of RPS19 in RPS19 mutated fibroblasts was reduced to 50% of that in controls, whereas RPS19 was decreased to 83% in RPS24 mutated fibroblast (Fig. 1). The level of RPS24 protein in both RPS24 mutated and RPS19 mutated DBA fibroblast is decreased to 70% of that in controls. This suggests that both mutations result in haploinsufficiency. The observed concomitant reduction of both small subunit proteins in mutant cell lines may be explained by a deficient assembly of the small subunit when one component is present in insufficient amounts. A similar coordinated down regulation of ribosomal proteins from the same subunit was recently reported in HeLa cells subject to siRNA knock-down of specific RPs [26] and in patient derived cell-lines [27].
Figure 1.
Levels of RPS19 and RPS24 proteins in primary fibroblasts as determined by western blot. Relative levels of RPS19 protein (white bar) and RPS24 protein (grey bar) in RPS19 (mut-RPS19) and RPS24 (mut-RPS24) mutant cells as compared to controls (100% expression). Asterix (*) indicates significant differences when compared to control cells (p<0.05).
3.2. Reduced proliferation and extended cell cycles in RPS19 and RPS24 mutant fibroblasts
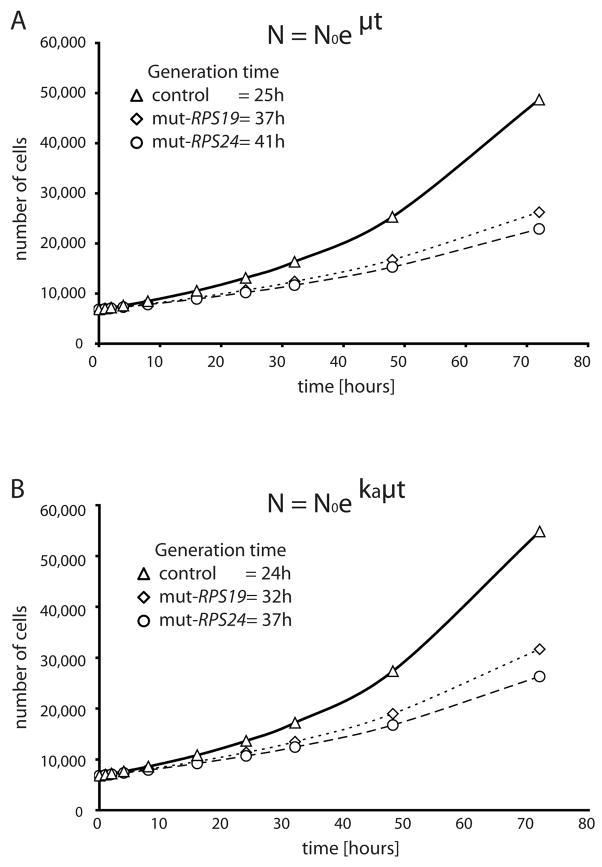
Increased apoptosis of erythroid precursors has been considered a major cause of anemia in DBA [19]. However, more recent data have also suggested proliferation defects and a specific cell cycle arrest in hematopoietic cells as contributing mechanisms in the disease [22, 24]. We performed a growth assay to clarify any defect in RPS19 and RPS24 mutant fibroblasts from the two DBA patients by comparing them to passage matched fibroblasts from two healthy individuals. Cells were seeded at a density of 6800 cells/cm2 and counted after 48 hours and 72 hours of exponential growth to deduce the generation time. The fibroblasts with RPS19 and RPS24 mutations showed a significantly decelerated growth corresponding to a generation time of 37 hours and 41 hours, respectively, as opposed to 25 h in the control fibroblasts (p<0.05; Fig. 2A). We used the estimated generation time to simulate growth curves for the mutant cell lines and controls, assuming exponential growth (for details see Materials and Methods). The simulated curves illustrate a difference in growth capacity of the mutant cells compared to the control fibroblasts (Fig. 2A).
Figure 2.
Growth curves of primary fibroblasts carrying RPS19 and RPS24 mutations, respectively, and three control fibroblasts. (A) 6,800 cells/cm2 were seeded in duplicates and counted after 48 h and 72 h to measure the generation time (tgen). A growth curve assuming exponential growth is simulated using equation N= Noeμt showing the number of cells as a function of time. μ= 0,693/tgen. (B) Replotted growth curves from (A) implementing an apoptosis correction factor (ka) to obtain the adjusted growth curve equation N=Noekaμt. RPS19 mutant cells (open squares), RPS24 mutant cells (open circles) and control fibroblasts (open triangles).
