Abstract
Summary: Array-based comparative genomic hybridization (CGH) technology is used to discover and validate genomic structural variation, including copy number variants, insertions, deletions and other structural variants (SVs). The visualization and summarization of the array CGH data outputs, potentially across many samples, is an important process in the identification and analysis of SVs. We have developed a software tool for SV analysis using data from array CGH technologies, which is also amenable to short-read sequence data.
Availability and implementation: SnoopCGH is written in java and is available from http://snoopcgh.sourceforge.net/
Contact: jg10@sanger.ac.uk; tc5@sanger.ac.uk
1 BACKGROUND
Genomic structural variants (SVs), including copy number variants (CNVs), can have important and pleiotropic effects on phenotype variation, increasingly being regarded as a significant type of genetic risk factor for monogenic and complex diseases (O'Donovan et al., 2008). However, detecting and analyzing structural variation remains challenging. Array comparative genomic hybridization (CGH) is a powerful tool for identifying copy number variation between DNA samples. In a typical array CGH experiment, DNA samples being compared (e.g. disease versus control) are differentially fluorescently labeled, pooled and hybridized to oligo probes spanning the genome of interest printed on a glass slide. The data outputs are log ratios of normalized fluorescence intensities reflecting the relative hybridization levels, and hence relative copy number levels between the samples at a given location in the genome. Thus, concentrated high or low log ratios of fluorescence intensities represent genomic regions of interest for CNVs. These regions can be very small, which makes the identification of biologically relevant events challenging. There is a growing catalog of software tools for determining and plotting CNV locations and breakpoints [see Wang et al., (2009) for a review and methodology]. However, there is a dearth of tools for interactive visualization of multiple samples with CGH data at varying degrees of resolution, with the ability to display genome annotation data, informative genomic tracks (e.g. GC content) and the results of SV break point analyses. Here, we present SnoopCGH, a software tool that facilitates the rapid analysis of normalized array CGH data. Its functionality includes assessment of data quality and normalization, detection of SVs and integration of useful annotation of features. We demonstrate SnoopCGH functionality using array CGH data comparing five laboratory-adapted strains of the human malaria species, Plasmodium falciparum.
2 FEATURES OF SNOOPCGH
SnoopCGH is a java-based standalone application that inputs CGH data in tab-, space- or comma-delimited format, containing columns with: chromosome number, probe name, probe starting and end positions, and a series of log intensity values corresponding to one or more comparisons or samples. It is possible to load more than one data file. The use of multiple window layers facilitates the visualization of subsets of data, with the ability to zoom in and out of regions of interest. SV breakpoint analysis methods are implemented and enable the rapid visualization and dissection of putative SV regions. In particular, data are smoothed using an algorithm based on Haar wavelets (Ben-Yaacov and Eldar, 2008), and islands of potential SVs are estimated using a Smith–Waterman algorithm (Price et al., 2005). The Haar wavelet approach has two smoothing parameters, namely start and end levels, that influence the sensitivity to the size of segments and trends, respectively. The default settings in SnoopCGH are based on suggested values in Ben-Yaacov and Eldar (2008). The breakpoint algorithms estimate levels of statistical significance and robustness of putative SVs using permutations. Prior to their application we may remove outliers to improve robustness using thresholds based on median absolute deviation statistics. The quantification of putative SVs leads to an ability to rank the regions of interest. We have also implemented a rank-based algorithm that considers differences in SVs between (groups of) samples (Laframboise et al., 2009). This extension may assist those working on association studies or multiple population studies considering differences in genetic variation. The strength of SnoopCGH is its ability to interface with downloadable annotation files (e.g. embl and gff formats) from genomic browsers, that include information on gene names and genomic features (e.g. GC content). It is also possible to read in other useful information, such as the results from breakpoint analyses run externally.
