Abstract
Summary: ANCHOR is a web-based implementation of an original method that takes a single amino acid sequence as an input and predicts protein binding regions that are disordered in isolation but can undergo disorder-to-order transition upon binding. The server incorporates the result of a general disorder prediction method, IUPred and can carry out simple motif searches as well.
Availability: The web server is available at http://anchor.enzim.hu. The program package is freely available for academic users.
Contact: zsuzsa@enzim.hu
1 INTRODUCTION
Many disordered proteins contain important functional elements involved in protein–protein interactions. Disordered binding regions play a critical role in various biological processes, involving regulation and signaling (Dyson and Wright, 2002). These segments differ from protein interaction sites of globular proteins due to their distinct structural properties (Mészáros et al., 2007). Such regions exist as a highly flexible structural ensemble in isolation and adopt a well-defined conformation only upon binding to their specific partner molecules. It was suggested that certain disorder prediction methods can be indicative of disordered binding regions (Garner et al., 1999). Specialized methods have been developed to regions adopting α-helical conformation in their bound state (Cheng et al., 2007) or for the binding partners of calmodulin (Radivojac et al., 2006). In contrast, ANCHOR is a general method for recognizing disordered binding regions.
ANCHOR aims to capture the basic biophysical properties of disordered binding regions using estimated energy calculations (Mészáros et al., 2009). Estimated energies can be assigned to each residue in a sequence and were shown to well-approximate the corresponding energies calculated from known structures of globular proteins (Dosztányi et al., 2005b). Generally, disordered regions can be discriminated from ordered proteins by unfavorable estimated energies. This concept is utilized in the IUPred server for the prediction of protein disorder (Dosztányi et al., 2005a). The estimated energies can also detect regions that are likely to gain energetically by interacting with globular proteins. Predictions in ANCHOR combine the general disorder tendency with the sensitivity to the structural environment (Mészáros et al., 2009). Because of this additional property, ANCHOR scores are relatively independent from IUPred scores.
The developed method was able to recognize disordered binding regions with almost 70% accuracy at the segment level on various datasets. We also ensured that disordered binding regions could be discriminated from generally disordered regions and that the false positive rate on a dataset of globular proteins was <5%. Since the publication of the original paper (Mészáros et al., 2009), we have found that the false positive rate can be further reduced by eliminating segments with IUPred scores too low to be compatible with disordered binding regions. Additionally, short predicted segments of length less than six residues are also filtered out.
ANCHOR predicts disordered binding regions without any information about the partner protein(s). A complementary approach identifies protein binding regions using motif searches. It was suggested that interaction with certain proteins or protein families are mediated through specific linear motifs that capture key residues responsible for binding. A growing number of such linear motifs are now being categorized in the ELM server (Puntervoll et al., 2003). The presence of sequence motifs reduces the complex task of finding putative protein binding sites to a simple pattern matching problem. However, such matches can contain many false positives, suggesting that the definition of the binding motif should include information about the specific structural context. Since several instances of linear motifs occur within disordered regions, disordered binding regions could help to filter out false positive matches. Therefore, complementing the prediction of disordered binding regions with specific motif searches can prove useful in many cases and help to explore other motifs.
2 THE ANCHOR SERVER
The minimum input of the web server is a single amino acid sequence. Sequences can also be specified by their corresponding UniProt IDs or ACs. A list of motifs can also be submitted, specified as regular expressions with or without their names. A few examples, including known eukaryotic linear motifs are given in the help to guide the user with the format. The motif search, however, is not restricted to known linear motifs, any kind of regular expression can be specified.
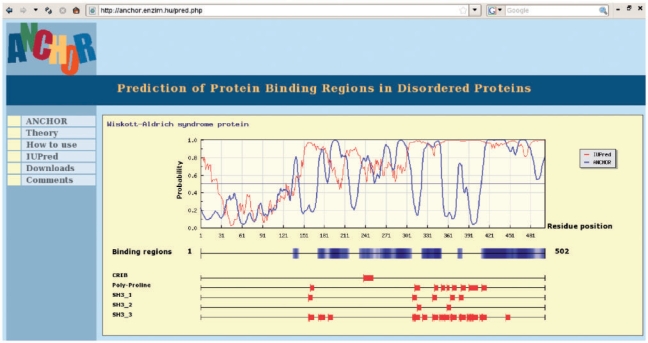
The basic output of our prediction method is a probability score, indicating the likelihood of the residue to be part of a disordered binding region along each position in the sequence. Regions that have a score >0.5 and pass the filtering criteria are predicted as disordered binding regions. The returned plot shows the prediction profile calculated by ANCHOR, the disordered binding region prediction method, together with IUPred, a general disorder prediction method. Predicted disordered binding regions and matched motifs are also indicated underneath the profile as horizontal bars. The graphical output is followed by a simple text output, summarizing the predicted and filtered binding regions, the location of the found motifs and the returned prediction profile. An example for the graphical output is presented in Figure 1. The core program of ANCHOR is written C, while motif searches are carried out by a Perl wrapper. This Perl program is called by the web server written in PHP. The graphical output is generated by the JpGraph software (JpGraph, 2005; http://www.aditus.nu/jpgraph/). The default option for graphical/text output is automatically determined by the browser type, but it can be changed by user. Additionally, list of sequences can also be submitted to generate simple text output on a larger scale.
Fig. 1.
An example of ANCHOR graphical output for the Wiskott–Aldrich Syndrome protein (WASp) with various motif searches. The N-terminal of the protein contains an ordered domain, otherwise it is largely disordered. Multiple disordered binding regions were predicted, and several of these can be confirmed experimentally [see Mészáros et al. (2009) for more details]. The results of the motif searches show regions containing various SH3 binding sites as specified in the ELM database. Additionally, proline rich regions and the CRIB motif implicated in binding to Cdc42 can also be located.
Funding: Hungarian Scientific Research Fund (OTKA-K72569); the National Office for Research and Technology, Hungary (NKTH07a-TB_INTER).
Conflict of Interest: none declared.
REFERENCES
- Cheng Y, et al. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry. 2007;46:13468–13477. doi: 10.1021/bi7012273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztányi Z, et al. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005a;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- Dosztányi Z, et al. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005b;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- Garner E, et al. Predicting binding regions within disordered proteins. Genome Inform. Ser. Workshop Genome Inform. 1999;10:41–50. [PubMed] [Google Scholar]
- Mészáros B, et al. Molecular principles of the interactions of disordered proteins. J. Mol. Biol. 2007;372:549–561. doi: 10.1016/j.jmb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Mészáros B, et al. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009;5:e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puntervoll P, et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, et al. Calmodulin signaling: analysis and prediction of a disorder-dependent molecular recognition. Proteins. 2006;63:398–410. doi: 10.1002/prot.20873. [DOI] [PubMed] [Google Scholar]