Abstract
Developmental mechanisms regulating gene expression and the stable acquisition of cell fate direct cytodifferentiation during organogenesis. Moreover, it is likely that such mechanisms could be exploited to repair or regenerate damaged organs. DNA methyltransferases (Dnmts) are enzymes critical for epigenetic regulation, and are used in concert with histone methylation and acetylation to regulate gene expression and maintain genomic integrity and chromosome structure. We carried out two forward genetic screens for regulators of endodermal organ development. In the first, we screened for altered morphology of developing digestive organs, while in the second we screed for the lack of terminally differentiated cell types in the pancreas and liver. From these screens, we identified two mutant alleles of zebrafish dnmt1. Both lesions are predicted to eliminate dnmt1 function; one is a missense mutation in the catalytic domain and the other is a nonsense mutation that eliminates the catalytic domain. In zebrafish dnmt1 mutants, the pancreas and liver form normally, but begin to degenerate after 84 hours post fertilization (hpf). Acinar cells are nearly abolished through apoptosis by 100 hpf, though neither DNA replication, nor entry into mitosis are halted in the absence of detectable Dnmt1. However, endocrine cells and ducts are largely spared. Surprisingly, dnmt1 mutants and dnmt1 morpholino-injected larvae show increased capacity for pancreatic beta cell regeneration in an inducible model of pancreatic beta cell ablation. Thus, our data suggest that Dnmt1 is dispensable for pancreatic duct or endocrine cell formation, but not for acinar cell survival. In addition, Dnmt1 may influence the differentiation of pancreatic beta cell progenitors or the reprogramming of cells toward the pancreatic beta cell fate.
Keywords: methylation, Dnmt1, pancreas, epigenetics, regeneration, zebrafish, ENU mutagenesis, mutant, beta cells, liver
INTRODUCTION
Organogenesis depends on the precise execution of discrete gene expression cascades and the acquisition of stable cell identities from multi-potent progenitors. DNA methylation on cytosine is a heritable epigenetic modification that is pivotal in regulating the transcriptional accessibility of the genome (reviewed by (Cedar, 1988)). Cytosine methylation is often inversely correlated with gene expression (reviewed by (Robertson and Jones, 2000)), and is required for the transcriptional silencing of imprinted genes and transposons, as well as the somatic inactivation of sex chromosomes (reviewed by (Goll and Bestor, 2005)). This regulation is effectuated in part by methylcytosine binding proteins that recruit chromatin modifying machinery and restrict transcription via heterochromatin formation (reviewed by (Bird and Wolffe, 1999)). Methylation extent is correlated with differential gene expression among tissue types (Futscher et al., 2002; Song et al., 2005), and there is increasing evidence that methylation influences gene expression during embryogenesis (Meissner et al., 2008; Nishino et al., 2004; Rai et al., 2006; Sanosaka et al., 2008; Shemer et al., 1991). These and other studies indicate that as embryogenesis proceeds, cells accumulate epigenetic marks that progressively alter developmental potential by regulating chromatin configuration.
In mammals, cytosine methylation patterns are generated and maintained by a family of DNA methyltransferases (Dnmts). Dnmt1 contains both a carboxyterminal domain that catalyzes methylation at CpG dinucleotides, as well as an N-terminal regulatory domain, which has independent transcriptional repressor activity (Dunican et al., 2008). Dnmt1 is regarded as a maintenance methyltransferase since it is targeted to the replication fork during S-phase by Uhrf1 (Bostick et al., 2007), where it preferentially catalyzes methyl group addition to the nascent, hemi-methylated DNA strand. Although its preference for hemi-methylated DNA is unique among Dnmts, Dnmt1 also has significant capacity for de novo methylation (Okano et al., 1998).
Several genetic studies have shown that Dnmt1 is dispensable in embryonic stem (ES) cells, but is required for the proliferation and survival of differentiated cell types (Jackson-Grusby et al., 2001; Lei et al., 1996; Li et al., 1992). Accordingly, Dnmt1 is required for embryonic development. Mice lacking Dnmt1 die during early organogenesis stages, and lack ∼90% of cytosine methylation (Lei et al., 1996; Li et al., 1992; Takebayashi et al., 2007). Reduced Dnmt1 activity in Xenopus (Stancheva et al., 2001) and zebrafish (Martin et al., 1999; Rai et al., 2006) has similar consequences. Multiple factors contribute to this lethality, including aberrant gene expression, genomic instability, and activation of cell cycle checkpoints and apoptosis (Chen et al., 2007; Jackson-Grusby et al., 2001; Stancheva et al., 2001; Unterberger et al., 2006). However, the molecular pathways connecting hypomethylation to cell death, as well as the relative importance of the catalytic and regulatory activities of Dnmt1, remain unclear.
Here, we analyze zebrafish dnmt1 mutants that were identified in genetic screens for regulators of endodermal organ formation and differentiation. Previous investigations of the developmental role of dnmt1 in zebrafish using a morpholino knockdown approach showed decreased levels of differentiated cell types in the exocrine pancreas and gut, while the endocrine cells of the primary islet remained intact (Rai et al., 2006). In contrast, dnmt1 mutants develop normally until 84 hpf when highly proliferative endodermal organs undergo a massive wave of apoptosis. These dnmt1 mutants retain the Dnmt1 regulatory domain, but are likely to be catalytically null, allowing specific interrogation of the role played by Dnmt1-mediated methylation during the growth, differentiation, and regeneration of the vertebrate pancreas.
RESULTS
Endodermal organ degeneration in dandelion (ddn) mutants
The suitability of zebrafish for embryological studies and mutagenesis screens has fostered its emergence as a powerful genetic system for studying vertebrate organ formation at high resolution. Importantly, major genetic cascades regulating pancreas formation appear to be conserved between zebrafish and mammalian species (Biemar et al., 2001; Delporte et al., 2008; Huang et al., 2001; Sun and Hopkins, 2001; Yee et al., 2001; Zecchin et al., 2004), as are major morphogenetic events (Field et al., 2003a). Briefly, in zebrafish, pdx1-expressing pancreatic progenitors evaginate from the primitive endodermal rod by 24 hours post fertilization (hpf), forming the dorsal pancreatic bud (Field et al., 2003b). Subsequently, a ventral bud emerges from the ventral intestine and encapsulates the dorsal bud by 52 hpf, establishing the mature pancreatic architecture (Field et al., 2003a). The dorsal bud is thought to give rise exclusively to the main cluster of endocrine cells, the primary islet (Field et al., 2003a). In contrast, the ventral bud generates the exocrine pancreas (acinar and duct cells), although it also contributes endocrine cells via progenitors in the pancreatic ducts (Dong et al., 2007; Field et al., 2003a).