In order to clarify the causes of reduced growth in RPS19 and RPS24 mutant cells, we analyzed the degree of apoptosis at 24 hours and 48 hours, respectively. The average apoptotic fractions of fibroblasts revealed by Annexin V staining in RPS19 and RPS24 mutant cells were 14.15% and 11.45%, respectively. The apoptosis in RPS19 and RPS24 mutant fibroblasts are significantly increased (p<0.05) when compared to the average apoptotic fraction in control cells (5.95%; Supplementary Fig. S1). This difference is similar to what has been shown after siRNA knock-down of RPS19 in TF-1 cells [22]. We then integrated the apoptosis rate for the mutant and control cells into the exponential growth equations by introducing an apoptosis constant (Ka) and we recalculated the proliferation curves (Fig. 2B). We assumed that the simulated curves for control and mutant cells would coincide if the increased apoptosis alone causes the reduced proliferative capacity in mutant cells. Interestingly, we observed a marked difference in the curve slopes between DBA derived fibroblasts and control cells. The corrected generation time was estimated to 32 hours for RPS19 mutant cells and 37 hours for RPS24 mutant cells when integrating apoptosis (Fig. 2B). The corresponding generation time for control cells was 24 hours. The prolonged generation time in the RPS19 and RPS24 mutant cells is significant (p<0.05) when compared to control cells. The results indicate that the reduced growth in DBA derived fibroblasts is mainly attributed to reduced proliferative capacity whereas the contribution from increased apoptosis is less significant.
3.3. Different cell-cycle distribution of RPS19 and RPS24 mutant fibroblasts
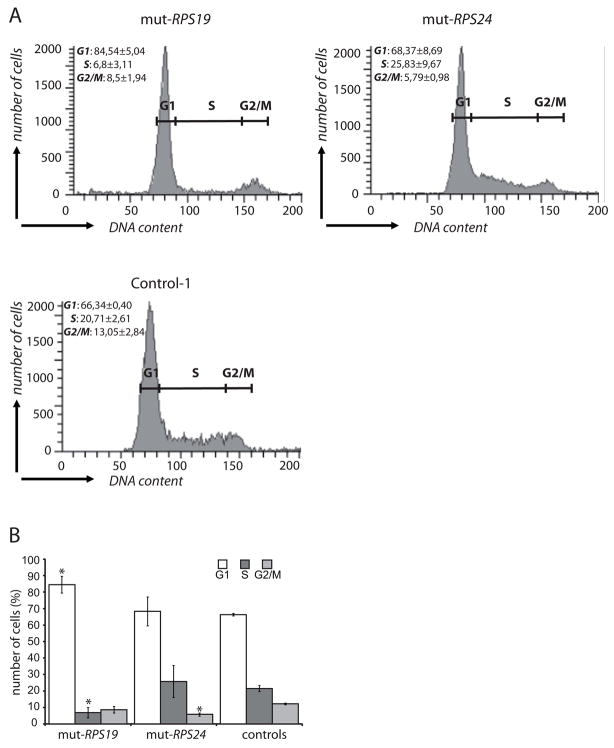
The prolonged cell cycles in mutant fibroblasts prompted us to investigate the proportion of cells in different phases (G1, S and G2/M) using propidium iodide (PI) staining (Fig. 3). The RPS19 mutant cells showed a significantly larger proportion of cells in the G1 phase (p=0.036) and a reduced number in the S phase (p=0.025) when compared to control cells (Fig’s 3A and 3B). The RPS24 mutant fibroblasts showed a distinct profile with a significant decrease in the number of cells in the G2/M phase (p=0.042). Furthermore, the number of RPS24 mutant cells decreased gradually throughout the S-phase (Fig. 3A). The results indicate that the RPS19 mutation results in an arrest of cells in the G1 phase whereas the RPS24 mutation leads to a perturbed S phase progression, possibly due to a defect already at the G1 to S transition.
Figure 3.
