Summary
Background
The monoamine oxidase-A (MAOA) gene plays a vital role in the metabolism of neurotransmitters, e.g, serotonin, norepinephrine, and dopamine. A polymorphism in the promoter region (MAOA-uVNTR) affects transcriptional efficiency. Allelic variation in MAOA-uVNTR has been associated with body mass index (BMI). We extended previous work by examining relations among this polymorphism and serum lipid levels.
Material/Methods
The sample consisted of 74 males enrolled in a study of caregivers for relatives with dementia. Regression models, adjusted for age, race, group status (caregiver/control), and cholesterol lowering medication (yes/no), were used to examine associations between high verses low MAOA-uVNTR activity alleles and total cholesterol, HDL, LDL, VLDL, LDL/HDL ratio, triglycerides, and BMI.
Results
Higher total cholesterol (p<0.03), LDL/HDL ratio (p<0.01), triglycerides (p<0.02), and VLDL (p<0.02) were associated with low activity MAOA-uVNTR alleles. HDL and LDL were modestly related to MAOA-uVNTR activity, however, they did not reach the conventional significance level (p<0.07 and p<0.10, respectively). BMI (p<0.74) was unrelated to MAOA-uVNTR transcription.
Conclusions
The present findings suggest that MAOA-uVNTR may influence lipid levels and individuals with less active alleles are at increased health risk.
Keywords: stress, genetics, lipoprotien, allelic variation
BACKGROUND
Monoamine oxidase A (MAOA) is a mitochondrial enzyme that degrades neurotransmitters involved with psychological and physical functioning. The gene that encodes MAOA is found on the X chromosome and contains a polymorphism (MAOA-uVNTR) located 1.2 kb upstream of the MAOA coding sequences [1]. In this polymorphism, consisting of a 30-base pair repeated sequence, six allele variants containing either 2-, 3-, 3.5-, 4-, 5-, or 6-repeat copies have been identified [2]. The 3- and 4-repeat alleles are the most commonly observed [2,3]. Functional studies indicating that alleles differ with respect to transcriptional efficiency suggest that the 3-repeat variant conveys lower transcriptional-efficiency and the 3.5- and 4-repeat alleles covey higher efficiency [1,4,5].
MAOA regulates monoamine turnover, and hence ultimately influences levels of norepinephrine, dopamine, and serotonin [6], making this gene a plausible candidate for affecting individual differences in the manifestation of psychological traits and psychiatric disorders [7]. Indeed, recent evidence indicates that the MAOA gene is associated with impulsivity and/or antisocial behavior e.g., [8–12], bipolar disorder [13], cluster B personality disorders [2], depressed suicide [14], alcohol dependence [11]. In prior work from our lab we have demonstrated associations among MAO activity and symptoms of depression and sleep quality [15]. Specifically, males with MAOA-uVNTR alleles that confer less activity had significantly more depressive symptoms and poorer sleep quality.
Moreover, MAOA activity has also been associated with physiological outcomes. Results from three studies have shown that lower transcriptional activity MAOA-uVNTR alleles are associated with higher body mass index (BMI) [16–18]. Related work from a family-based association study of the MAOA gene has demonstrated a preferential transmission of a low activity allele MAOA polymorphism in obese subjects [19]. Although we are unaware of any studies that have examined the association, it is plausible that MAOA-uVNTR may also be associated with other physiological outcomes associated with obesity, such as lipid metabolism.
We examined the relation between allelic variation (low vs high activity alleles) in the MAOA-uVNTR polymorphism, and lipid levels and BMI in a group of 74 males who were enrolled in a case/control study of caregiving for a relative with dementia. Specifically, we examined the main effect of MAOA-vUNTR on the following: total cholesterol, HDL, LDL, VLDL, LDL/HDL ratio, triglycerides, and BMI. Each model was adjusted for age, race, group status (caregiver/control), and cholesterol lowering medication (yes/no).