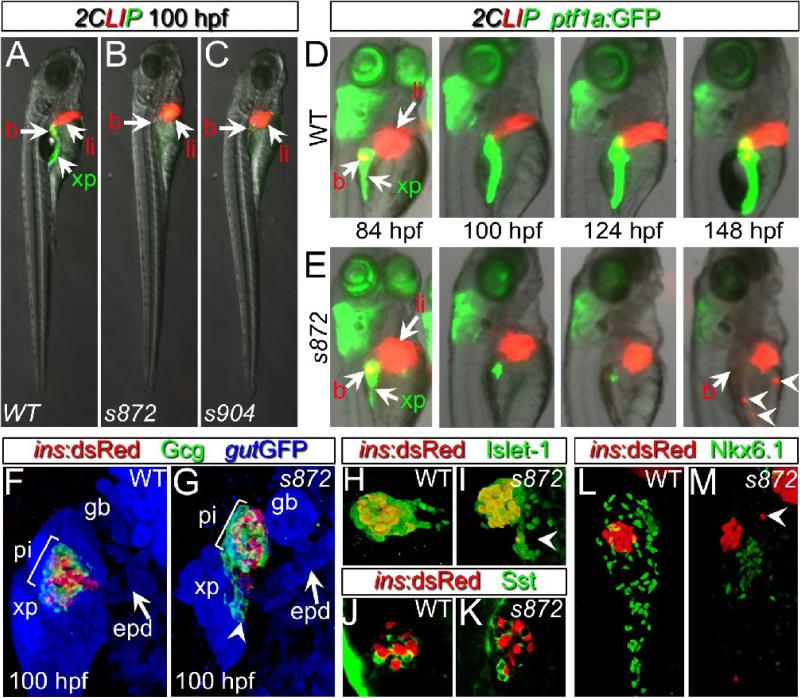
To identify novel regulators of pancreas morphogenesis and cytodifferentiation, we completed two ethylnitrosourea genetic screens. The first (Ober et al., 2006) was conducted using the Tg(XlEef1a1:GFP)s854 (aka gut:GFP) line, which expresses GFP throughout the endoderm, illuminating the morphogenesis of digestive organs (Field et al., 2003b). The second was conducted using the Tg(ins:dsRed)m1081;Tg(fabp10:dsRed;ela3l:GFP)gz12 (Farooq et al., 2008) line which we refer to as 2CLIP (2-Color Liver Insulin acinar Pancreas) for simplicity. This transgenic combination of red fluorescence in pancreatic beta cells and hepatocytes, and green fluorescence in pancreatic acinar cells permitted assessment of pancreas and liver cytodifferentiation. Two phenotypically indistinguishable mutants were recovered from the screens, s872 and s904, and were determined to be allelic by complementation test (27% mutant, n=272). The mutant phenotype was fully penetrant, and exhibited little variation. At 100 hpf, the mutants display a markedly smaller exocrine pancreas, a smaller liver, smaller eyes, and dysmorphic branchial arches (Fig. 1A-C and data not shown). Mutant larvae generally die at 8 dpf. We examined individual wild-type (WT) and mutant animals from fertilization to 148 hpf, and found that they were indistinguishable until 84 hpf, at which point the amount of Tg(ptf1a:GFP)jh1-expressing acinar tissue (Godinho et al., 2007) progressively diminished (Fig 1D,E). Early steps of liver differentiation and morphogenesis also appeared to be unaffected, as indicated by expression at 100 hpf of Tg(fabp10:dsRed)gz12 (n>100) and immunostaining for Prox1 (pan-hepatic marker) and Alcam (intrahepatic biliary network marker; (Sakaguchi et al., 2008); n=3) although the overall liver mass was reduced (Fig 2A,C,E,F). In addition, in all ddn mutants, Tg(fabp10:dsRed)gz12-expressing cell fragments and/or aggregates, could be observed throughout the vasculature, most often accumulating in the caudal vein network (Fig 2 B,D,G-J). In these fragments, dsRed fluorescence did not co-localize with DNA/nuclei (data not shown). In addition, fabp10 and ceruloplasmin mRNAs, normally present in differentiated hepatocytes, were not detected in these clusters (data not shown). These results suggest that the ectopic fluorescence results from persistence of dsRed protein in fragments of dead hepatocytes (perhaps engulfed by macrophages), rather than from live, ectopic hepatocytes. To reflect this phenotype — the rapid growth and subsequent dispersal/death of endodermal tissues—we named this mutant dandelion (ddn).
Figure 1. Initial formation and degeneration of the exocrine pancreas in dandelion (ddn) mutants.
(A-C) Lateral views of WT (A), ddns872 (B), and ddns904 mutant (C) larvae at 100 hpf in the 2-color Liver, Insulin, acinar Pancreas (2CLIP) transgenic background (see methods). Acinar cells of the exocrine pancreas (xp) are green and hepatocytes (li) and pancreatic beta cells (b) are red. ddn mutants exhibit minimal acinar tissue, but overall larva morphology is only moderately affected. (D,E) Successive images of individual WT (D) and ddns872 mutant (E) larvae between 84 and 148 hpf in the 2CLIP; Tg(ptf1a:EGFP)jh1 background. The exocrine pancreas and liver grow larger in WT, but degenerate in ddn mutants. Hepatocyte fragments are found in the circulation (arrowheads). (F-K) 3-D projections of confocal stacks showing composition of WT (F,H,J) and ddns872 mutant (G,I,K) endocrine pancreas at 100 hpf in Tg(ins:dsRed)m1081 background. (F,G) A core of Tg(ins:dsRed)m1081+ cells (red) is surrounded by a mantle of Glucagon+ (Gcg) cells (green). Extrapancreatic duct (epd) structure is intact. (H,I) Isl-1, and (J,K) Somatostatin (Sst) positive cells appear unaffected. Arrowheads in G, I, and M point to endocrine cells outside of the primary islet. (L,M) Nkx6.1 immunostaining reveals the presence of intrapancreatic duct cells in both WT and dnmt1s872 mutant larvae.
