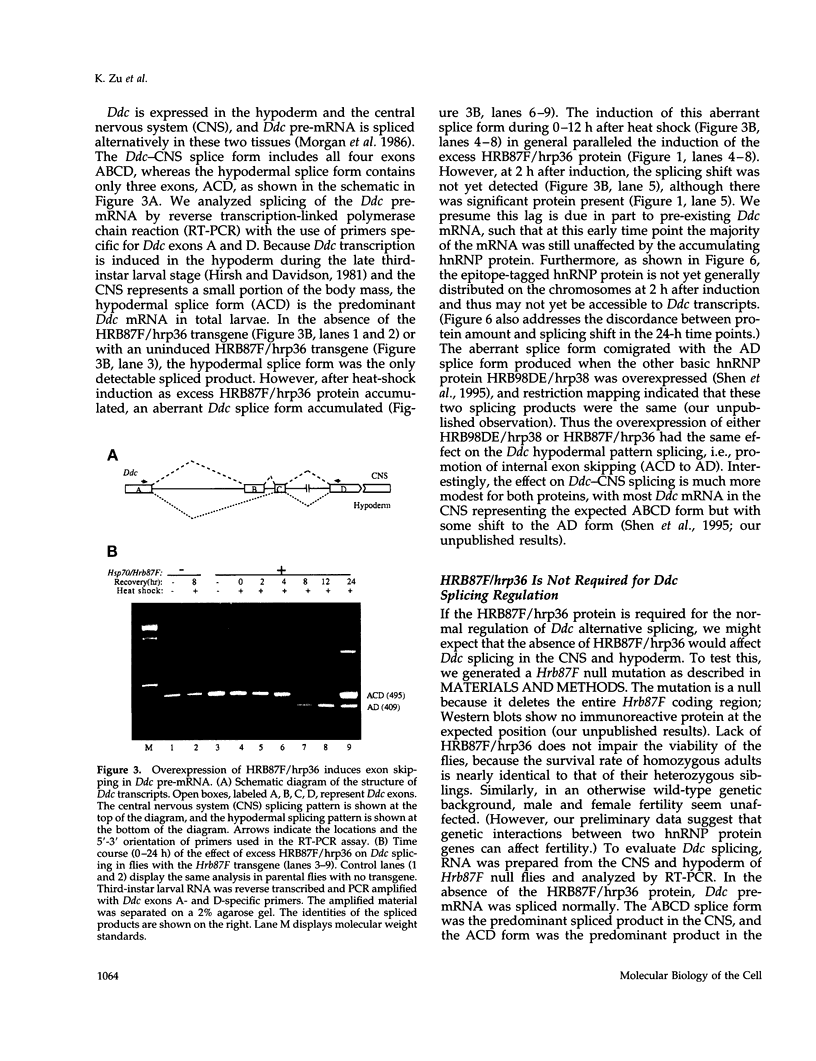
Abstract
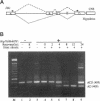
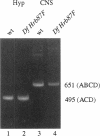
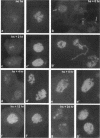
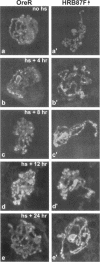
The Drosophila melanogaster genes Hrb87F and Hrb98DE encode the fly proteins HRB87F and HRB98DE (also known as hrp36 and hrp38, respectively) that are most similar in sequence and function to mammalian A/B-type hnRNP proteins. Using overexpression and deletion mutants of Hrb87F, we have tested the hypothesis that the ratio of A/B hnRNP proteins to SR family proteins modulates certain types of alternative splice-site selection. In flies in which HRB87F/hrp36 had been overexpressed 10- to 15-fold above normal levels, aberrant internal exon skipping was induced in at least one endogenous transcript, the dopa decarboxylase (Ddc) pre-mRNA, which previously had been shown to be similarly affected by excess HRB98DE/hrp38. In a second endogenous pre-mRNA, excess HRB87F/hrp36 had no effect on alternative 3' splice-site selection, as expected from mammalian hnRNP studies. Immunolocalization of the excess hnRNP protein showed that it localized correctly to the nucleus, specifically to sites on or near chromosomes, and that the peak of exon-skipping activity in Ddc RNA correlated with the peak of chromosomally associated hnRNP protein. The chromosomal association and level of the SR family of proteins were not significantly affected by the large increase in hnRNP proteins during this time period. Although these results are consistent with a possible role for hnRNP proteins in alternative splicing, the more interesting finding was the failure to detect significant adverse effects on flies with a greatly distorted ratio of hnRNPs to SR proteins. Electron microscopic visualization of the general population of active genes in flies overexpressing hnRNP proteins also indicated that the great majority of genes seemed normal in terms of cotranscriptional RNA processing events, although there were a few abnormalities consistent with rare exon-skipping events. Furthermore, in a Hrb87F null mutant, which is viable, the normal pattern of Ddc alternative splicing was observed, indicating that HRB87F/hrp36 is not required for Ddc splicing regulation. Thus, although splice-site selection can be affected in at least a few genes by gross overexpression of this hnRNP protein, the combined evidence suggests that if it plays a general role in alternative splicing in vivo, the role can be provided by other proteins with redundant functions, and the role is independent of its concentration relative to SR proteins.