MATERIAL AND METHODS
Patient population
As previously described [15], participants were recruited to be part of a study designed to examine the underlying biological and behavioral mechanisms whereby stressful social and physical environments might lead to health disparities. Caregivers were recruited using flyers, ads in the local media, and community outreach efforts. Non-care giver controls were recruited by asking caregivers to nominate two to five friends who live in their neighborhood and are similar with respect to demographic factors (i.e., gender, age, and race). The study was approved by the Duke University Medical Center Institutional Review Board and all subjects gave written informed consent prior to their participation in the study. Individuals who were experiencing any acute major medical or psychiatric disorders were excluded from the study. Data were collected during a visit to the General Clinical Research Center at Duke University Medical Center, at which time a physical examination was performed and blood was drawn for assessment of lipid levels and genetic analysis.
The full study sample consisted of 85 males and 259 females. Because the MAOA gene is located on the X chromosome (Xp11.23) the heterozygosity in females complicates matters when assigning individuals to a low or high transcriptional category. Specifically, evidence suggests that it is unclear whether levels of MAOA transcription in females is the product of one or both copies of the gene [20,21]. Therefore, as with many studies examining the MAOA u-VNTR, only the 85 males were included in the present study. Eleven individuals had incomplete genetic data, leaving the present sample with 74 males. One or more lipid measures were unavailable for up to 10 individuals of the 74 participants in the present study. Participants excluded due to missing lipid variables did not differ significantly with respect MAOA gene activity (i.e, high vs low activity alleles). Analyses were conducted to include the maximum number of individuals per lipid measure (i.e., 64–74).
Measures
Genotyping
As described in Brummett et al.[15], fresh blood samples were obtained and signed into the Center for Human Genetics DNA Bank. Deoxyribonucleic acid (DNA) was extracted and stored according to methods and quality checks as previously reported [22]. An aliquot of DNA was used for MAOA genotyping. The MAOA-u VNTR region was amplified with primers: Forward-FAM-ACAGCCTGACCGTGGAGAAG and Reverse-GAACGGACGCTCCATTCGGA [1]. Polymerase chain reaction products were separated by polyacrylamide gel electrophoresis and visualized on a Hitachi FMBIO IIT Multi-View Scanner (San Francisco, CA). A greater than 95% genotyping efficiency was required before data were submitted for further analysis.
Lipid assessment
Total cholesterol, triglycerides, LDL, VLDL, HDL, and LDL/HDL ratio were assessed by the CDC approved laboratory facility at Lab Corp. located in Burlington, NC. Values are represented in mg/dL.
Cholesterol medication
Participants were asked to provide a list of all medications they were taking. Of the 74 participants, 14 were currently prescribed a cholesterol lowering medication; these 14 individual were given a code of 1 to reflect this medication status and the remaining participants were coded as 0.
Body Mass Index (BMI)
Participants were weighed and measured during their physical examination. BMI was expressed as weight in kilograms divided by height in meters squared (kg/m2) [23].
Statistical analysis
General Linear Models (GLM) were used to assess the relation among MAOA-uVNTR genotype and measures of total cholesterol, triglycerides, LDL, VLDL, HDL, LDL/HDL ratio, and BMI. MAOA-uVNTR alleles were categorized according to transcriptional functionality, with the 3-repeat variant defined as low-activity (coded 0) and longer alleles (3.5- and 4-repeat) defined as high activity (coded 1). In the present sample, there were no individuals with other MAOA-uVNTR alleles, i.e., 2-, 5-, and 6-repeat variants. Distributions of dependent constructs were examined and measures that were found to be skewed (i.e., triglycerides) were transformed by taking their logarithm. Analyses modeled the potential main effect of MAOA-uVNTR on each lipid outcome and BMI. Age, race, group (caregiver vs control), and cholesterol lowering medication (yes/no) were included in all regression analyses as adjustment variables. SAS 9.1 (Cary, NC) was used to conduct all analyses. We estimated the power to detect differences between the high activity and the low activity allele groups for our largest and smallest given sample size (i.e., n=64–74, depending on dependent variable). Assuming a two-tailed test with alpha =05, we had the ability to detect an effect size of 0.71 SD for our maximum sample size and of 0.77 SD for our minimum sample size, with power of 0.80.
