Abstract
Summary: FBA-SimVis is a VANTED plug-in for the constraint-based analysis of metabolic models with special focus on the visual exploration of metabolic flux data resulting from model analysis. The program provides a user-friendly environment for model reconstruction, constraint-based model analysis, and interactive visualization of the simulation results. With the ability to quantitatively analyse metabolic fluxes in an interactive and visual manner, FBA-SimVis supports a comprehensive understanding of constraint-based metabolic flux models in both overview and detail.
Availability: Software with manual and tutorials are freely available at http://fbasimvis.ipk-gatersleben.de/
Contact: grafahr@ipk-gatersleben.de
Supplementary information: Examples and supplementary data are available at http://fbasimvis.ipk-gatersleben.de/
1 INTRODUCTION
Mathematical modelling of metabolism offers new approaches to improve the understanding of complex biological processes. Flux balance analysis (FBA) is a constraint-based modelling approach which allows the prediction of optimal metabolic yield and steady state flux distributions under different environmental conditions and genetic backgrounds (Varma and Palsson, 1994). Due to its predictive power and the advantage of not requiring the knowledge of kinetic parameters, FBA has successfully been applied to a variety of biological systems to study different aspects of metabolism (Kauffman et al., 2003; Reed and Palsson, 2003).
Several software tools have been developed to perform FBA, including packages freely available for academic research such as CellNetAnalyzer (Klamt et al., 2007), the COBRA Toolbox (Becker et al., 2007) and FBA (http://gcrg.ucsd.edu/Downloads/Flux_Balance_Analysis).
Although most of these tools offer a variety of constraint-based analysis techniques, they have been mainly focused on model analysis while comparative data examination and interpretation in terms of interactive visualization of FBA results has only been considered to a minor extent.
To provide a visual analysis of FBA results in an interactive way, aimed at facilitating the analysis and interpretation of metabolic fluxes in response to genetic and/or environmental conditions, we developed FBA-SimVis, a framework for integrated constraint-based model analysis and visualization. It has been implemented as a plug-in for the open source program VANTED (Junker et al., 2006).
2 METHODS AND IMPLEMENTATION
FBA-SimVis is a user-friendly tool for the visual exploration and analysis of constraint-based metabolic models. It is implemented as a plug-in for VANTED, an analysis and visualization software for biological networks containing experimental data. The plug-in extends the Java-based VANTED system by integrating methods for constraint-based model analysis and interactive flux visualization. The analysis techniques are implemented in MATLAB using the CLP solver (COIN-OR Linear Program Solver) for linear and non-linear optimization. The MATLAB routines are based on the free library of the COBRA Toolbox, which has been modified to meet our demands, for example, by integrating a non-linear optimization procedure to handle the problem of alternate optimal solutions possibly produced by linear optimization offered by the toolbox. Due to its underlying quadratic optimization, the non-linear optimization routine does not produce multiple optima, thus having the advantage of reducing redundant scenarios. To provide a MATLAB environment-independent application of the software, the MATLAB routines are integrated as standalone executables, making the plug-in readily available to users free of charge. The main features of FBA-SimVis are described below.
2.1 Model reconstruction
FBA-SimVis and the VANTED system provide a model reconstruction environment. Via the graphical user interface, the user can create and edit a metabolic model by using a simple drag-and-drop mechanism for network creation and a text menu for network refinement. Data content in the text fields and in the interactive network map can be edited, stored and modified individually by the user.
To reconstruct a metabolic model the user can either create a new model in the visual manner described above (i.e. using the graphical model editor to edit existing or dropping new elements onto the canvas), or import a model via SBML from files (e.g. by dropping the file onto the canvas) or model databases, and layout the corresponding network map by using the different layout algorithms provided by VANTED. Once the metabolic model is reconstructed, the model can be exported in the form of a network map and a SBML file, which together form the basis for subsequent model analysis and flux visualization.
2.2 Constraint-based model analysis
FBA-SimVis integrates various methods for the constraint-based analysis of metabolic models:
Flux balance analysis is based on the optimization of an objective function, which is used as an evaluation criterion to identify an optimal flux distribution among all possible steady state flux distributions that meets the objective. FBA can be used to predict optimal metabolic yield and steady state flux distributions under different environmental conditions and genetic backgrounds.