Cell cycle analysis of human primary fibroblasts. The cell cycle phases were measured by propidium iodide labeling. (A) Representative histograms from cell cycle analysis of the RPS19 mutated (mut-RPS19), RPS24 mutated (mut-RPS24) cells and three controls. The cell cycle phases G1, S and G2/M are indicated. (B) Diagram showing the distribution of cells in distinct cell cycle phases from two independent experiments. The average percentages of cells in the different cell cycle phases (G1, S and G2/M) are shown with standard deviations. Asterix (*) indicates significant differences when compared to control cells in the same phase, respectively (p<0.05).
3.4. Impaired rRNA maturation in RPS19 and RPS24 mutant cells
Ribosomal proteins are pivotal for the maturation and assembly of ribosomal subunits. Previous reports have shown that RPS19 mutations cause impaired 18S rRNA synthesis leading to reduced 40S assembly [13, 14]. We analyzed rRNA intermediates (45S, 41S, 30S, 21S, 18SE) in the mutant fibroblast cell lines and control cells by northern blot analysis using βand γ probes complementary to 18S rRNA [14]. Analysis of RPS19 mutant cells showed a significant increase in the relative levels of 21S and 41S rRNA with increased 21S/18SE (p=0.029) and 41S/30S (p=0.040) ratios when compared to controls. The RPS24 mutant cells showed increased relative levels of 30S rRNA resulting in a reduced 41S/30S ratio (p=0.047) as well as reduced amounts of 21S and 18S-E pre-rRNA leading to an increased 30S/21S ratio (p=0.029) when compared to the control cells (Supplementary Fig’s 2A and 2B). These results are consistent with the extended cell cycles observed in our study as well as previous studies on cells with RPS19 and RP24 mutations [14, 27, 28].
3.5. Dysregulated cell cycle proteins in RPS19 and RPS24 deficient fibroblasts
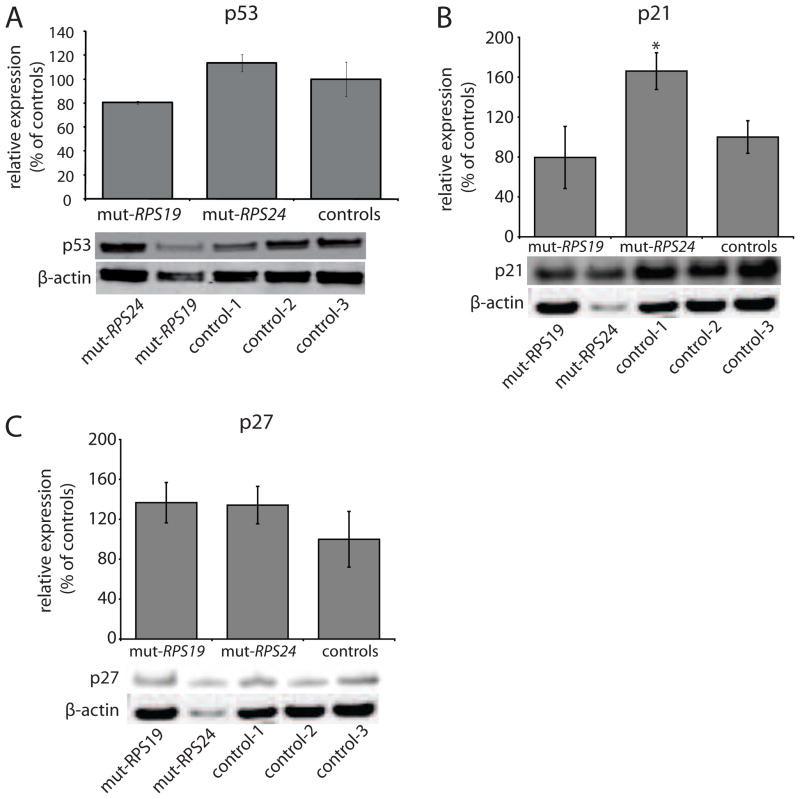
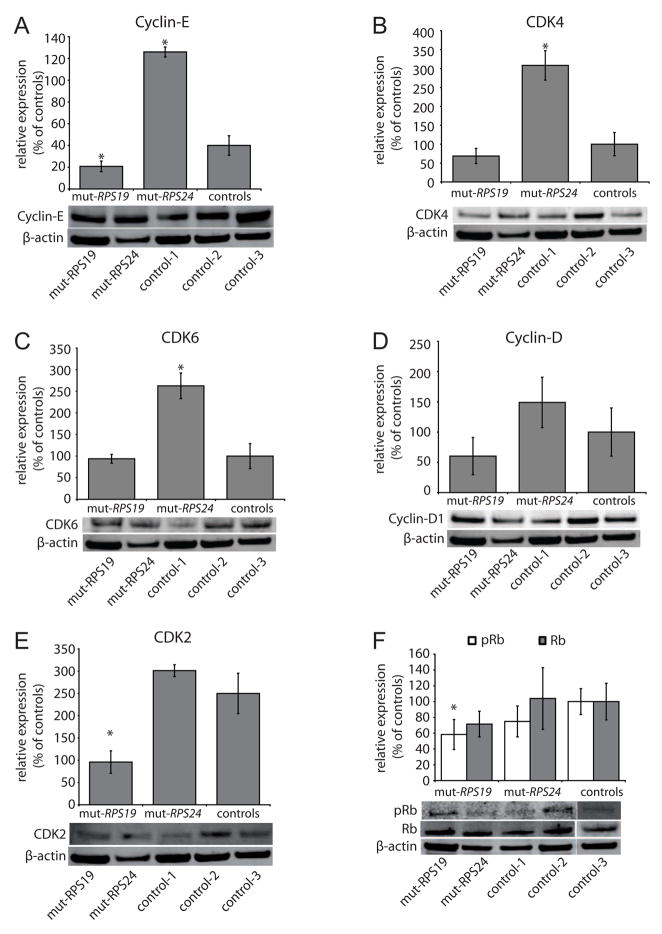
The reduced growth and the aberrant cell cycle distribution of the mutant fibroblasts prompted us to analyze specific proteins involved in cell cycle regulation. We selected distinct cell cycle regulators in the G1 and S phases. We first analysed the protein levels of p53 in RPS19 and RPS24 mutant cells. The levels were within the normal range when compared to control fibroblasts (Fig. 4A). We then analysed the two G1 check-point proteins p21 and p27. The levels of p21 were found significantly increased in RPS24 mutant fibroblasts suggesting cellular stress in the G1 phase (Fig. 4B). The p21 levels were found normal in RPS19 mutant