Abstract
Summary: The current fast growth of genome-wide association studies (GWAS) combined with now common computationally expensive imputation requires the online access of large user groups to high-performance computing resources capable of analyzing rapidly and efficiently millions of genetic markers for ten thousands of individuals. Here, we present a web-based interface—called GRIMP—to run publicly available genetic software for extremely large GWAS on scalable super-computing grid infrastructures. This is of major importance for the enlargement of GWAS with the availability of whole-genome sequence data from the 1000 Genomes Project and for future whole-population efforts.
Contact: ta.knoch@taknoch.org; f.rivadeneira@erasmusmc.nl
1 INTRODUCTION
By 2008 more than 150 associations between common genetic variants and human complex traits and disease have been successfully identified through the use of GWAS (Altshuler et al., 2008). It rapidly became evident that very large samples sizes are required to detect variants with modest genetic effects (e.g. a study requires ∼8600 samples to have 90% of power to find genetic variants with a frequency of 0.20, an odds ratio of 1.2 and a genome-wide significance of 10−8). Such study sizes are achieved by meta-analysis of data shared collaboratively in consortia analyzing 100 s of traits in greater than ∼40 000 individuals (e.g. Psaty et al., 2009). Since they use different genotyping platforms (e.g Affymetrix, Illumina), imputation of millions of markers from a reference (e.g. a HapMap population) is required (de Bakker et al., 2008; International HapMap Consortium et al., 2007). Statistical methods as linear or logistic regressions measure marker wise the actual association of the genetic variants with quantitative and binary diseases and traits. Freely available software like MACH2QTL/DAT (Li et al., 2006), SNPTEST (Marchini et al., 2007) or ProbABEL (Aulchenko et al., 2007) perform similarly well for these analyses and allow trivial parallelization for distributed computing: the computation time on a regular computer for one continuous trait (∼2.5 million markers, ∼6000 samples) is currently ∼6 h. Assuming linear scaling future studies with ∼50 million markers from genome sequencing in 105–106 samples and even low (1%) allele frequencies can result in approximately >85–850 days of analysis. Thus, secure, fast accessible web services and scalable high-performance computing grid infrastructures as the Erasmus Computing Grid (de Zeeuw et al., 2007) or the German MediGRID (Krefting et al., 2008) are required to make this analysis feasible.
Here, we present a web-based interface and application to run publicly available genetic software for extremely large GWAS on such super-computing grid infrastructures. Consequently, we provide a solution to analyze GWAS in very large populations.
2 IMPLEMENTATION
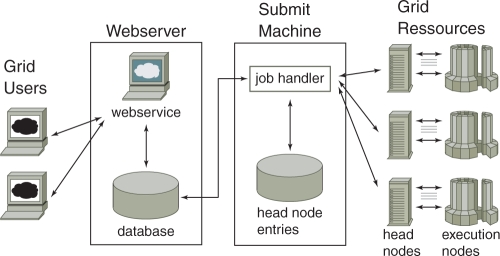
To achieve high-speed result delivery, the work is split and distributed on different grid processors by trivial parallelization depending on the total data amount. The complete system consists of (i) the user remote access computer; (ii) a web server with user webservices and a data/application database; (iii) a submit machine with a job handler and a grid resource database; and (iv) grid resources with head nodes and execution nodes. The implementation consists of a hardened Linux system, which has a hardened apache2 web server and a PostgreSQL database. Php is used for the web site and the job-handler is scripted in Perl. Concerning security, data transmission is encrypted and complete user separation is applied. Currently, the system administrator manages user accounts and monitors user access, job status and statistics. He also uploads the GWA imputed data to all available grid head nodes for each genotyped cohort, since it is of large size and the same for all cohort phenotypes. Thus, only the phenotype information has to be uploaded by the GRIMP user to the system, which controls the detailed workflow (Fig. 1).
Fig. 1.
Structure of the work flow of GRIMP consisting of (i) remote user access, (ii) a web server with web services and a data/application database, (iii) a submit machine with job handler and grid resource database and (iv) grid resources with head nodes and execution nodes.
2.1 User package submission
After logging into the system the users manually specify the analysis details: they label the analysis and select a regression model (currently, linear and logistic models), dataset and optionally a gender stratified or combined analysis. Additional individual-phenotype links and phenotype specifying annotation files can now be uploaded to the database as well. Further covariates (specified in the phenotype file) can also be annotated. After choosing the progress notification scheme, the user submits the process package.