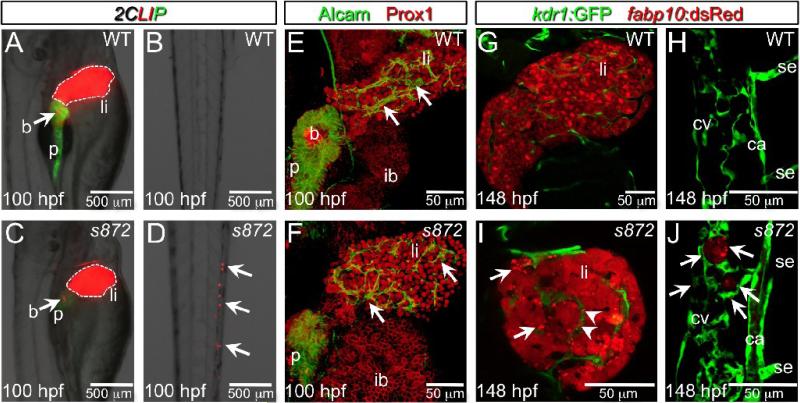
Figure 2. Liver degeneration in dandelion (ddn) mutants.
(A-D) Lateral aspect of trunk (A,C) and tail (B,D) regions of 100 hpf WT (A,B) and ddn mutant (C,D) larvae in the 2CLIP transgenic background. The liver is smaller in ddn mutants relative to WT, and cell fragments emitting dsRed fluorescence, derived from the liver, aggregate in clusters in the tail (D, arrows). (E,F) 3-D projections of confocal stacks showing Alcam and Prox1 antibody staining of 100 hpf WT (E) and ddn mutants (F). Initial specification, morphogenesis, and differentiation of the liver appear to be unaffected. Intrahepatic ductal network is indicated by arrows. (G-J) Confocal slices through the liver (G,I) and tail vasculature (H,J) of Tg(kdrl:GFP)s843; Tg(fabp10:dsRed)gz12 WT (G,H) and ddn mutant (I,J) larvae at 6 dpf. Although never present in WT, bright dsRed+ degenerating hepatocyte fragments are observed throughout the ddn mutant liver (I, arrows) and within the hepatic vasculature (I, arrowheads). These particles accumulate in the network of the caudal vein (cv; J, arrows). Other abbreviations: ca, caudal aorta; se, intersegmental vessel; ib, intestinal bulb; p, pancreas; li, liver; b, beta cells.
Formation of the endocrine pancreas and ductal system
After the loss of most pancreatic acinar cells in ddn mutants, an apparently WT cluster of beta cells consistently remained in the primary islet (Fig. 1B,C). Therefore, the composition and architecture of the islet was examined using immunostaining for the pan-endocrine transcription factor Islet-1 (n=3), and the delta and alpha cell hormones Somatostatin (n=2), and Glucagon (n=6), respectively. At 100 hpf, we found that ddn mutants retained the WT complement and arrangement of endocrine cells: a core of beta cells surrounded by a mantle of alpha and delta cells (Fig. 1 F-K). However, we often observed small clusters of endocrine cells outside of the islet (n=5; arrowheads, Fig. 1G, I, M), suggesting the generation of endocrine cells from the ventral pancreatic bud as in WT, but impaired migration/morphogenesis due to the deterioration of pancreatic acinar tissue. To further investigate which tissues degenerate in ddn mutants, we examined the formation of the pancreatic ducts. By Tg(gut:GFP)s854 expression, the extra-pancreatic duct and gall bladder appeared to be intact in ddn mutants at 100 hpf (Fig.1G, n=3). We also examined Nkx6.1 distribution, which normally marks intra-pancreatic ducts at 100 hpf (Fig 1K, n=3). Nkx6.1 expression was maintained in ddn mutants, but at lower levels than in WT, which indicated the persistence of disorganized duct cells. Altogether, these data show that the ddn mutation differentially affects the cell types that comprise the pancreas, with the acinar cells being the most sensitive.
dandelion mutants lack Dnmt1 catalytic activity
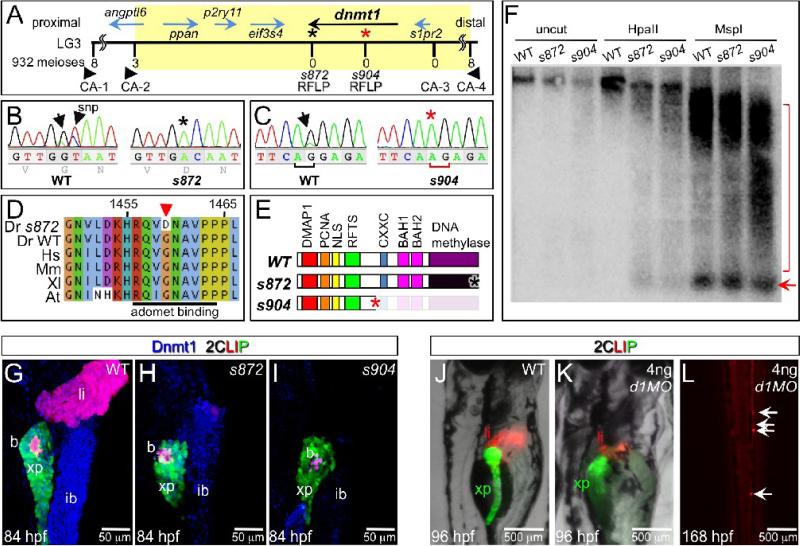
To elucidate the developmental mechanisms responsible for this phenotype, we isolated the gene affected by the ddn mutations. ddn was genetically mapped to a 1.2 cM interval on chromosome three, which contains six genes (Fig. 3A). Sequencing of these candidates revealed mutations only in dnmt1. In s872, a mis-sense mutation caused a G1459D substitution in the DNA methyltransferase motif X (Fig. 3B,D). This residue lies within the S-adenosyl l-methionine (AdoMet) binding domain, which is conserved in all eukaryotic and bacterial methyltransferases (reviewed by (Goll and Bestor, 2005)). Importantly, AdoMet is the methyl donor, and its binding to Dnmt1 is essential for cytosine methylation; thus, this mutation likely abolishes the catalytic activity of Dnmt1 without affecting other functional domains. In s904, an exon 15 splice acceptor mutation causes a 1-bp frameshift (Fig. 3C). This frameshift is predicted to truncate Dnmt1 at Arg476, after appending 31 missense residues; thus, in s904 mutants, Dnmt1 lacks the entire catalytic domain as well as the CXXC, BAH1, and BAH2 domains (Fig. 3E).
