Abstract
Summary: SIMBioMS is a web-based open source software system for managing data and information in biomedical studies. It provides a solution for the collection, storage, management and retrieval of information about research subjects and biomedical samples, as well as experimental data obtained using a range of high-throughput technologies, including gene expression, genotyping, proteomics and metabonomics. The system can easily be customized and has proven to be successful in several large-scale multi-site collaborative projects. It is compatible with emerging functional genomics data standards and provides data import and export in accepted standard formats. Protocols for transferring data to durable archives at the European Bioinformatics Institute have been implemented.
Availability: The source code, documentation and initialization scripts are available at http://simbioms.org.
Contact: support@simbioms.org; mariak@ebi.ac.uk
1 INTRODUCTION
The growing use of high-throughput technologies in biomedical studies and the volume and complexity of data generated in such experiments have created a need for dedicated software systems to collect, store and manage these data. Moreover, essential information about biomedical research subjects (patients) and samples have to be recorded and linked to the data. Projects are often collaborative, include many researchers and laboratories and may be spread across different sites. Personal information must be managed in a secure manner, the data access rights should be consistent with ethical requirements. Generic laboratory information management systems are not always appropriate for these purposes. The existing open source software systems (e.g. Reich et al., 2006; Saal et al., 2002; Saeed et al., 2003) have been primarily designed for use in a single laboratory.
To address these issues, we have developed a web-based System for Information Management in BioMedical Studies—SIMBioMS. It was originally implemented for needs of a particular multi-site project (MolPAGE, www.molpage.org). Since later it proved to be easily customizable and scalable for other applications, including population genomics studies, we generalized the system as open source software.
SIMBioMS provides an interface for data entry via web forms, upload facilities of pre-formatted datasets from files, data export facilities (including configurable export definable by XML templates) as well as advanced data access and user rights management. The system can be configured to support the minimum information requirements MIBBI (Taylor et al., 2008), data can be imported/exported in accepted standard formats MAGE-TAB (Rayner et al., 2006) and ISA-TAB (Sansone et al., 2008), as well as custom-made XML and tab-delimited formats, allowing for easy data import and export from users' own tools, and generic tools such as Excel. A simple browsing and customizable data filtering options allow for the essential content exploration and report construction on metadata level. Selected data can be imported into analysis tools, such as Bioconductor.
2 SYSTEMS DESIGN, IMPLEMENTATION AND CUSTAMIZATION
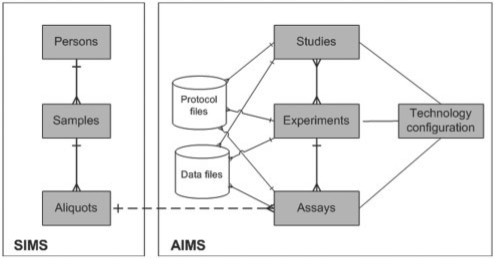
The system consists of two components—Sample Information Management System (SIMS) and Assay data and Information Management System (AIMS) (Fig. 1). As the names suggest, SIMS is designed to collect phenotypical, environmental and technical information about samples, while AIMS handles the experimental data from high-throughput assays. SIMS provides a simple solution for data anonymization by creating identifiers linked to person's information in a separate module. SIMS extends a previously published system (PASSIM; Viksna et al., 2007). The main new features include customizability and compatibility with data formats MAGE-TAB and ISA-TAB. While, PASSIM was designed to manage patient and sample data, it did not have any means for linking it to data from high-throughput assays.
Fig. 1.
High level class diagram of SIMS and AIMS.
AIMS is a new system filling this gap, designed for adoptability for any technological platform, and for easy extraction of captured data for analysis. It is linked to SIMS through a three-level hierarchy: a person can be linked to one or more samples, a sample can have one or more aliquots. Each aliquot can have one or more assays performed on it, and each assay can be linked to one or more data files. Assays are grouped in experiments and studies, each of which can have one or more data files attached. For instance, raw microarray data files would be normally linked to individual assays, while normalized gene expression matrices to experiments. Assays are technology-specific; the current AIMS configurations include genotyping, sequencing, proteomics and metabonomics.
The two systems can be installed and used independently, or jointly—if a laboratory already has a local informatics system for sample or assay data, it can be used jointly with AIMS or SIMS, respectively.
SIMBioMS run in Apache Tomcat servlet containers, or other application servers. The data are stored in PostgreSQL databases, but other popular database management systems have been tested and can be used with minimal changes. The systems are platform independent, and have been tested on several MS Windows and Linux. Several preconfigured versions, including ones for type 2 diabetes, metabolic syndrome and autoimmune diseases are packed into .war web-application archives. AIMS/SIMS can be installed either as local (e.g. on a laptop) or as centralized databases. Installation for local use is a simple two-step procedure that does not require special database software (java light database h2 is used). Filtering functionality is customizable, for enumerated value fields a drop-down list can be provided, fields are defined as parameters.
3 RESULTS AND DISCUSSION
The systems development effort up to now is ∼8 person-years. To the best of our knowledge, this is the only open source web-based system that integrates capturing of rich phenotypic data with management of high-throughput data from multiple platforms for needs of multi-site collaborative projects and that has already proven its usefulness. We are currently running three SIMBioMS instances to support collaborative projects, including an instance containing data from over 25 000 assays on nine different technology platforms, and an instance for population-wide epidemiology studies. We have implemented protocols for data transfer to the permanent data archives: ArrayExpress (Parkinson et al., 2008) and European Genotype Archive (EGA) and data from over 6500 assays have been transferred. In the future, the system will be extended to include next-generation sequencing data. Source code, documentation, initialization scripts, templates for metadata configuration, links to demo instances and user guide are available at http://simbioms.org.
ACKNOWLEDGEMENTS
We would like to thank Anthony Maher, Derek Crockford, Magnus Åberg, Severine Zirah, Marc E Dumas, Anna Asplund, Erik Björling, Susanne Schwonbeck, Jens Lamerz, Andreas Petri, Kristian Almstrup, Matthias Schuster, Dimo Dietrich, Florian Eckhardt, Leena Peltonen, Huei-Yi Shen, Inga Prokopenko, Cecilia Lindgren, Samuli Ripatti, Erkki Raulo, Juha Muilu and Jan Eric Litton and ArrayExpress curators.
Funding: EU projects LSHG-CT-2004 Nr.512066 MolPAGE; HEALTH-F4-2007- 201413; ENGAGE.
Conflict of Interest: none declared.
REFERENCES
- Parkinson H, et al. ArrayExpress update—from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2008;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner T, et al. A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB. BMC Bioinformatics. 2006;7:489. doi: 10.1186/1471-2105-7-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, et al. GenePattern 2.0. Nat. Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Saal L, et al. BioArray Software Environment (BASE): a platform for comprehensive management and analysis of microarray data. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-8-software0003. software0003.1–software0003.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Sansone S, et al. The First RSBI (ISA-TAB) Workshop: “Can a Simple Format Work for Complex Studies?”. OMICS. 2008;12:143–149. doi: 10.1089/omi.2008.0019. [DOI] [PubMed] [Google Scholar]
- Taylor C, et al. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nat. Biotechnol. 2008;26:889–896. doi: 10.1038/nbt.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viksna J, et al. PASSIM—an open source software system for managing information in biomedical studies. BMC Bioinformatics. 2007;8:52. doi: 10.1186/1471-2105-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]