RESULTS
Table 1 presents the characteristics of the present sample. The average participant had lipid levels within the normal range. Regarding MAOA-uVNTR frequencies, 67.6% of the sample had alleles that indicate higher transcription of MAOA; this distribution is similar to those from participants without existing psychiatric disorders from two other U.S. samples [2,24] e.g., approximately 60% and 65%, respectively.
Table 1.
Sample Characteristics (n=64–74).
| Characteristic | ||
|---|---|---|
| Age, mean (years) (SD) | 68.4 | (13.9) |
| Caucasian/African American | 59 (79.7%)/15 (20.3%) | |
| Total Cholesterol, mean (SD) | 180.7 | (27.1) |
| HDL, mean (SD) | 44.8 | (11.1) |
| LDL, mean (SD) | 111.6 | (25.3) |
| VLDL, mean (SD) | 22.1 | (9.5) |
| LDL/HDL ratio, mean (SD) | 2.5 | (8.9) |
| Triglycerides, mean (SD) | 130.5 | (98.9) |
| Body Mass Index, mean (SD) | 27.3 | (5.1) |
| MAOA-uVNTR Frequencies | ||
| 3 Repeat (Low Activity) | 24 | (32.4%) |
| 3.5 Repeat & 4 Repeat (High Activity) | 50 | (67.6%) |
Means and standard deviations presented are unadjusted. None of the participants in the present sample had the less common 2-, 5-, or 6-repeat alleles.
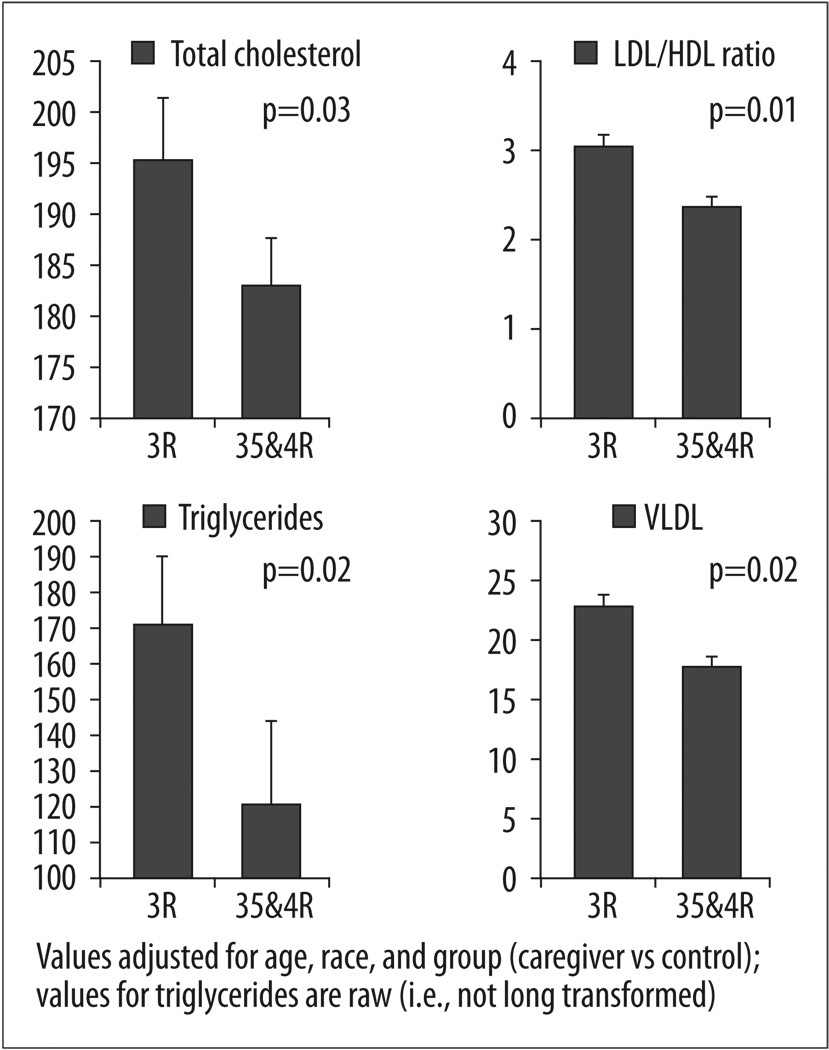
Main effect analyses for MAOA-uVNTR were statistically significant for the following lipid measures: total cholesterol, LDL/HDL ratio, triglycerides, and VLDL. Low activity alleles were associated with a worse lipid profile, in a range suggesting increased health-risk, as compared to higher activity alleles. Results of regression analyses are presented in Table 2, and statistically significant findings are graphically depicted in Figure 1. The main effect of MAOA-uVNTR expression was not significant with respect to HDL, nor LDL; however the trend was in the same direction – such that lower activity alleles were associated with lipid levels indicative of increased risk for poor health outcomes. Effect sizes for significant findings, as reflected in Cohen’s d [25], were as follows: total cholesterol d=0.54; LDL/HDL ratio d=0.81; triglycerides d=0.59; and VLDL d=0.64. BMI was not significantly related to MAOA-uVNTR alleles.