Figure 3. The dandelion phenotype is caused by mutations in dnmt1.
(A) Genetic map of the ddn region on linkage group 3; the numbers above the SSLP/RFLP markers indicate the number of recombinants in 932 meioses analyzed. Six genes lie within the critical region (shaded). (B) Sequence traces of pooled phenotypically WT and pooled dnmt1s872 mutant cDNAs; a G to A transversion at nucleotide 4376 results in a G1459D substitution in Dnmt1. (C) Sequence traces of pooled phenotypically WT and pooled dnmt1s904 mutant genomic DNA; a mutation in the exon 15 splice acceptor results in a single base pair frameshift and premature termination following 31 mis-sense substitutions. (D) Peptide sequence alignment of D. rerio (Dr) Dnmt1 from dnmt1s872 mutant and WT with H. sapiens (Hs), M. musculus (Mm), X. laevis (Xl), and A. thaliana (At). (E) Domain map of WT and ddn alleles of Dnmt1. (F) Southern blot of WT and dnmt1s872 and dnmt1s904 mutant genomic DNA either uncut or digested with methylation-sensitive HpaII or methylation insensitive MspI, and hybridized with a radiolabeled probe to the DANA consensus sequence. Appearance of DANA band (arrow) in dnmt1 mutant DNA digested with HpaII indicates hypomethylation of this transposable element, while smearing of high molecular weight DNA indicates widespread genomic hypomethylation (compare bracketed regions). (G-I) Dnmt1 antibody staining in 2CLIP transgenic line. Dnmt1 (blue) is observed in endodermal organs of WT (G) and dnmt1s872 mutants (H), but not dnmt1s904 mutants (I). (J-L) Injection of 4ng of dnmt1 morpholino (d1MO) into the 2CLIP line phenocopies ddn mutants. Compared to WT (J), d1MO-injected larvae (K) show smaller mass of differentiated pancreatic acinar cells (green) and hepatocytes (red) at 96 hpf. Ectopic dsRed fluorescence is observed in the posterior vasculature of d1MO-injected larvae at 168 hpf (L).
To test methyltransferase activity in dnmt1 mutants, we characterized global DNA methylation using methylation-sensitive DNA restriction analysis. Southern blotting of HpaII-digested genomic DNA with a probe for DANA, a short interspersed nuclear element (SINE) that comprises ∼10% of the zebrafish genome (Izsvak et al., 1996), revealed hypomethylation of this transposon sequence in dnmt1s872 and dnmt1s904 mutants (Fig. 3F). In addition, using an antibody generated against the Dnmt1 catalytic domain, we examined the distribution of Dnmt1 in endodermal organs at 84 hpf, when pancreatic degeneration begins. In WT larvae, Dnmt1 was observed in the exocrine pancreas, liver, and intestine (Fig. 3G; n>10). Dnmt1 was also detectable in s872 mutants (n=3), indicating that the mutant form of the protein is not degraded (Fig 3H). Dnmt1 was not detectable in most endodermal cells in s904 mutants (n=8), although we consistently observed a few weakly labeled cells scattered throughout the endodermal organs. Altogether, these data imply that the indistinguishable phenotypes of the dnmt1s872 and dnmt1s904 mutants result from a lack of methyltransferase activity.
Finally, we inhibited production of Dnmt1 using a morpholino (d1MO) that targets the translation start site (Rai et al., 2006). Injection of 4 ng of d1MO recapitulated the previously reported phenotype: small eyes, small pharyngeal arches, and ventral body curvature (Rai et al., 2006). More specifically, d1MO injection reduced, but did not eliminate the mass of hepatocytes, acinar cells, and pancreatic duct cells, as assessed in the 2CLIP (n=8), and Tg(−3.5nkx2.2a:GFP)ia3 (n=6; (Pauls et al., 2007)) backgrounds (Fig 3J,K and data not shown). In addition, d1MO injections had no effect on the early wave of beta cell production, as previously reported ((Rai et al., 2006); data not shown). The similarity of phenotypes in d1MO-injected embryos and dnmt1s872 and dnmt1s904 mutants supports the assertion that loss of Dnmt1 function in these mutants and morphants results in defects in endodermal organ growth/maintenance.
Lack of methyltransferase activity results in apoptosis
The loss of pancreatic acinar cell markers in dnmt1 mutants could be due to cell dedifferentiation, cell death, or both. To assess the extent of each, we first examined the expression of sox17, a transcription factor gene that is expressed throughout the early endoderm, but not in differentiated pancreatic tissue. Since silencing of the SOX17 promoter in cancer cell lines is associated with its CpG island hypermethylation (Zhang et al., 2008), we hypothesized that re-expression of sox17 would indicate a reversion to a more primitive identity. However, we did not detect sox17 expression in the pancreas of dnmt1 mutants at 84 or 100 hpf (n=10). Next, since repression of transposon activity is one of the major characterized functions of cytosine methylation, we investigated the expression of the repetitive SINE element DANA. While DANA sequences were hypomethylated in dnmt1 mutants (see above), we did not detect ectopic expression of the DANA sequence by in situ hybridization (n=12).