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amero S. A., Raychaudhuri G., Cass C. L., van Venrooij W. J., Habets W. J., Krainer A. R., Beyer A. L. Independent deposition of heterogeneous nuclear ribonucleoproteins and small nuclear ribonucleoprotein particles at sites of transcription. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8409–8413. doi: 10.1073/pnas.89.18.8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David Y., Bani M. R., Chabot B., De Koven A., Bernstein A. Retroviral insertions downstream of the heterogeneous nuclear ribonucleoprotein A1 gene in erythroleukemia cells: evidence that A1 is not essential for cell growth. Mol Cell Biol. 1992 Oct;12(10):4449–4455. doi: 10.1128/mcb.12.10.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M., Michaud S., Kingston J., Reed R. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 1992 Oct;6(10):1986–2000. doi: 10.1101/gad.6.10.1986. [DOI] [PubMed] [Google Scholar]
- Beyer A. L., Christensen M. E., Walker B. W., LeStourgeon W. M. Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell. 1977 May;11(1):127–138. doi: 10.1016/0092-8674(77)90323-3. [DOI] [PubMed] [Google Scholar]
- Beyer A. L., Osheim Y. N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988 Jun;2(6):754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Beyer A., Sikes M., Osheim Y. EM methods for visualization of genetic activity from disrupted nuclei. Methods Cell Biol. 1994;44:613–630. doi: 10.1016/s0091-679x(08)60935-8. [DOI] [PubMed] [Google Scholar]
- Birney E., Kumar S., Krainer A. R. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993 Dec 25;21(25):5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvoli M., Cobianchi F., Biamonti G., Riva S. Recombinant hnRNP protein A1 and its N-terminal domain show preferential affinity for oligodeoxynucleotides homologous to intron/exon acceptor sites. Nucleic Acids Res. 1990 Nov 25;18(22):6595–6600. doi: 10.1093/nar/18.22.6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celis J. E., Bravo R., Arenstorf H. P., LeStourgeon W. M. Identification of proliferation-sensitive human proteins amongst components of the 40 S hnRNP particles. Identity of hnRNP core proteins in the HeLa protein catalogue. FEBS Lett. 1986 Jan 1;194(1):101–109. doi: 10.1016/0014-5793(86)80059-x. [DOI] [PubMed] [Google Scholar]