Table 2.
Regression results: associations of MAOA-uVNTR with lipid levels and body mass index.
| Lipid Value | Degrees of freedom |
F value | p value |
|---|---|---|---|
| Total Cholesterol | (1.66) | 4.90 | p=0.030 |
| LDL/HDL ratio | (1.63) | 9.44 | p=0.003 |
| Triglycerides | (1.66) | 5.86 | p=0.019 |
| VLDL | (1.69) | 6.17 | p=0.016 |
| LDL | (1.63) | 2.81 | p=0.099 |
| HDL | (1.73) | 3.35 | p=0.072 |
| Body Mass Index | (1.73) | 0.11 | p=0.743 |
All models were adjusted for age, race, statin medication status (yes/no), and group (caregiver or control); triglyceride values were log transformed.
Figure 1.
MAOA-uVNTR Genotypes (less transcriptionally active 3-repeat vs more active 3.5- & 4-repeat) and lipids related measures (adjusted means and standard errors).
Because BMI levels are associated with lipid metabolism, we conducted post hoc analyses that included BMI as a covariate in all main effect models predicting lipid related outcomes. The results were essentially unaltered, such that all significant findings remained so when adjustment for BMI was included. However the evidence for association with HDL was slightly improved and this measure became statistically significant (p<0.03) in this post hoc analysis.
DISCUSSION
The major conclusion from the present results suggests that MAOA-uVNTR genotypes conferring lower rates of transcription are associated with a poorer profile of lipid levels. Allelic variation in MAOA-uVNTR transcriptional efficiency was associated with 6% of the variance in total cholesterol, 8% in triglycerides, 13% in the ratio of low-density lipoprotein to high-density lipoprotein, and 9% in very low-density lipoprotein. This is consistent with results from related research in this area that has examined other psychological and physical constructs and found that the presence of alleles conferring less transcriptional activity is related to negative health outcomes. For example, low activity MAOA alleles have been significantly related to; alcoholism and symptoms of antisocial personality [11]; cluster B personality disorders [2]; conduct disorder [10,12]; poorer sleep and increased symptoms of depression [15]; and higher levels of BMI and/or obesity [16–19]. Paralleling the association we found between low activity MAOA alleles is research that has found decreased tissue MAO levels associated with elevated lipid levels. Obese males have been found to have a 50% reduction in MAO activity in adipose tissue, as well as increased triglyceride levels, in comparison to non-obese men [26]. Adolescents whose platelet MAO activity decreases over a three-year period also exhibit increasing total cholesterol levels over the same period [27].