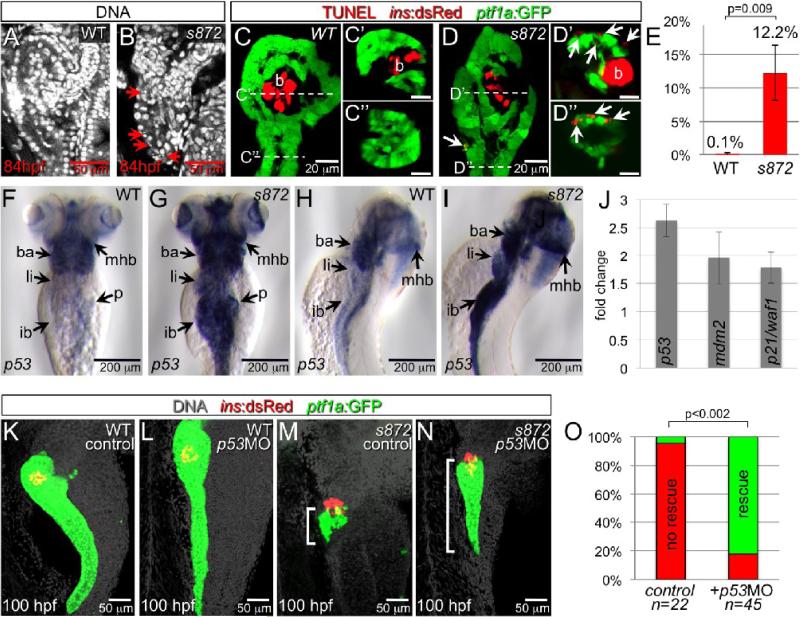
In dnmt1 mutants, but not WT, we frequently observed pyknotic nuclei throughout the pancreas during degeneration (Fig. 4A,B), suggesting cell loss via death. To determine whether this phenotype was caused by programmed cell death, we performed TUNEL assays. In 84 hpf WT embryos, virtually no TUNEL+ cells were observed in Tg(ptf1a:GFP)+ acinar tissue, while more than 12% of acinar cells were labeled in dnmt1 mutants (Fig 4 C-E). In addition, widespread TUNEL labeling was observed in the mutant liver and intestine, which in WT exhibit esssentially no apoptosis at this stage (data not shown).
Figure 4. Acinar cell death in dnmt1 mutants occurs in part by p53-dependent apoptosis.
(A,B) DRAQ5 staining of DNA in 84 hpf WT (A) and dnmt1s872 mutant (B) larvae. Pyknotic nuclei (arrows) are frequently observed in the pancreas of dnmt1 mutants, but not WT. (C,D) Apoptosis detection by TUNEL labeling of 84 hpf WT (C) and dnmt1 mutant (D) larvae in Tg(ptf1a:EGFP)jh1; Tg(ins:dsRed)m1081 background. C’/C’’ and D’/D’’ are transverse sections of similar samples at the approximate planes indicated in panels C and D, respectively. (E) Quantification of TUNEL+ acinar cells. (F-I) Expression of p53 mRNA in WT (F,H) and dnmt1s872 mutants (G,I); expression appears dramatically increased in mutants. (J) Fold-change of p53 and target genes mdm2 and p21/waf1 in dnmt1s872 mutants vs. WT by real-time RT-PCR. (K-N) 100 hpf WT (K,L) and dnmt1s872 mutant (M,N) larvae, which are either uninjected controls (K,M) or p53 morpholino-injected (p53MO; L,N). p53MO-injected dnmt1s872 mutants show increased persistence of acinar tissue. (O) More than 82% of p53MO-injected dnmt1s872 mutants showed rescue (green); n=22 (control), n=45 (p53MO). Error bars = SEM. Significance assessed by student's t-test.
To determine whether the observed apoptosis could be p53-dependent, we examined the expression of p53 by in situ hybridization. In 84 hpf dnmt1 mutants, clear up-regulation of p53 expression was apparent in affected tissues, including pancreas, liver, intestine, branchial arches, and eyes (Fig 4F-I). We confirmed this result using real-time RT-PCR, which showed up-regulation of p53 and its targets mdm2 and p21/waf1 (Fig 4J). To test the significance of p53 up-regulation, we inhibited production of P53 using an antisense splice morpholino (p53MO) (Chen et al., 2005). At 108 hpf, we observed a considerable rescue of exocrine pancreas morphology in 82% of p53MO-injected dnmt1 mutants (37/45, p<0.0002) as compared to uninjected dnmt1 mutants (1/22; Fig 4K-O). This result suggests that in dnmt1 mutants, hypomethylation is sensed as DNA damage, which results in some p53-dependent apoptosis. While we cannot rule out an incomplete knockdown of P53 following p53MO injection, the incomplete suppression of the degeneration phenotype suggests that some cell death in dnmt1 mutants is mediated by a p53-independent response. Other mutant phenotypes, including small liver, circulating hepatocyte fragments, and small eyes, which become apparent later than the exocrine pancreas phenotype, were not measurably rescued by p53MO injections, perhaps due to depletion of the morpholino.