Knock-out analysis (i.e. the deletion of a given enzyme or gene) is performed by setting the flux trough a particular reaction to zero and calculating the objective function. In silico knock-out analysis provides an efficient method to study the essentiality of a reaction in a metabolic network and to gain insight into metabolic changes caused by the deletion.
Robustness analysis is performed by varying a particular flux over a specified range of values and recalculating the objective function. As the resulting curve depicts the sensitivity of the objective function to that particular flux, robustness analysis can be used to assess the effect of reducing flux trough particular reaction on a given objective.
Flux variability analysis is performed by constraining the objective function to the optimal value and computing the minimal and maximal flux through every reaction in the network (range of fluxes). Flux variability analysis can be used to study the redundancies of the metabolic model under investigation.
In addition to the constraint-based analysis techniques provided by FBA-SimVis, the original VANTED tool offers a variety of graph theoretical analysis techniques (e.g. shortest path length, connectivity analysis, cycle detection) and statistical analysis techniques (e.g. t-test, correlation analysis, cluster analysis) which can also be used for an in-depth topological and statistical analysis of the modelling results.
2.3 Model and flux visualization
The visualization of the constraint-based analysis results is done in a highly interactive way. Instead of just visualizing a fixed picture showing the computed flux distribution, FBA-SimVis provides an interactive graphical user interface where user interactions directly affect the visualization. Automatic mapping of the computed net flux onto the network map is done by scaling the width of the reaction edges according to the flux and displaying the flux values next to the corresponding reaction edges.
FBA-SimVis allows users to compare metabolic fluxes in response to varying genetic and/or environmental conditions in an interactive and comparative way by providing a dynamic parameter analysis. It offers different visualizations for the various constraint-based analysis results:
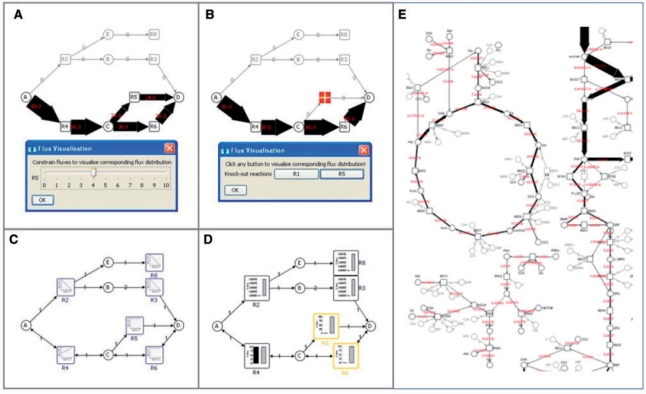
To investigate the influence of varying environmental conditions, up to three different parameters specified by the user can be varied by slider interactions, with the resulting flux distribution being automatically displayed on the network map, see Figure 1A.
The dynamic variation of the genetic background is performed by interacting with a button panel which comprises the enzymatic reactions to be knocked-out, and which results in the automatic display of the flux distribution of the specified knock-out simulation, see Figure 1B. In addition, an overview perspective allows the quantification of the sensitivity of a given objective function to the knock-out of any of the network reactions by expressing the optimal value (i.e. the numerical value of the objective function) of the enzymatic deletion as a fraction of the optimal value of the ‘reference scenario’ (i.e. the simulation with the complete enzyme set). The interactive parameter analysis supports a fast and intuitive understanding of the simulated flux data, allowing insights into metabolic and physiological changes of metabolism in response to varying genetic and/or environmental conditions.
The visualization of simulation results obtained from robustness analysis is offered in two ways, either displaying the sensitivity curve for each of the network reactions within the respective reaction nodes (Fig. 1C), or by displaying the flux distribution of a particular enzymatic reaction, which can be varied by slider interactions. While the first option provides a rapid and comparative overview of all reactions included in the network, the second option offers the user the possibility to obtain detailed insights into specific enzyme/objective function dependencies.