cells and the p27 levels were within the normal range in both RPS19 and RPS24 mutant cell lines (Fig. 4C). Two major targets for p21 in the G1 phase are the Cyclin-D dependent kinase (CDK) 4 and CDK6 complex in early G1 phase and the Cyclin-E-CDK2 complex in mid to late G1 required for phosphorylation of the retinoblastoma protein (Rb) and entry into the S phase [29–33]. In the RPS24 mutant cells, the analysis revealed increased levels of Cyclin-E, CDK4 and CDK6 whereas the levels of Cyclin-D and CDK2 were normal with a tendency for increased levels (Fig’s 5A–E). This suggests that RPS24 mutant cells progress through the G1 phase and transit into the S-phase. This is consistent with the observed distribution of cells within the different cell cycle phases, showing a normal proportion of cells in G1 phase. The increased p21 level in RPS24 mutant cells, indicating cellular stress, is also consistent with a relative S phase blockage and may explain the gradual decrease in cell numbers during the S phase and the significant decrease in number of cells in G2/M phase observed on cell cycle histograms (Fig. 3A). In contrast, the RPS19 mutant cells showed reduced levels of Cyclin-E and CDK2 whereas the Cyclin-D, CDK4 and CDK6 appeared normal (Fig’s 5A–E). These results indicate that RPS19 mutant cells display a G1 block preceding the arrest in RPS24 mutant cells. This is also supported by the reduced level of phosphorylated Rb restricted to RPS19 mutant cells indicating impaired progression into the S-phase (Fig. 5F). Thus, the marked prolongation of the cell cycle in the RPS19 and RPS24 mutant fibroblasts with their abnormal and distinct phase distributions are consistent with alterations in cell cycle regulators. The combined results from the analysis of Cyclin-E, Cyclin-D, CDK2/4/6 and Rb indicate that the RPS19 mutation results in an arrest in the G1 phase in fibroblasts whereas the RPS24 mutation results in a defect in the S phase. Interestingly, this is associated with normal levels of p53, a key regulator of p21 and a gate keeper in the G1 progression. The cell cycle inhibitor p53 has previously been found to accumulate in a Rps19 mutant mouse and zebrafish models [34, 35]. This indicates that mutations in ribosomal protein genes cause cellular stress and activates cell cycle inhibitors. Activated cell cycle inhibitors may also explain the reduced growth and, with acquired mutations in e.g p53, the tumor predisposition observed in zebrafish and humans with mutations in ribosomal protein genes [6, 20, 34–36]
Figure 4.