2.2 Package preprocessing
The phenotype file is transformed to fit the format required by the analytical application implemented (currently, mach2qtl and mach2dat for linear and logistic regression, are freely available at http://www.sph.umich.edu/csg/abecasis/MACH/download/). In principle, any GWA analysis software can be used here and installed in the application database.
2.3 Job submission to the grid infrastructure
An implemented job handler periodically checks the database for newly submitted packages and also checks for the workload on the grid head nodes for available capacity to split the packages properly into jobs to be distributed to an individual grid part. To avoid queue overflow, each head node has a predefined amount of jobs that can be queued. Thereafter, the job handler creates a submit file and packages to be uploaded to the individual grid head node. The local respective grid middleware will handle the jobs of the package for these specific grid infrastructures. Currently, we use here the Globus toolkit, but in principle any grid driving middleware can be used here. For high-speed delivery the individual jobs have highest priority compared with other and filler jobs.
2.4 Job/package monitoring
The job handler checks every 5 min the database for sent jobs and verifies the current status of the individual jobs distributed to a CPU through the middleware on the specific grid head node. An individual failed job is resubmitted up to three times. After all individual jobs of a package are completed, the results are uploaded to the database and the package on the head node is removed. In case of complete failure, the job handler will remove all jobs of the package on the head node including the uploaded package and a failure notification is sent.
2.5 Package post-processing and notification
Once all jobs of a package were finished, all individual result files are combined into one file together with additional marker annotations such as chromosome, position, allele frequency, sample size and quality of the imputed markers. The results are archived in the database for later analysis and the result files are compressed to save disk space. Depending on the choice of notification the user is now informed—e.g. by email.
3 RESULTS AND CONCLUSIONS
Through a web-based interface the successful implementation of GRIMP allows to use publicly available genetic software for very large GWAS on scalable super-computing grid infrastructures such as the Erasmus Computing Grid or the German MediGRID within an hour. The analysis of ∼2.5 million markers and ∼6000 samples now takes ∼12 min in contrast with ∼6 h. For ∼107 markers and ∼105 samples, we achieve ∼10–20 min, in contrast with ∼400 h, i.e. ∼17 days for a single CPU. Thus, GRIMP will improve the learning curve for new users and will reduce human errors involved in the management of large databases. Consequently, researchers and other users with little experience will largely benefit from the use of high-performance grid computing infrastructures. Since each Grid infrastructure has different middleware setups, adjustments might be needed for each particular GRIMP implementation. Currently, we have successfully setup GRIMP for the Rotterdam Study, a prospective population-based cohort study of chronic disabling conditions in >12 000 Dutch elderly individuals (http://www.epib.nl/ergo.htm; Hofman et al., 2007). Thus, with its user-friendly interface GRIMP gives access to distributed computing to primarily biomedical researchers with or without experience, but with extreme computational demands. This is of major importance for the enlargement of GWAS with the availability of whole-genome sequence data from the 1000 Genomes Project and for future whole-population efforts.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Luc V. de Zeeuw, Rob de Graaf (Erasmus Computing Grid, Rotterdam, The Netherlands) and the National German MediGRID and Services@MediGRID, German D-Grid for access to their grid resources.
Funding: German Bundesministerium für Forschung und Technology (# 01 AK 803 A-H, # 01 IG 07015 G). European Commission (HEALTH-F2-2008-201865-GEFOS). Netherlands Organization of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012).
Conflict of Interest: none declared.
REFERENCES
- Altshuler D, et al. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, et al. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zeeuw LV, et al. Het bouwen van een 20 TeraFLOP virtuelle supercomputer. NIOC proceedings 2007. 2007:52–59. Avaliable at URL: http://www.nioc2007.nl/content/files/nioc%20proceedings%202007.pdf. [Google Scholar]
- Hofman A, et al. The Rotterdam Study: objectives and design update. Eur. J. Epidemiol. 2007;22:819–829. doi: 10.1007/s10654-007-9199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krefting D, et al. MediGRID – towards a user friendly secured grid infrastructure. Future Gener. Comput. Syst. 2008;25:326–336. [Google Scholar]
- Li Y, et al. Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am. J. Hum. Genet. 2006;S79:2290. [Google Scholar]
- Marchini J, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Psaty B, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium. Design of prospective meta-analysis of genome-wide association studies from 5 cohorts. Circ. Cardiovasc. Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.