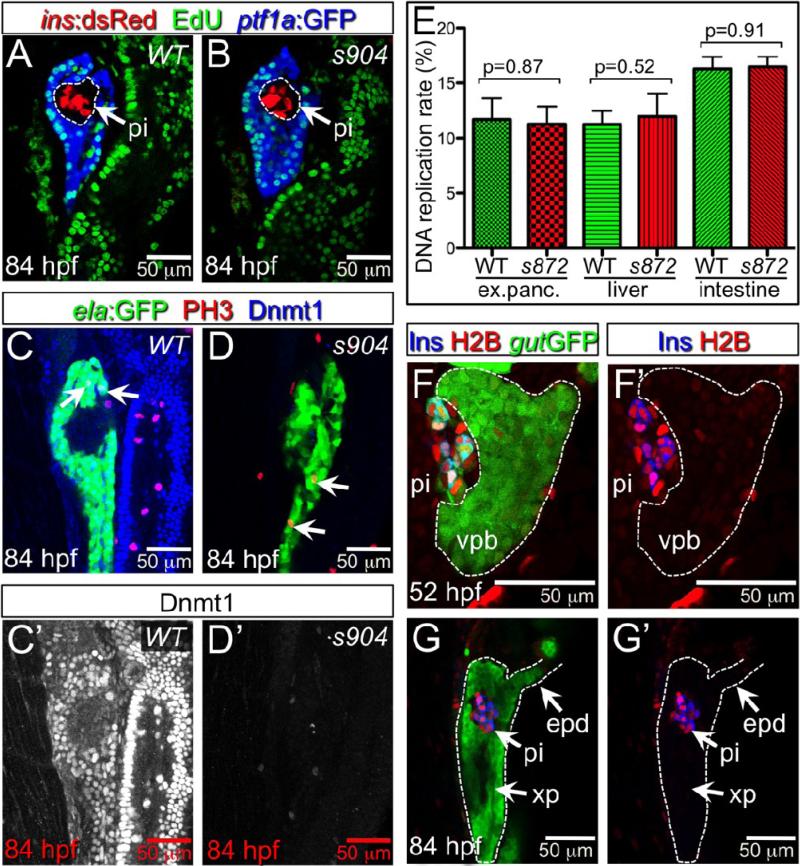
The depletion of Dnmt1 in cultured cells causes inhibition of replication origin initiation and intra-S-phase arrest (Knox et al., 2000; Milutinovic et al., 2004) via activation of Chk1, Chk2, and ATR checkpoint kinases (Unterberger et al., 2006); prolonged arrest at cell cycle checkpoints may initiate apoptotic pathways. To determine whether dnmt1 mutant acinar cells arrest during S-phase, we examined the incorporation of the thymidine analog EdU as a measure of DNA synthesis (Fig 5 A,B). We observed no significant changes in the number of labeled pancreatic acinar cells between WT and dnmt1 mutants at 84 hpf ( p=0.87; Fig 5E). We also examined the EdU incorporation rate in other highly proliferative endodermal tissues. The rate of labeling also appeared to be unaffected in liver (p=0.52) and intestine (p= 0.91; Fig 5E). Next, to determine whether acinar cells were blocked at the G2/M checkpoint, we used phosphorylated histone H3 (PH3) to mark cells in late G2 through M phase (Fig 5C,D). Again, we observed no significant difference in the number of PH3+ cells between WT and dnmt1 mutants at 84 hpf (44 sections from 4 WT embryos, 46 sections from 4 s904 mutants, p=0.73). Finally, we tested whether inhibition of checkpoint kinases could rescue the degeneration phenotype, and therefore reveal whether activation of checkpoints was involved. WT and dnmt1 mutant embryos were treated with SB 203580 (p38 MAP kinase inhibitor; (Takenaka et al., 1998)) or 2-Morpholin-4-yl-6-thianthren-1-yl-pyran-4-one (ATM inhibitor (ATMi); (Hickson et al., 2004)) from 48 to 108 hpf. Neither of these treatments affected the degeneration of the exocrine pancreas (data not shown; n>25 dnmt1s872 mutants). Together, these results suggest that loss of Dnmt1 catalytic activity per se does not prohibit DNA replication or entry into mitosis, and that the majority of cell death is unlikely to be due to prolonged arrest at cell cycle checkpoints.
Figure 5. Lack of Dnmt1 activity does not arrest the cell cycle.
(A,B) Confocal planes showing EdU incorporation in 84 hpf WT (A) and dnmt1 mutant (B) Tg(ptf1a:EGFP)jh1; Tg(ins:dsRed)m1081 larvae. (C,D) Phospho-histone 3 (late G2 interphase through anaphase marker) and Dnmt1 staining of 84 hpf WT and dnmt1s904 mutant Tg(fabp10:DsRed; ela:GFP)gz12 larvae. (C’,D’) Dnmt1 channel only. (E) EdU incorporation rate in pancreatic acinar cells, hepatocytes, and intestine of dnmt1 mutants appears indistinguishable from WT (p=0.87, p=0.52, p=0.91, respectively). Error bars = SEM. Significance assessed by student's t-test. (F,G) H2BRFP label retention analysis of endodermal organ forming region in WT Tg(gut:GFP)s854 animals at 52 (F) and 84 (G) hpf. The ventral pancreatic bud (vpb) is demarcated by a dashed line. H2BRFP is diminished in the highly proliferative vpb relative to the dorsal pancreatic bud during outgrowth (F) and pancreas tail formation (G). (F’,G’) H2BRFP (red) and Ins (blue) channels only. Primary islet (pi).
Since Dnmt1 is targeted to the replication fork during S phase, we reasoned that the different responses of acinar and endocrine cells to the loss of Dnmt1 catalytic activity might be based on differences in proliferation rates between these two populations. In other words, if DNA is passively demethylated during replication in the absence of Dnmt1, then cells which have divided more frequently should be more affected. In WT embryos, endocrine cells in the primary islet are rarely labeled with EdU, indicating a very low proliferation rate (dashed circle, Fig. 5A). In contrast, the acinar cells are generated from the ventral pancreatic bud, which grows dramatically between 34 and 84 hpf, and exhibits a high level of proliferation (Fig 5A). We devised an H2BRFP label retention assay to examine the cumulative proliferative histories of pancreatic cells. We presumed that H2BRFP would behave similarly to H2BGFP, a stable fusion protein that is localized to chromatin (Megason and Fraser, 2003); because of its stability, the intensity of nuclear fluorescence is diminished over time primarily by its stoichiometric dilution via cell division (Brennand et al., 2007). As such, fluorescent signal should be retained in non-proliferating cells and lost in highly proliferative cells. H2BRFP mRNA was injected into one-cell stage embryos, leading to uniform labeling through 24 hpf. By 52 hpf, after the fusion of the dorsal and ventral pancreatic buds, clear differences in fluorescence intensity were observed between the two buds (Fig. 5F), with the dorsal bud showing strong fluorescence and the ventral bud showing weaker fluorescence; this difference was more pronounced at 84 hpf after the outgrowth of the pancreatic tail (Fig 5G). These data show that pancreatic cells originating from the dorsal and ventral buds have vastly different proliferative histories. Furthermore, since dnmt1 mRNA is maternally provided in zebrafish (Martin et al., 1999; Mhanni and McGowan, 2002), these results suggest that the dorsal bud-derived endocrine cells in the primary islet are not affected in dnmt1 mutants because they are formed by 24 hpf, when maternal messages or proteins may still be present in the embryo. Moreover, these islet cells are largely quiescent and may not require Dnmt1 after their differentiation.