Analysis of p53, p21 and p27 protein levels in fibroblasts carrying RPS19 mutation (mut-RPS19), RPS24 mutation (mut-RPS24) and controls (100% expression). Western blot analysis were normalised to beta-actin. (A) p53 protein expression. The diagram illustrates the average of two independent measurements and standard deviations without significant differences. Representative analysis (bottom) using antibodies against p53 and β-actin, respectively. (B) Relative levels of p21 in RPS19 and RPS24 mutant cells from two different experiments and controls (100% expression). Asterix (*) denotes significant differences compared to controls (p<0.05). Representative analysis (bottom) using antibodies against p21 and β-actin, respectively. (C) Relative levels of p27 in RPS19 and RPS24 mutant cells as compared to controls (100% expression). Representative analysis (bottom) using antibodies against p27 and β-actin, respectively.
Figure 5.
Western blot analysis of the cell cycle regulators Cyclin-E, CDK4, CDK6, Cyclin-D1, CDK2 and retinoblastoma protein (Rb) in fibroblasts carrying RPS19 mutation (mut-RPS19), RPS24 mutation (mut-RPS24) and three controls (100% expression). Averages of at least two independent experiments are shown with standard deviations. The western analysis was normalized to β-actin and representative blots are illustrated below each diagram. Asterix (*) denotes significant differences (p<0.05) compared to controls. (A) Relative levels of Cyclin-E. (B) Relative levels of CDK4. (C) Relative levels of CDK6. (D) Relative levels of Cyclin-D showing tendencies for variations in RPS19 and RPS24 mutant cells but these are non-significant. (E) Relative levels of CDK2. (F) Relative levels of phosphorylated Rb (pRb; white bars) and non-phosphorylated Rb (Rb; grey bar) proteins.
The regulatory mechanism by which impaired ribosome biogenesis results in prolonged cell cycle with distinct arrests is yet unclear. Incomplete ribosome assembly may directly contribute to this process through activation of cell cycle regulators and different cellular stress responses. It has been suggested that perturbed ribosome assembly destabilises the nucleolus and affects the coresident cell cycle machinery leading to cell cycle progression defects [37]. Impaired ribosomal function may also exert downstream effects on protein translation, which in turn are responsible for the complex DBA phenotype. Previous reports have shown that reduced levels of ribosomal proteins trigger apoptosis and cell cycle arrest in a p53 dependent manner [34–36]. Repeated analysis of total p53 levels by western blot and mRNA analysis using qRT/PCR (data not shown) in this study showed no significant differences between RPS19 and RPS24 mutant cells and control fibroblasts (Fig. 4A). This could possibly be explained by the assumption that fibroblasts are less sensitive to stress induced by the mutations in ribosomal protein genes or that mutant cells bypass p53 and directly activate down-stream targets of importance for a cellular stress response and/or cell cycle regulation. Both these possibilities are compatible with the results presented herein.
Our combined results indicate that primary fibroblasts with RPS19 and RPS24 gene mutations and perturbed ribosome biogenesis are associated with a marked proliferative defect, distinct cell cycle arrests and dysregulated cell-cycle proteins. The increased apoptosis has little effect on the overall cell growth. Our observations are restricted to two available cell lines but may have implications for non-hematopoietic features in DBA such as short stature, developmental abnormalities resulting in various malformations and the predisposition to malignant disease. Haploinsufficiency for several ribosomal proteins in other species such as Drosophila and mice are associated with reduced body size [38]. Further studies are required to more precisely clarify the mechanisms involved in the cellular growth reduction and cell cycle control caused by RPS19 and RPS24 mutations.