- Champlin D. T., Lis J. T. Distribution of B52 within a chromosomal locus depends on the level of transcription. Mol Biol Cell. 1994 Jan;5(1):71–79. doi: 10.1091/mbc.5.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway G., Wooley J., Bibring T., LeStourgeon W. M. Ribonucleoproteins package 700 nucleotides of pre-mRNA into a repeating array of regular particles. Mol Cell Biol. 1988 Jul;8(7):2884–2895. doi: 10.1128/mcb.8.7.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres J. F., Stamm S., Helfman D. M., Krainer A. R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994 Sep 16;265(5179):1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis M. J., Piñol-Roma S., Burd C. G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Swanson M. S., Piñol-Roma S. Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem Sci. 1988 Mar;13(3):86–91. doi: 10.1016/0968-0004(88)90046-1. [DOI] [PubMed] [Google Scholar]
- Fu X. D., Mayeda A., Maniatis T., Krainer A. R. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5' and 3' splice site selection. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995 Sep;1(7):663–680. [PMC free article] [PubMed] [Google Scholar]
- Gattoni R., Stevenin J., Jacob M. Comparison of the nuclear ribonucleoproteins containing the transcripts of adenovirus-2 and HeLa cell dna. Eur J Biochem. 1980;108(1):203–211. doi: 10.1111/j.1432-1033.1980.tb04713.x. [DOI] [PubMed] [Google Scholar]
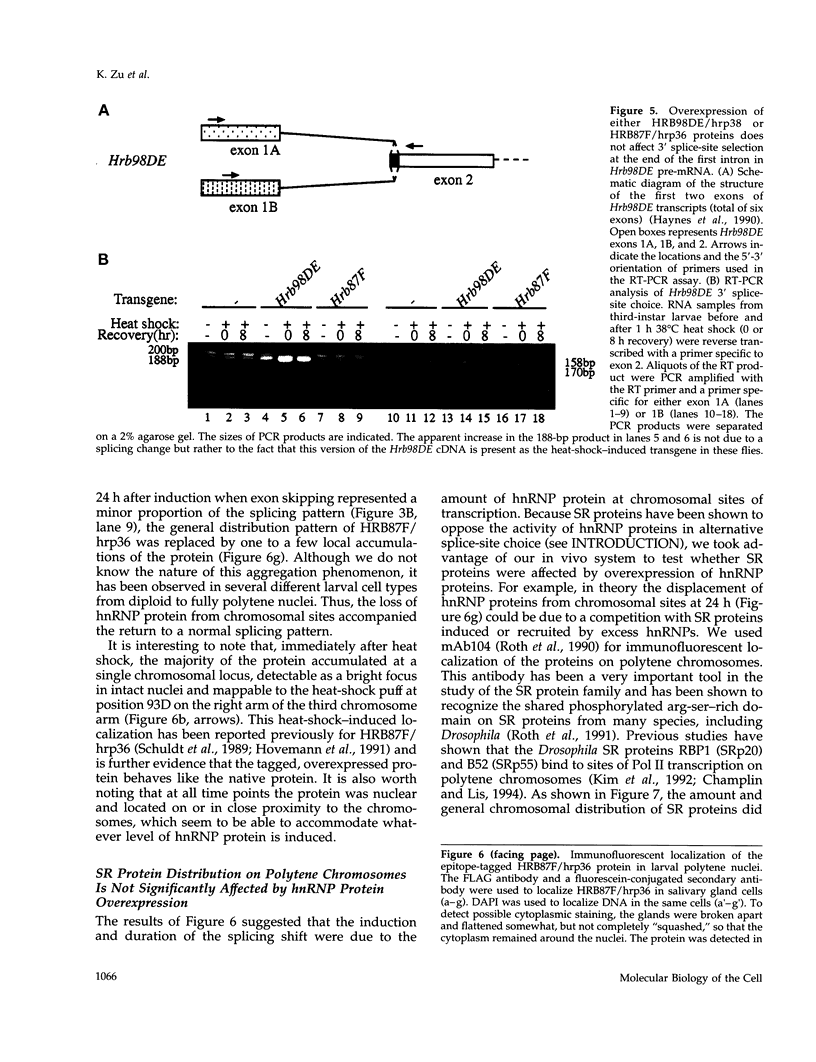
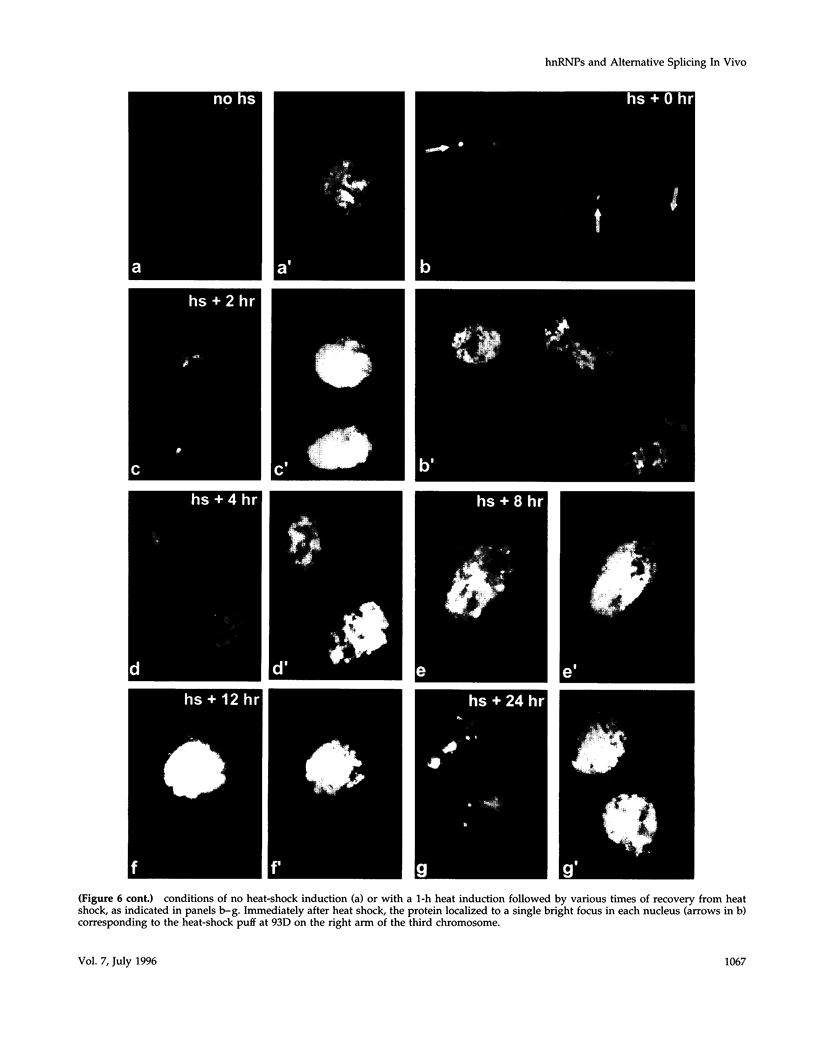
- Haynes S. R., Johnson D., Raychaudhuri G., Beyer A. L. The Drosophila Hrb87F gene encodes a new member of the A and B hnRNP protein group. Nucleic Acids Res. 1991 Jan 11;19(1):25–31. doi: 10.1093/nar/19.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes S. R., Raychaudhuri G., Beyer A. L. The Drosophila Hrb98DE locus encodes four protein isoforms homologous to the A1 protein of mammalian heterogeneous nuclear ribonucleoprotein complexes. Mol Cell Biol. 1990 Jan;10(1):316–323. doi: 10.1128/mcb.10.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
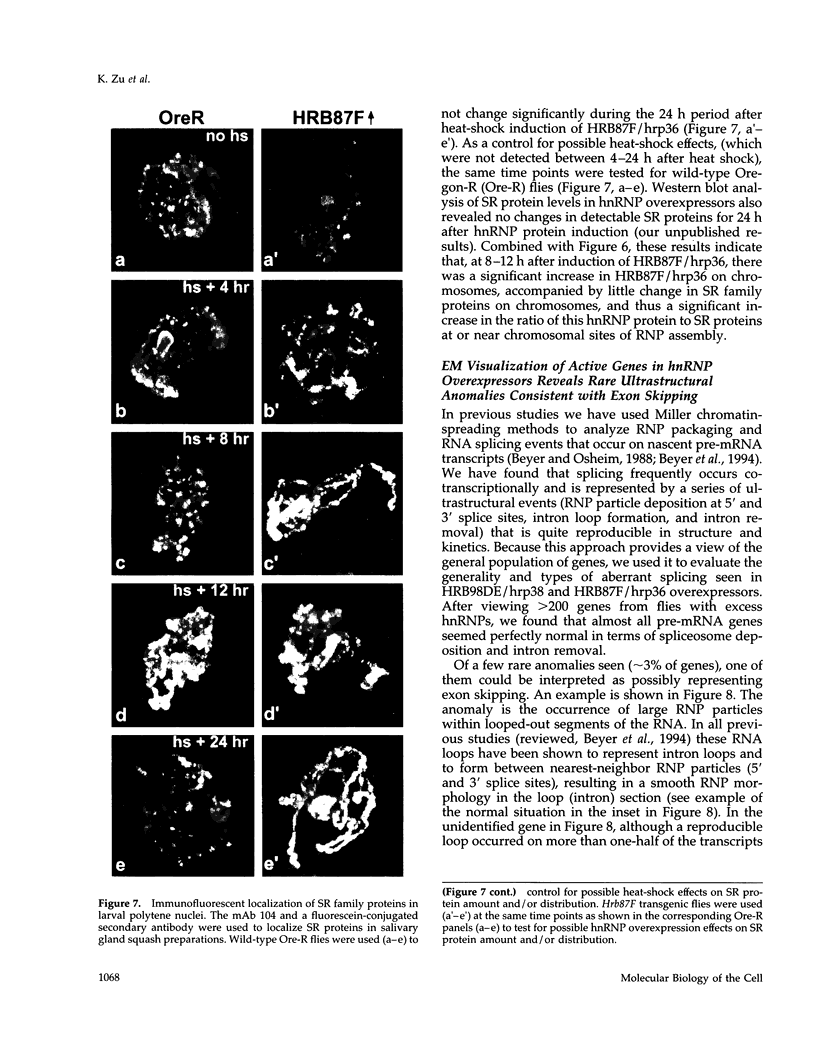
- Haynes S. R., Rebbert M. L., Mozer B. A., Forquignon F., Dawid I. B. pen repeat sequences are GGN clusters and encode a glycine-rich domain in a Drosophila cDNA homologous to the rat helix destabilizing protein. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1819–1823. doi: 10.1073/pnas.84.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelspach M., Cavaloc Y., Chebli K., Stévenin J., Gattoni R. Titration of serine/arginine (SR) splicing factors during adenoviral infection modulates E1A pre-mRNA alternative splicing. RNA. 1995 Oct;1(8):794–806. [PMC free article] [PubMed] [Google Scholar]