Regeneration of beta cells in dnmt1 mutants and morphants
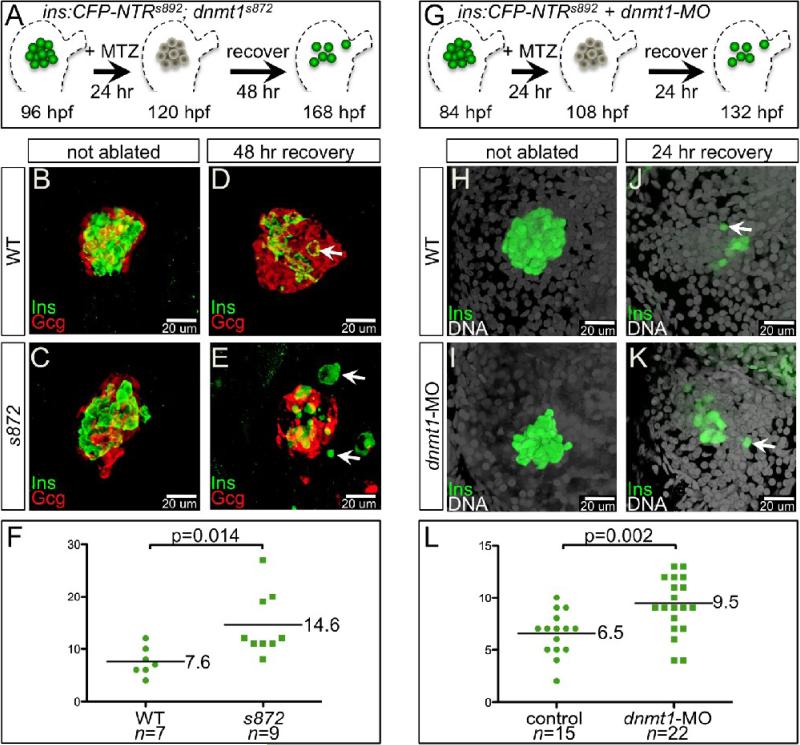
The highly proliferative ventral pancreatic bud generates all three types of pancreatic tissue: acinar, duct, and endocrine. Intriguingly, our data show that acinar cells are more severely affected by the loss of Dnmt1 catalytic activity than are endocrine or duct cells. Indeed, the organization of the primary islet remains unaffected in dnmt1 mutants: a core of beta cells surrounded by a mantle of alpha cells (Fig. 6B,C). To further investigate the role of dnmt1 and methylation in the later development of endocrine cells from progenitors, we examined beta cell neogenesis in dnmt1 mutants during regeneration of the primary islet. In WT larval zebrafish pancreas, we and others have shown that the beta cell mass has the capacity to recover following targeted ablation (Curado et al., 2007; Pisharath et al., 2007). Using this strategy, we ablated beta cells in WT and dnmt1 mutants using a nitroreductase transgene (Tg(ins:CFP-NTR)s892) together with metronidazole (MTZ) exposure from 96−120 hpf, followed by a recovery period in the absence of MTZ for 48 hrs (Fig. 6A). The ablated WT islets exhibited recovery of beta cells during the washout period (7.6 Ins+ cells/islet, n=7; Fig. 6D). Remarkably, we observed a significantly greater number of beta cells generated de novo in recovering dnmt1 mutant larvae (14.6 Ins+ cells/islet, n=9, p=0.014; Fig. 6F). Many of the newly produced beta cells in recovering dnmt1 mutants were morphologically abnormal possibly due to the deteriorating health of 7 dpf dnmt1 mutants, which die around 8 dpf. Therefore, we depleted Dnmt1 in WT embryos using d1MO. With injection of 4 ng, we oberved the reported phenotype: small endodermal organs, small eyes, and curved body axis (see above and data not shown; (Rai et al., 2006). However, injection of 2 ng of d1MO did not have an overt effect. We reasoned that a moderate, and likely temporary, reduction in Dnmt1 would result in modest changes in cytosine methylation, and might influence regeneration of beta cells from progenitors. Thus, we injected 2 ng of d1MO into one-cell stage Tg(ins:CFP-NTR)s892;Tg(−4.0ins:GFP)zf5 embryos, and then treated them with MTZ from 84−108 hpf to ablate beta cells. After MTZ washout and a 24 hour recovery period, beta cells were counted (Fig 6G). In WT, we observed an average of 6.7 new cells per pancreas (Fig 6J, n=6), while in morphants, an average of 10.8 cells were observed (Fig 6K, n=9, p=0.008). Importantly, in dnmt1 morphants, the morphology of the new beta cells appeared WT-like.
Figure 6. Enhanced recovery of beta cell mass in dnmt1 mutants and morphants.
(A) Pancreatic beta cell ablation scheme in dnmt1s872 mutants. 96 hpf Tg(ins:CFP-NTR)s892 WT and dnmt1 mutant larvae were exposed to MTZ for 24 hr, then washed for 48 hr and analyzed at 168 hpf. (B-E) Three-dimensional projections of the primary islet at 168 dpf in WT (B,D) and dnmt1 mutant (C,E) larvae that were untreated (B,C), or ablated with MTZ (D,E). (B,C) WT and dnmt1 mutant islets have a core of Ins+ beta cells surrounded by a mantle of Gcg+ alpha cells. (D) In recovering WT larvae, new beta cells are observed in the islet (arrow). (E) In recovering dnmt1 mutant islets, nearly double the number of beta cells are present, though some appear morphologically abnormal (arrows; compare with panel D). (F) 7.6 ± 1.0 beta cells were observed in recovering WT islets (n=7), and 14.6 ± 2.0 beta cells were observed in recovering dnmt1 mutant larvae (n=9; p=0.014). (G) Pancreatic beta cell ablation scheme in Tg(ins:CFPNTR)s892 embryos injected with 2 ng of dnmt1 morpholino (dnmt1-MO). 84 hpf larvae were exposed to MTZ for 24 hr, then washed for 24 hr and analysed at 132 hpf. (H-K) Three-dimensional projections of the primary islet at 132 hpf in WT (H,J) and dnmt1-MO injected (I,K) larvae that were untreated (H,I), or ablated with MTZ (J,K). (K) dnmt1-MO injected larvae showed a greater degree of beta cell recovery. (L) 6.5 ± 0.5 beta cells were observed in recovering WT larvae (n=15), and 9.5 ± 0.6 beta cells were observed in recovering dnmt1-MO injected larvae (n=22; p=0.002). Error bars = SEM. Significance assessed by student's t-test.