Supplementary Material
Apoptotic fractions of fibroblasts revealed by Annexin V staining in fibroblasts with the RPS19 mutation (mut-RPS19), RPS24 mutation (mut-RPS24) and controls, respectively. (A) Representative histograms from flow cytometry analysis. The average proportion of apoptotic cells from independent measurements at 24 h and 48 h are given (14.15%; 11.45% and 5.95%, respectively). (B) Diagram showing the average apoptotic fractions in RPS19 and RPS24 mutant fibroblasts compared to control fibroblasts. Apoptosis is significantly increased in both RPS19 and RPS24 mutant cell lines (*; p<0.05).
Analysis of pre-rRNA processing in total RNA from fibroblasts with RPS19 and RPS24 mutations as compared to control fibroblasts. (A) Autoradiograph of a Northern blot analysis using γ-probe to detect rRNA intermediates in total RNA isolated from RPS19 mutated (mut-RPS19), RPS24 mutated (mut-RPS24) and control fibroblasts (Control 1–3). Distinct rRNA intermediates are indicated to the left. (B) Relative levels of rRNA intermediates illustrated as ratios between 41S/30S, 30S/21S and 21S/18S pre-rRNA, respectively. Averages of three independent northern blots are shown with standard deviations. Asterix (*) denotes significant differences compared to controls. Light grey bars=controls; dark grey bars=RPS24 mutant cells; white bars=RPS19 mutant cells.
Acknowledgments
We thank the patients who provided samples for this study. We acknowledge Dr. Joakim Klar for technical assistance and the Swedish Human Proteome Research Project (HPR) for providing ribosomal protein antibodies. This work was supported by National Institutes of Health, USA (grant number 5R01-HL079567-04), Swedish Research Council, Swedish Cancer foundation, Swedish Children’s Cancer foundation and Uppsala University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Young NS, Alter B. Inherited bone marrow failure syndromes: Introduction. In: Young, Alter, editors. Aplastic anemia acquired and inherited. WB saunders Campany; Philadelphia, PA: 1994. [Google Scholar]
- 2.Halperin DS, Freedman MH. Diamond-blackfan anemia: etiology, pathophysiology, and treatment. Am J Pediatr Hematol Oncol. 1989;11:380–394. [PubMed] [Google Scholar]
- 3.Willig TN, Draptchinskaia N, Dianzani I, Ball S, Niemeyer C, Ramenghi U, Orfali K, Gustavsson P, Garelli E, Brusco A, Tiemann C, Perignon JL, Bouchier C, Cicchiello L, Dahl N, Mohandas N, Tchernia G. Mutations in ribosomal protein S19 gene and diamond blackfan anemia: wide variations in phenotypic expression. Blood. 1999;94:4294–4306. [PubMed] [Google Scholar]
- 4.Gripp KW, McDonald-McGinn DM, La Rossa D, McGain D, Federman N, Vlachos A, Glader BE, McKenzie SE, Lipton JM, Zackai EH. Bilateral microtia and cleft palate in cousins with Diamond-Blackfan anemia. Am J Med Genet. 2001;101:268–274. doi: 10.1002/ajmg.1329. [DOI] [PubMed] [Google Scholar]
- 5.Gazda HT, Sheen MR, Vlachos A, Choesmel V, O’Donohue MF, Schneider H, Darras N, Hasman C, Sieff CA, Newburger PE, Ball SE, Niewiadomska E, Matysiak M, Zaucha JM, Glader B, Niemeyer C, Meerpohl JJ, Atsidaftos E, Lipton JM, Gleizes PE, Beggs AH. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vlachos A, Ball S, Dahl N, Alter BP, Sheth S, Ramenghi U, Meerpohl J, Karlsson S, Liu JM, Leblanc T, Paley C, Kang EM, Leder EJ, Atsidaftos E, Shimamura A, Bessler M, Glader B, Lipton JM. Diagnosing and treating Diamond Blackfan anaemia: results of an international clinical consensus conference. Br J Haematol. 2008;142:859–876. doi: 10.1111/j.1365-2141.2008.07269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Dijken PJ, Verwijs W. Diamond-Blackfan anemia and malignancy. A case report and a review of the literature. Cancer. 1995;76:517–520. doi: 10.1002/1097-0142(19950801)76:3<517::aid-cncr2820760324>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 8.Lipton JM, Federman N, Khabbaze Y, Schwartz CL, Hilliard LM, Clark JI, Vlachos A. Osteogenic sarcoma associated with Diamond-Blackfan anemia: a report from the Diamond-Blackfan Anemia Registry. J Pediatr Hematol Oncol. 2001;23:39–44. doi: 10.1097/00043426-200101000-00009. [DOI] [PubMed] [Google Scholar]