3 APPLICATION TO A PARASITE CGH ARRAY
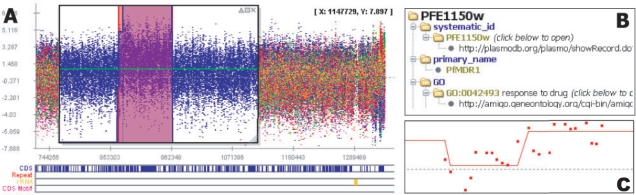
Plasmodium falciparum (Pf) malaria has an enormous morbidity and mortality burden in sub-Saharan Africa. The Pf genome is AT rich (80%), and contains some CNVs associated with drug resistance and erythrocyte invasion (Nair et al., 2008). The Pf CGH array was designed at the Wellcome Trust Sanger Institute and consists of ∼2 million 25 bp probes (many overlapping, but all mapping uniquely). It is being applied in an ongoing SV discovery study involving five Pf laboratory strains: 3D7 (the reference, African), DD2 (Indonesian), HB3 (Honduran), IT (South American or South East Asian) and PFCLIN (Ghanaian). We demonstrate the usefulness of SnoopCGH using screenshots of chromosome 5 data. Figure 1A shows the log2 intensities in a ∼600 kb region normalized using an average of all five strains. A separate window layer highlights a region of the IT genome that could contain increased copy number variation. This region includes coding sequence (CDS). Gene information (e.g. name, ontology, GC content) uploaded into SnoopCGH (Fig. 1B) indicates that this region contains the multi-drug resistance CNV (PfMDR1); (Price et al., 2004). Applying the Smith–Waterman algorithm to the IT data, highlighted (only) the region containing PfMDR1 as being both highly statistically significant (P <0.0001) and robust to a sensitivity analysis (Fig. 1A). It is possible to change the analysis settings and methods, and move the resulting window layers across the genome. Changing the resolution of the frame also facilitates rapid dissection of data quality and analysis results. For example, Figure 1C presents the Haar wavelet smooth for the intensities from a subset of 23 probes within the PfMDR1, part of a (4 kb) region with every smoothed value in excess of zero, indicative of a putative CNV.
Fig. 1.
(A) The log2 intensities of the five strains for ∼600 kb of chromosome 5, with a separate window highlighting the IT data (blue), and a subregion (pink) that is both statistically significant (P <0.0001) and > 90% robust in a sensitivity analysis. Annotation tracks are presented at the bottom, and (B) shows the annotation navigator window for the PfMDR1 gene located in the subregion; (C) shows the Haar wavelet smoothed values for part of the PfMDR1 gene.
4 DISCUSSION
SnoopCGH enables the visual assessment of genomes for SVs, with an estimation of their locations and statistical significance, as well as the ability to cross-check with external information (e.g. sequence annotation). Although we have implemented several fast breakpoint analytical methods, more sophisticated and computationally expensive approaches are being developed. These may be incorporated into a SnoopCGH analysis by either reading in the results from file or incorporating the method itself into our flexible software architecture. Ongoing work involves implementing new breakpoint detection methods, and incorporating tools to highlight the concordance of results from alternative SV detection methods. SVs may be detected using data from new sequencing technologies by considering differences in nucleotide coverage between target and reference genomes. It is possible to use SnoopCGH on such data, where (log transformed) ratios of normalized coverage (within or between samples) substitute for log ratios of normalized fluorescence intensities. However, sufficient consideration should be given to issues in the preprocessing steps, such as the uniqueness of read mappings, sequencing and assembly errors, and normalization accounting for GC content. In conclusion, SnoopCGH is a powerful visualization and analysis tool for those analyzing CGH data and discovering SVs genomewide, and has potential utility for those using new sequencing technologies for the same purpose.
Funding: Bill and Melinda Gates Foundation; Wellcome Trust; Medical Research Council UK.
Conflict of Interest: none declared.
REFERENCES
- Ben-Yaacov E, Eldar YC. A fast and flexible method for the segmentation of ACGH data. Bioinformatics. 2008;24:1139–1145. doi: 10.1093/bioinformatics/btn272. [DOI] [PubMed] [Google Scholar]
- Laframboise T, et al. A flexible rank-based framework for detecting copy number aberrations from array data. Bioinformatics. 2009;25:722–728. doi: 10.1093/bioinformatics/btp063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, et al. Adaptive copy number evolution in malaria parasites. PLoS Genet. 2008;4:e1000243. doi: 10.1371/journal.pgen.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan MC, et al. Phenotypic variations on the theme of CNVs. Nat. Genet. 2008;40:1392–1393. doi: 10.1038/ng1208-1392. [DOI] [PubMed] [Google Scholar]
- Price RN, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price TS, et al. SW-ARRAY: a dynamic programming solution for the identification of copy-number changes in genomic DNA using array comparative genome hybridization data. Nucleic Acids Res. 2005;33:3455–3464. doi: 10.1093/nar/gki643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LY, et al. MSB: a mean-shift-based approach for the analysis of structural variation in the genome. Genome Res. 2009;19:106–117. doi: 10.1101/gr.080069.108. [DOI] [PMC free article] [PubMed] [Google Scholar]