- Hirsh J., Davidson N. Isolation and characterization of the dopa decarboxylase gene of Drosophila melanogaster. Mol Cell Biol. 1981 Jun;1(6):475–485. doi: 10.1128/mcb.1.6.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz D. S., Krainer A. R. Mechanisms for selecting 5' splice sites in mammalian pre-mRNA splicing. Trends Genet. 1994 Mar;10(3):100–106. doi: 10.1016/0168-9525(94)90233-x. [DOI] [PubMed] [Google Scholar]
- Hovemann B. T., Dessen E., Mechler H., Mack E. Drosophila snRNP associated protein P11 which specifically binds to heat shock puff 93D reveals strong homology with hnRNP core protein A1. Nucleic Acids Res. 1991 Sep 25;19(18):4909–4914. doi: 10.1093/nar/19.18.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R. L. Initial organization of the Drosophila dorsoventral axis depends on an RNA-binding protein encoded by the squid gene. Genes Dev. 1993 Jun;7(6):948–960. doi: 10.1101/gad.7.6.948. [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Zuo P., Manley J. L., Baker B. S. The Drosophila RNA-binding protein RBP1 is localized to transcriptionally active sites of chromosomes and shows a functional similarity to human splicing factor ASF/SF2. Genes Dev. 1992 Dec;6(12B):2569–2579. doi: 10.1101/gad.6.12b.2569. [DOI] [PubMed] [Google Scholar]
- Klemenz R., Weber U., Gehring W. J. The white gene as a marker in a new P-element vector for gene transfer in Drosophila. Nucleic Acids Res. 1987 May 26;15(10):3947–3959. doi: 10.1093/nar/15.10.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer A. R., Conway G. C., Kozak D. The essential pre-mRNA splicing factor SF2 influences 5' splice site selection by activating proximal sites. Cell. 1990 Jul 13;62(1):35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- Kraus M. E., Lis J. T. The concentration of B52, an essential splicing factor and regulator of splice site choice in vitro, is critical for Drosophila development. Mol Cell Biol. 1994 Aug;14(8):5360–5370. doi: 10.1128/mcb.14.8.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Casas-Finet J. R., Luneau C. J., Karpel R. L., Merrill B. M., Williams K. R., Wilson S. H. Mammalian heterogeneous nuclear ribonucleoprotein A1. Nucleic acid binding properties of the COOH-terminal domain. J Biol Chem. 1990 Oct 5;265(28):17094–17100. [PubMed] [Google Scholar]