- 9.Alter BP. Diagnosis, genetics, and management of inherited bone marrow failure syndromes, Hematology/the Education Program of the American Society of Hematology. American Society of Hematology. 2007;2007:29–39. doi: 10.1182/asheducation-2007.1.29. [DOI] [PubMed] [Google Scholar]
- 10.Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, Tentler D, Mohandas N, Carlsson B, Dahl N. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 11.Farrar NMJE, Caywood E, McDevitt MA, Kowalski J, Takemoto CM, Talbot CC, Jr, Meltzer P, Esposito D, Beggs AH, Schneider HE, Grabowska A, Ball SE, Niewiadomska E, Sieff CA, Vlachos A, Atsidaftos E, Ellis SR, Lipton JM, Gazda HT, Arceci RJ. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 2008:1582–1592. doi: 10.1182/blood-2008-02-140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat. 2007;12:1178–1182. doi: 10.1002/humu.20608. [DOI] [PubMed] [Google Scholar]
- 13.Flygare J, Aspesi A, Bailey JC, Miyake K, Caffrey JM, Karlsson S, Ellis SR. Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 2007;109:980–986. doi: 10.1182/blood-2006-07-038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choesmel V, Bacqueville D, Rouquette J, Noaillac-Depeyre J, Fribourg S, Cretien A, Leblanc T, Tchernia G, Da Costa L, Gleizes PE. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2007;109:1275–1283. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cmejlova J, Dolezalova L, Pospisilova D, Petrtylova K, Petrak J, Cmejla R. Translational efficiency in patients with Diamond-Blackfan anemia. Haematologica. 2006;91:1456–1464. [PubMed] [Google Scholar]
- 16.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 17.Lindstrom MS. Emerging functions of ribosomal proteins in gene-specific transcription and translation. Biochemical and biophysical research communications. 2009;379:167–170. doi: 10.1016/j.bbrc.2008.12.083. [DOI] [PubMed] [Google Scholar]
- 18.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Molecular cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellis SR, Lipton JM. Diamond Blackfan anemia: a disorder of red blood cell development. Curr Top Dev Biol. 2008;82:217–241. doi: 10.1016/S0070-2153(07)00008-7. [DOI] [PubMed] [Google Scholar]
- 20.McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, de Angelis MH, Myers RM, Attardi LD, Barsh GS. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohene-Abuakwa Y, Orfali KA, Marius C, Ball SE. Two-phase culture in Diamond Blackfan anemia: localization of erythroid defect. Blood. 2005;105:838–846. doi: 10.1182/blood-2004-03-1016. [DOI] [PubMed] [Google Scholar]
- 22.Miyake K, Utsugisawa T, Flygare J, Kiefer T, Hamaguchi I, Richter J, Karlsson S. Ribosomal protein S19 deficiency leads to reduced proliferation and increased apoptosis but does not affect terminal erythroid differentiation in a cell line model of Diamond-Blackfan anemia. Stem Cells. 2008;26:323–329. doi: 10.1634/stemcells.2007-0569. [DOI] [PubMed] [Google Scholar]
- 23.Hamaguchi I, Flygare J, Nishiura H, Brun AC, Ooka A, Kiefer T, Ma Z, Dahl N, Richter J, Karlsson S. Proliferation deficiency of multipotent hematopoietic progenitors in ribosomal protein S19 (RPS19)-deficient diamond-Blackfan anemia improves following RPS19 gene transfer. Mol Ther. 2003;7:613–622. doi: 10.1016/s1525-0016(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 24.Kuramitsu M, Hamaguchi I, Takuo M, Masumi A, Momose H, Takizawa K, Mochizuki M, Naito S, Yamaguchi K. Deficient RPS19 protein production induces cell cycle arrest in erythroid progenitor cells. Br J Haematol. 2008;140:348–359. doi: 10.1111/j.1365-2141.2007.06930.x. [DOI] [PubMed] [Google Scholar]