We are not aware of any direct mechanism that may link the MAOA gene to lipid metabolism. Although speculative, it is possible that indirect pathways may play a role in this association. Decreased breakdown of epinephrine and nor-epinephrine in persons with less active MAOA-uVNTR alleles and hence reduced MAOA expression could result in increased adrenergic stimulation of adipocytes and hence increase lipid mobilization from fat stores. Monoamines are part of a complex system that regulates food intake and energy output [28]. Thus, the neurobiological mechanisms involved with food consumption and with the pathological characteristics associated with MAOA activity may overlap with mechanisms that control lipid metabolism. Likewise, increased stress may be found among individuals who manifest psychosocial constructs that have been related to lower transcriptional activity MAOA alleles (e.g., alcoholism, personality disorders, poor sleep and increased depression), and importantly, lipid metabolism has been associated with increased stress. For example, excessive stress-induced adrenergic stimulated lipolysis may lead to increases in lipids [29]; stress may interfere with lipid clearance by inhibiting lipoprotein lipase secretion [30]; and stress may contribute to insulin resistance and hypercortisolemia resulting in a down regulation of the hepatic LDL receptor and impairments in LDL clearance from the blood [31,32]. Therefore, it is possible that decreased MAOA-uVNTR transcriptional activity is associated with poorer lipid metabolism via its association with constructs that may lead to increased stress.
Based on prior research [16–19] we might have expected BMI to be related to MAOA-uVNTR transcriptional activity. The failure to replicate other findings in this area may be related, in part, to sample differences. Unlike the present sample, all participants in the Need et al. study [16] were female; the Ducci et al. [17] sample consisted of Finnish criminal alcoholics vs controls; the Camarena et al. study [19] was conducted in a population of primarily Spanish decent; and finally, the participants in the Fuemmeler et al. [18] study were adolescents. Thus, it is possible that gender, racial, or other sample differences may account for the discrepant findings.
Certain limitations should be noted with respect to the present findings. The sample consisted of 74 participants, therefore these results should be interpreted with proper caution and replication in additional samples will be required in order to determine their validity. We were unable to examine the ethnicity by gene interaction due to insufficient numbers of participants in certain cells (e.g., n=2 for African Americans classified in the low MAOA activity group). Thus, our results may not apply to individuals of different racial backgrounds. It should also be noted that the lipid measures evaluated are by nature correlated and our analyses included no special adjustment for this fact. Despite this overlap we thought it of interest to present the full range of measures that clinicians are likely to see in an applied setting. Finally, the original study from which this sample was drawn was not designed specifically to test the association between MAOA activity and lipid levels, and the heterogeneity of the sample (caregivers and non caregivers) may have influenced the present results.
CONCLUSIONS
We have found in men with both high and low chronic stress levels the less active alleles of the MAOA-uVNTR polymorphism are associated with an adverse lipid profile – higher total cholesterol, triglycerides, VLDL cholesterol, and LDL/HDL ratio, with a trend toward lower HDL cholesterol that becomes significant if BMI is controlled. These data add to the growing body of literature suggesting that there is a connection between neurotransmitter and lipid regulation. Taken together with findings in other studies showing the less active alleles are also associated with alcoholism, antisocial personality, poorer sleep, increased depressive symptoms, higher BMI/obesity levels, these findings suggest the testable hypothesis that persons with the less active alleles should be at higher risk of developing coronary heart disease.