- Kumar A., Wilson S. H. Studies of the strand-annealing activity of mammalian hnRNP complex protein A1. Biochemistry. 1990 Dec 4;29(48):10717–10722. doi: 10.1021/bi00500a001. [DOI] [PubMed] [Google Scholar]
- Kuo H. C., Nasim F. H., Grabowski P. J. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science. 1991 Mar 1;251(4997):1045–1050. doi: 10.1126/science.1825520. [DOI] [PubMed] [Google Scholar]
- LeStourgeon W. M., Beyer A. L., Christensen M. E., Walker B. W., Poupore S. M., Daniels L. P. The packaging proteins of core hnRNP particles and the maintenance of proliferative cell states. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):885–898. doi: 10.1101/sqb.1978.042.01.090. [DOI] [PubMed] [Google Scholar]
- Lin C. H., Patton J. G. Regulation of alternative 3' splice site selection by constitutive splicing factors. RNA. 1995 May;1(3):234–245. [PMC free article] [PubMed] [Google Scholar]
- Matunis E. L., Kelley R., Dreyfuss G. Essential role for a heterogeneous nuclear ribonucleoprotein (hnRNP) in oogenesis: hrp40 is absent from the germ line in the dorsoventral mutant squid. Proc Natl Acad Sci U S A. 1994 Mar 29;91(7):2781–2784. doi: 10.1073/pnas.91.7.2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis E. L., Matunis M. J., Dreyfuss G. Association of individual hnRNP proteins and snRNPs with nascent transcripts. J Cell Biol. 1993 Apr;121(2):219–228. doi: 10.1083/jcb.121.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis E. L., Matunis M. J., Dreyfuss G. Characterization of the major hnRNP proteins from Drosophila melanogaster. J Cell Biol. 1992 Jan;116(2):257–269. doi: 10.1083/jcb.116.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis M. J., Matunis E. L., Dreyfuss G. Isolation of hnRNP complexes from Drosophila melanogaster. J Cell Biol. 1992 Jan;116(2):245–255. doi: 10.1083/jcb.116.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Helfman D. M., Krainer A. R. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol Cell Biol. 1993 May;13(5):2993–3001. doi: 10.1128/mcb.13.5.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Krainer A. R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992 Jan 24;68(2):365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Munroe S. H., Cáceres J. F., Krainer A. R. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994 Nov 15;13(22):5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Zahler A. M., Krainer A. R., Roth M. B. Two members of a conserved family of nuclear phosphoproteins are involved in pre-mRNA splicing. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1301–1304. doi: 10.1073/pnas.89.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael W. M., Choi M., Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995 Nov 3;83(3):415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Min H., Chan R. C., Black D. L. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 1995 Nov 1;9(21):2659–2671. doi: 10.1101/gad.9.21.2659. [DOI] [PubMed] [Google Scholar]