Simulation results of flux variability analysis are provided by displaying the minimal and maximal flux for each of the network reactions within the respective reaction nodes, see Figure 1D.
To further support comparative data evaluation (i.e. the comparison of flux distributions) and automatic layout, VANTED provides different chart types (e.g. pie and bar plots) facilitating direct data comparison and offers several graph layout algorithms (e.g. force-directed, hierarchical and circular layouts) to automatically arrange the network elements. Moreover, the user has the possibility to save and reload flux distribution patterns for subsequent analysis. In addition to the interactive visualization of simulated flux data computed by FBA-SimVis, the software supports the visualization of any kind of flux data provided by external analysis. Details about data formats supported by the system can be found on the FBA-SimVis website.
Fig. 1.
This picture shows example visualisations for (A) flux balance analysis, (B) knock-out analysis, (C) robustness analysis, and (D) flux variability analysis for a small example network and (E) visualisation of a partial flux map of a large model of barley seed metabolism.
3 EXAMPLE
An application example of how to use FBA-SimVis is provided for a small metabolic network. The metabolic model as well as a step-by-step description of model analysis and interactive flux visualization are available at the FBA-SimVis website.
Figure 1A–D summarize the visualization of the constraint-based analysis results of the small example network, demonstrating the usefulness of the interactive visualization routines for comparative evaluation of the simulation results which support a fast and intuitive understanding of metabolic fluxes in both overview and detail. The different views have been discussed in the previous section.
In addition, an application example of a large metabolic model of barley seed metabolism is given in Figure 1E. This model comprises more than 250 reactions and shows the usability of the analysis and visualization routines for large real world models.
4 DISCUSSION
FBA-SimVis is a user-friendly tool for the visual exploration and analysis of constraint-based metabolic models. Due to the integration of constraint-based analysis techniques with interactive visualization routines, FBA-SimVis offers a holistic analysis of stoichiometric models of metabolism. Although different tools such as CellNetAnalyzer (Klamt et al., 2007) or the COBRA Toolbox (Becker et al., 2007) provide a variety of constraint-based analysis techniques, these programs mainly focus on model analysis, while model reconstruction and visualization of analytical results are only partly supported. Other tools offering divers routines for model reconstruction and flux visualization are MetVis (Noack et al., 2007) and YANAsquare (Schwarz et al., 2007). These tools refer to external programs to perform FBA, nevertheless, the tools and external programs are also independent of commercial software and freely available. Besides providing a comprehensive analysis of constraint-based metabolic models by combining methods for model reconstruction, analysis and visualization, FBA-SimVis offers a unique combination of features not present in other analytical tools including easy change of parameters and optimization routines to facilitate model analysis and highly interactive visualization, and specific statistical analysis routines to support the comparative evaluation of simulation results.
Conflict of Interest: none declared.
REFERENCES
- Becker SA, et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nature Protocols. 2007;2:727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- Junker BH, et al. VANTED: a system for advanced data analysis and visualisation in the context of biological networks. BMC Bioinformatics. 2006;7:109.1–109.13. doi: 10.1186/1471-2105-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman KJ, et al. Advances in flux balance analysis. Curr. Opin. Biotechnol. 2003;14:491–496. doi: 10.1016/j.copbio.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Klamt S, et al. Structural and functional analysis of cellular networks with CellNetAnalyzer. BMC Syst. Biol. 2007;1:1–13. doi: 10.1186/1752-0509-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noack S, et al. Visualizing regulatory interactions in metabolic networks. BMC Biol. 2007;5:46.1–46.17. doi: 10.1186/1741-7007-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JL, Palsson B.Ø. Thirteen years of building constraint-based in silico models of Escherichia coli. J. Bacteriol. 2003;185:2692–2699. doi: 10.1128/JB.185.9.2692-2699.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz R, et al. Integrated network reconstruction, visualization and analysis using YANAsquare. BMC Bioinformatics. 2007;8:313.1–313.10. doi: 10.1186/1471-2105-8-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, Palsson B.Ø. Review: metabolic flux balancing: basic concepts, scientific and practical. Bio/Technology. 1994;12:994–998. [Google Scholar]