Acknowledgments
Source of support: This research was supported by the National Institute on Aging, with co-funding by National Institute of Environmental Health Sciences and National Institute of Mental Health grant R01AG19605; by the Clinical Research Unit grant M01RR30; by the National Heart Lung and Blood Institute grant 5P01 HL036587; and by the Duke Behavioral Medicine Research Center
Abbreviations
- BMI
Body Mass Index
- MAOA
Monoamine oxidase-A
- HDL
High-Density Lipoprotein
- LDL
Low-Density Lipoprotein
- VLDL
Very Low-Density Lipoprotein
- MAOA-uVNR
Variable number of tandem repeats in the MAOA linked polymorphic region
REFERENCES
- 1.Sabol SZ, Hu S, Hamer D. A functional polymorphism in the monoamine oxidase A gene promoter. Human Genetics. 1998;103:273–279. doi: 10.1007/s004390050816. [DOI] [PubMed] [Google Scholar]
- 2.Jacob CP, Muller J, Schmidt M, et al. Cluster B personality disorders are associated with allelic variation of monoamine oxidase A acitivity. Neuropsychopharmacology. 2005;4:1–8. doi: 10.1038/sj.npp.1300737. [DOI] [PubMed] [Google Scholar]
- 3.Huang Y, Cate SP, Battistuzzi C, et al. An association between a functional polymorphism in the monoamine oxidase a gene promoter, impulsive traits and early abuse experiences. Neuropsychopharmacology. 2004;29:1498–1505. doi: 10.1038/sj.npp.1300455. [DOI] [PubMed] [Google Scholar]
- 4.Deckert J, Catalano M, Syagailo YV, et al. Excess of high activity monoamine oxidase A gene promoter alleles in female patients with panic disorder. Human Molecular Genetics. 1999;8:621–624. doi: 10.1093/hmg/8.4.621. [DOI] [PubMed] [Google Scholar]
- 5.Denney RM, Koch H, Craig IW. Association between monoamine oxidase A activity in human male skin fibroblasts and genotype of the MAOA promoter-associated variable number tandem repeat. Human Genetics. 1999;105:542–551. doi: 10.1007/s004399900183. [DOI] [PubMed] [Google Scholar]
- 6.Youdim MBH, Finberg JPM, Tipton KF. Monoamine oxidase. In: Trendelengurg U, Weiner U, editors. Catecholamine II. Berlin: Springer-Verlag; 1988. pp. 119–192. [Google Scholar]
- 7.Shih JC, Chen K, Ridd MJ. Monoamine oxidase: from genes to behavior. Annu Rev Neurosci. 1999;22:197–217. doi: 10.1146/annurev.neuro.22.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manuck SB, Flory JD, Ferrell RE, et al. A regulatory polymorphism of the monoamine oxidase-A gene may be associated with variability in aggression, impulsivity, and central nervous system serotonergic responsivity. Psychiatry Res. 2000;95:9–23. doi: 10.1016/s0165-1781(00)00162-1. [DOI] [PubMed] [Google Scholar]
- 9.Caspi A, McClay J, Moffitt TE, et al. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–854. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- 10.Foley DL, Eaves LJ, Wormley B, et al. Childhood adversity, monoamine oxidase A genotype, and risk for conduct disorder. Arch Gen Psychiatry. 2004;61:738–744. doi: 10.1001/archpsyc.61.7.738. [DOI] [PubMed] [Google Scholar]
- 11.Contini V, Marques FZC, Garcia CED, et al. MAOA-uVNTR Polymorphism in a Brazilian Sample: Further support for the association with impulsive behaviors and alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:305–308. doi: 10.1002/ajmg.b.30290. [DOI] [PubMed] [Google Scholar]
- 12.Lawson DC, Turic D, Langley K, et al. Association analysis of monoamine oxidase A and attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2003;116B:84–89. doi: 10.1002/ajmg.b.10002. [DOI] [PubMed] [Google Scholar]
- 13.Preisig M, Ferrero F, Malafosse A. Monoamine oxidase a and trytophan hydroxylase gene polymorphisms: are they associated with bipolar disorder? Am J Pharmacogenomics. 2005;5:45–52. doi: 10.2165/00129785-200505010-00004. [DOI] [PubMed] [Google Scholar]
- 14.Du L, Faludi G, Palkovits M, et al. High activity-related allele of MAOA gene associated with depressed suicide in males. Neuroreport. 2002;13:1195–1198. doi: 10.1097/00001756-200207020-00025. [DOI] [PubMed] [Google Scholar]