- Morgan B. A., Johnson W. A., Hirsh J. Regulated splicing produces different forms of dopa decarboxylase in the central nervous system and hypoderm of Drosophila melanogaster. EMBO J. 1986 Dec 1;5(12):3335–3342. doi: 10.1002/j.1460-2075.1986.tb04648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan G. J., Guo W., Wormsley S., Helfman D. M. Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of beta-tropomyosin pre-mRNA. J Biol Chem. 1992 Dec 15;267(35):25480–25487. [PubMed] [Google Scholar]
- Munroe S. H., Dong X. F. Heterogeneous nuclear ribonucleoprotein A1 catalyzes RNA.RNA annealing. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):895–899. doi: 10.1073/pnas.89.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Mount S. M. Genetic enhancement of RNA-processing defects by a dominant mutation in B52, the Drosophila gene for an SR protein splicing factor. Mol Cell Biol. 1995 Nov;15(11):6273–6282. doi: 10.1128/mcb.15.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planck S. R., Listerud M. D., Buckley S. D. Modulation of hnRNP A1 protein gene expression by epidermal growth factor in Rat-1 cells. Nucleic Acids Res. 1988 Dec 23;16(24):11663–11673. doi: 10.1093/nar/16.24.11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontius B. W., Berg P. Renaturation of complementary DNA strands mediated by purified mammalian heterogeneous nuclear ribonucleoprotein A1 protein: implications for a mechanism for rapid molecular assembly. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8403–8407. doi: 10.1073/pnas.87.21.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman D. S., Dreyfuss G. RNA annealing activities in HeLa nuclei. EMBO J. 1994 Jan 1;13(1):213–221. doi: 10.1002/j.1460-2075.1994.tb06251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri G., Haynes S. R., Beyer A. L. Heterogeneous nuclear ribonucleoprotein complexes and proteins in Drosophila melanogaster. Mol Cell Biol. 1992 Feb;12(2):847–855. doi: 10.1128/mcb.12.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring H. Z., Lis J. T. The SR protein B52/SRp55 is essential for Drosophila development. Mol Cell Biol. 1994 Nov;14(11):7499–7506. doi: 10.1128/mcb.14.11.