- 25.Ellis SR, Massey AT. Diamond Blackfan anemia: A paradigm for a ribosome-based disease. Medical hypotheses. 2006;66:643–648. doi: 10.1016/j.mehy.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA. 2008;14:1918–1929. doi: 10.1261/rna.1132008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Badhai J, Frojmark AS, Razzaghian HR, Davey E, Schuster J, Dahl N. Posttranscriptional down-regulation of small ribosomal subunit proteins correlates with reduction of 18S rRNA in RPS19 deficiency. FEBS letters. 2009;583:2049–2053. doi: 10.1016/j.febslet.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choesmel V, Fribourg S, Aguissa-Toure AH, Pinaud N, Legrand P, Gazda HT, Gleizes PE. Mutation of ribosomal protein RPS24 in Diamond-Blackfan anemia results in a ribosome biogenesis disorder. Hum Mol Genet. 2008;17:1253–1263. doi: 10.1093/hmg/ddn015. [DOI] [PubMed] [Google Scholar]
- 29.Koff A, Giordano A, Desai D, Yamashita K, Harper JW, Elledge S, Nishimoto T, Morgan DO, Franza BR, Roberts JM. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 30.Reed SI, Bailly E, Dulic V, Hengst L, Resnitzky D, Slingerland J. G1 control in mammalian cells. J Cell Sci Suppl. 1994;18:69–73. doi: 10.1242/jcs.1994.supplement_18.10. [DOI] [PubMed] [Google Scholar]
- 31.Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 32.Abukhdeir AM, Park BH. P21 and p27: roles in carcinogenesis and drug resistance. Expert reviews in molecular medicine. 2008;10:e19. doi: 10.1017/S1462399408000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blain SW. Switching cyclin D-Cdk4 kinase activity on and off. Cell cycle (Georgetown, Tex) 2008;7:892–898. doi: 10.4161/cc.7.7.5637. [DOI] [PubMed] [Google Scholar]
- 34.MacInnes AW, Amsterdam A, Whittaker CA, Hopkins N, Lees JA. Loss of p53 synthesis in zebrafish tumors with ribosomal protein gene mutations. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10408–10413. doi: 10.1073/pnas.0805036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008 doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- 36.Lai K, Amsterdam A, Farrington S, Bronson RT, Hopkins N, Lees JA. Many ribosomal protein mutations are associated with growth impairment and tumor predisposition in zebrafish. Dev Dyn. 2009;238:76–85. doi: 10.1002/dvdy.21815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pederson T. Ribosomal protein mutations in Diamond-Blackfan anemia: might they operate upstream from protein synthesis? Faseb J. 2007;21:3442–3445. doi: 10.1096/fj.07-8766hyp. [DOI] [PubMed] [Google Scholar]
- 38.Saeboe-Larssen LMS, Merriam J, Oksvold MP, Lambertsson A. Ribosomal protein insuffiency and the minute syndrome in drosophila: a dose response relationship. Genetics. 1998:1215–1224. doi: 10.1093/genetics/148.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Apoptotic fractions of fibroblasts revealed by Annexin V staining in fibroblasts with the RPS19 mutation (mut-RPS19), RPS24 mutation (mut-RPS24) and controls, respectively. (A) Representative histograms from flow cytometry analysis. The average proportion of apoptotic cells from independent measurements at 24 h and 48 h are given (14.15%; 11.45% and 5.95%, respectively). (B) Diagram showing the average apoptotic fractions in RPS19 and RPS24 mutant fibroblasts compared to control fibroblasts. Apoptosis is significantly increased in both RPS19 and RPS24 mutant cell lines (*; p<0.05).
Analysis of pre-rRNA processing in total RNA from fibroblasts with RPS19 and RPS24 mutations as compared to control fibroblasts. (A) Autoradiograph of a Northern blot analysis using γ-probe to detect rRNA intermediates in total RNA isolated from RPS19 mutated (mut-RPS19), RPS24 mutated (mut-RPS24) and control fibroblasts (Control 1–3). Distinct rRNA intermediates are indicated to the left. (B) Relative levels of rRNA intermediates illustrated as ratios between 41S/30S, 30S/21S and 21S/18S pre-rRNA, respectively. Averages of three independent northern blots are shown with standard deviations. Asterix (*) denotes significant differences compared to controls. Light grey bars=controls; dark grey bars=RPS24 mutant cells; white bars=RPS19 mutant cells.