- 15.Brummett BH, Krystal AD, Siegler IC, et al. Associations of a regulatory polymorphism of the monoamine oxidase-A gene promoter (MAOA-uVNTR) with symptoms of depression and sleep quality. Psychosom Med. 2007;69:396–401. doi: 10.1097/PSY.0b013e31806d040b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Need AC, Ahmadi KR, Spector TD, Goldstein DB. Obesity is associated with genetic variants that alter dompamine availability. Ann Hum Genet. 2006;70:293–303. doi: 10.1111/j.1529-8817.2005.00228.x. [DOI] [PubMed] [Google Scholar]
- 17.Ducci F, Newman TK, Funt S, et al. A functional polymorphism in the MAOA gene promoter (MAOA-LPR) predicts central dopamine function and body mass index. Mol Psychiatry. 2006;11:856–866. doi: 10.1038/sj.mp.4001856. [DOI] [PubMed] [Google Scholar]
- 18.Fuemmeler BF, Agurs-Collins T, McClernon FJ, et al. Genes implicated in serotonergic and dopaminergic functioning interact with gender and ethnicity to predict BMI categories. Obes Res. doi: 10.1038/oby.2007.65. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camarena B, Santiago H, Aguilar A, et al. Family-based association study between the monoamine oxidase A gene and obesity: Implications for psychopharmacogenetic studies. Neuropsychobiology. 2004;49:126–129. doi: 10.1159/000076720. [DOI] [PubMed] [Google Scholar]
- 20.Benjamin D, Van Bakel I, Craig IW. A novel expression based approach for assessing the inactivation status of human X-linked genes. Eur J Hum Genet. 2000;8:103–108. doi: 10.1038/sj.ejhg.5200427. [DOI] [PubMed] [Google Scholar]
- 21.Carrel L, Willard H. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 22.Rimmler J, McDowell JG, Slotterback BD, et al. Development of a data coordinating center (DCC): Data quality control for complex disease studies. Am J Hum Genet. 1998;63:240. [Google Scholar]
- 23.Bray GA. Contemporary Diagnosis and Management of Obesity. Newton, PA: Handbooks in Health Care Co.; 1998. [Google Scholar]
- 24.Williams RB, Marchuk DA, Gadde KM, et al. Serotonin-related gene polymorphisms and central nervous system serotonin function. Neuropsychopharmacology. 2003;28:533–541. doi: 10.1038/sj.npp.1300054. [DOI] [PubMed] [Google Scholar]
- 25.Cohen J. 2nd ed. Hillsdale, NJ: Lawerence Erlbaum Associates; 1988. Statistical power analyses for the behavioral sciences. [Google Scholar]
- 26.Visentin V, Prevot D, De Saint Front VD, et al. Alteration of amine oxidase activity in the adipose tissue of obese subjects. Obes Res. 2004;12:547–555. doi: 10.1038/oby.2004.62. [DOI] [PubMed] [Google Scholar]
- 27.Kiive E, Merenakk L, Harro M, Harro J. Changes in platelet monoamine oxidase activity, cholesterol levels and hyperactive behavior in adolescents over a period of three years. Neuroscience Letters. 2005;384:310–315. doi: 10.1016/j.neulet.2005.04.093. [DOI] [PubMed] [Google Scholar]
- 28.Ramos EJ, Meguid MM, Campos AC, Coelho JC, Neuropeptide Y. alpha-melanocyte-stimulating hormone, and monoamines in food intake regulation. Nutrition. 2005;21:269–279. doi: 10.1016/j.nut.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 29.Brindley DN, McCann BS, Niaura R, et al. Stress and lipoprotein metabolism: Modulators and mechanisms. Metabolism: Clinical & Experimental. 1993;42:3–15. doi: 10.1016/0026-0495(93)90255-m. [DOI] [PubMed] [Google Scholar]
- 30.Stoney CM, Hughes JW, Kuntz KK, et al. Cardiovascular stress responses among Asian Indian and European American women and men. Ann Behav Med. 2002;24:113–121. doi: 10.1207/S15324796ABM2402_08. [DOI] [PubMed] [Google Scholar]
- 31.Brindley DN, Salter AM. Hormonal regulation of the hepatic low density lipoprotein receptor and the catabolism of low density lipoproteins: Relationship with the secretion of very low density lipoproteins. Prog Lipid Res. 1991;30:349–360. doi: 10.1016/0163-7827(91)90003-n. [DOI] [PubMed] [Google Scholar]
- 32.Salter AM, Fisher SC, Brindley DN. Binding of low-density lipoprotein to monolayer cultures of rat hepatocytes is increased by insulin and decreased by dexamethasone. FEBS Lett. 1987;220:159–162. doi: 10.1016/0014-5793(87)80895-5. [DOI] [PubMed] [Google Scholar]