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. M., Preston C. R., Phillis R. W., Johnson-Schlitz D. M., Benz W. K., Engels W. R. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988 Mar;118(3):461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M. B., Murphy C., Gall J. G. A monoclonal antibody that recognizes a phosphorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J Cell Biol. 1990 Dec;111(6 Pt 1):2217–2223. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M. B., Zahler A. M., Stolk J. A. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991 Nov;115(3):587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldt C., Kloetzel P. M., Bautz E. K. Molecular organization of RNP complexes containing P11 antigen in heat-shocked and non-heat-shocked Drosophila cells. Eur J Biochem. 1989 Apr 15;181(1):135–142. doi: 10.1111/j.1432-1033.1989.tb14704.x. [DOI] [PubMed] [Google Scholar]
- Shen J., Beall C. J., Hirsh J. Tissue-specific alternative splicing of the Drosophila dopa decarboxylase gene is affected by heat shock. Mol Cell Biol. 1993 Aug;13(8):4549–4555. doi: 10.1128/mcb.13.8.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Hirsh J. cis-regulatory sequences responsible for alternative splicing of the Drosophila dopa decarboxylase gene. Mol Cell Biol. 1994 Nov;14(11):7385–7393. doi: 10.1128/mcb.14.11.7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Zu K., Cass C. L., Beyer A. L., Hirsh J. Exon skipping by overexpression of a Drosophila heterogeneous nuclear ribonucleoprotein in vivo. Proc Natl Acad Sci U S A. 1995 Mar 14;92(6):1822–1825. doi: 10.1073/pnas.92.6.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel C. W., Fresco L. D., Rio D. C. The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: multiprotein complexes at an exon pseudo-5' splice site control U1 snRNP binding. Genes Dev. 1992 Aug;6(8):1386–1401. doi: 10.1101/gad.6.8.1386. [DOI] [PubMed] [Google Scholar]
- Siebel C. W., Kanaar R., Rio D. C. Regulation of tissue-specific P-element pre-mRNA splicing requires the RNA-binding protein PSI. Genes Dev. 1994 Jul 15;8(14):1713–1725. doi: 10.1101/gad.8.14.1713. [DOI] [PubMed] [Google Scholar]
- Singh R., Valcárcel J., Green M. R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995 May 26;268(5214):1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- Siomi H., Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995 May;129(3):551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982 Oct 22;218(4570):341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Staknis D., Reed R. Direct interactions between pre-mRNA and six U2 small nuclear ribonucleoproteins during spliceosome assembly. Mol Cell Biol. 1994 May;14(5):2994–3005. doi: 10.1128/mcb.14.5.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staknis D., Reed R. SR proteins promote the first specific recognition of Pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994 Nov;14(11):7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Mayeda A., Hampson R. K., Krainer A. R., Rottman F. M. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993 Dec;7(12B):2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- Swanson M. S., Dreyfuss G. RNA binding specificity of hnRNP proteins: a subset bind to the 3' end of introns. EMBO J. 1988 Nov;7(11):3519–3529. doi: 10.1002/j.1460-2075.1988.tb03228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talerico M., Berget S. M. Effect of 5' splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990 Dec;10(12):6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Manley J. L. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995 May;1(3):335–346. [PMC free article] [PubMed] [Google Scholar]
- Weighardt F., Biamonti G., Riva S. Nucleo-cytoplasmic distribution of human hnRNP proteins: a search for the targeting domains in hnRNP A1. J Cell Sci. 1995 Feb;108(Pt 2):545–555. doi: 10.1242/jcs.108.2.545. [DOI] [PubMed] [Google Scholar]
- Wu Z. A., Murphy C., Callan H. G., Gall J. G. Small nuclear ribonucleoproteins and heterogeneous nuclear ribonucleoproteins in the amphibian germinal vesicle: loops, spheres, and snurposomes. J Cell Biol. 1991 May;113(3):465–483. doi: 10.1083/jcb.113.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Bani M. R., Lu S. J., Rowan S., Ben-David Y., Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5' splice site selection in vivo. Proc Natl Acad Sci U S A. 1994 Jul 19;91(15):6924–6928. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost H. J., Lindquist S. RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell. 1986 Apr 25;45(2):185–193. doi: 10.1016/0092-8674(86)90382-x. [DOI] [PubMed] [Google Scholar]
- Zahler A. M., Lane W. S., Stolk J. A., Roth M. B. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992 May;6(5):837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zink B., Paro R. In vivo binding pattern of a trans-regulator of homoeotic genes in Drosophila melanogaster. Nature. 1989 Feb 2;337(6206):468–471. doi: 10.1038/337468a0. [DOI] [PubMed] [Google Scholar]