DISCUSSION
In this paper, we report the identification of two mutations in the gene encoding the maintenance methyltransferase Dnmt1 that disrupt its catalytic activity. We have used these new reagents to specifically investigate the roles of DNA methylation by Dnmt1 during pancreatic development. Our data reveal two novel developmental roles for cytosine methylation in the formation of the pancreas: the survival of differentiated acinar cells, and control of de novo beta cell formation.
The pancreas is comprised of three major endodermal cell types, endocrine, duct, and acinar cells, that are differentially affected in dnmt1 mutants. The formation of endocrine and duct cells is largely unaffected, while acinar cell survival is severely compromised, with the majority of cells degenerating by 100 hpf. Because dnmt1 is supplied maternally (Martin et al., 1999; Mhanni and McGowan, 2002), it is possible that these differential effects reflect different proliferative histories of each tissue, and thus differential dilution of maternal Dnmt1. Indeed, our label retention analysis shows that in the highly proliferative ventral bud cells, H2BRFP is diluted much more than the largely quiescent dorsal bud cells. This result suggests that the endocrine cells, which predominantly arise from the dorsal bud, are spared either because they do not require dnmt1, or because maternal dnmt1 contribution is retained in these cells. Since the degenerating tissues in dnmt1 mutants (pancreatic acinar cells, hepatocytes, eye, branchial arches) exhibit high proliferation rates, these findings strongly suggest that Dnmt1 is required for the survival of highly proliferative cell types. However, the ventral bud also contributes a portion of endocrine cells and all duct cells, and these appear largely unaffected in dnmt1 mutants. Thus, in the pancreas, the requirement for dnmt1 may be specific to proliferating acinar cells, rather than all ventral bud derivatives.
Our results expand upon those of Rai and colleagues (Rai et al., 2006) who showed that knockdown of Dnmt1 with translation-inhibiting MOs resulted in reduced acinar cell mass, as measured by reduced trypsin expression. In our hands dnmt1-MO injections did not eliminate the differentiation of acinar cells, beta cells, or pancreatic ducts. Rather, dnmt1-MO injections reduced the mass of the ventral pancreatic bud derived tissues with no effect on the early wave of beta cell production from the dorsal pancreatic bud. Since dnmt1 is maternally and zygotically contributed, it is likely that Dnmt1 protein is present during some stages of ventral pancreas growth and differentiation in both dnmt1 mutants, and dnmt1 morphants. Nonetheless, our findings and those of Rai et al. (Rai et al., 2006) are consistent with a model in which Dnmt1 activity may not be absolutely required for the differentiation of pancreatic acinar cells, but rather for their proliferative expansion.
Since cytosine methylation is inversely correlated with gene expression, we examined dnmt1 mutants for reactivation of the primitive endodermal marker sox17 in the pancreas, and expression of the SINE element DANA. Expression of neither gene was significantly altered at either 84 or 100 hpf. Thus, although deletion of Dnmt1 in cultured mouse fibroblasts resulted in increased transcription of up to 10% of detectably expressed genes (Jackson-Grusby et al., 2001), and loss of dnmt1 in ddn mutants can reactivate the expression of silenced transgenes (Goll et al., 2009), our limited survey argues against a major role for large-scale mis-regulation of gene expression in causing the pancreatic acinar tissue degeneration in dnmt1 mutants.
Initiation of apoptosis results from a wide variety of cellular stresses, including DNA damage and aberrant activation of oncogenes, and is often mediated by the tumor suppressor p53. Moreover, prior studies indicate that apoptosis may be influenced by the concerted action of Dnmt1 and p53; for instance, loss of Dnmt1 catalytic activity in mouse ES cells results in phosphorylation of p53 (Takebayashi et al., 2007), and loss of Dnmt1 function in cultured mouse fibroblasts causes apoptosis that is largely rescued by p53 inactivation (Jackson-Grusby et al., 2001). We found moderately increased survival of acinar cells in dnmt1 mutants when we reduced p53 levels using an antisense morpholino, but not a complete rescue. Thus, it is possible that p53-independent apoptotic pathways are also initiated, although it is also possible that the p53 levels were not sufficiently reduced, or were recovering at later stages of development as the morpholino was diluted with cell division.
When DNMT1 is inactivated in HCT116 cancer cells, a transient arrest at the G2/M cell cycle checkpoint is enacted then resolved, resulting in dramatic chromosomal aberrations, mitotic catastrophe, and cell death (Chen et al., 2007). This observation prompted speculation that Dnmt1 may have multiple roles which are progressively uncovered by reduction of Dnmt1 protein levels (Brown and Robertson, 2007). The canonical role of Dnmt1 is revealed by moderate reduction of protein levels, such as is found in hypomorphs and partial knockdowns, leading to phenotypes such as DNA hypomethylation, genomic instability, and decreased replication potential. In contrast, the total depletion of Dnmt1 reveals non-canonical roles that manifest as massive DNA damage and chromosomal instability. For instance, Dnmt1 associates with peri-centromeric heterochromatin DNA during G2 and M phases, indicating a post-DNA replication function in centromere methylation (Easwaran et al., 2004). Methylation influences kinetochore assembly at centromeres (Mitchell et al., 1996), and demethylation causes chromatin remodeling at pericentric regions (Ma et al., 2005). Thus, cytosine methylation plays many roles in maintaining the integrity of the genome and the structure of chromosomes. Since zebrafish dnmt1 mutants lack functional zygotic Dnmt1, but are initially endowed with a maternal supply, the functional complement of Dnmt1 is diminished with each cell division. It is likely that such “non-canonical” roles of Dnmt1 would first become apparent in rapidly replicating cell populations, such as the pancreatic acinar cells, and might result in genomic damage during cell division. The induction of p53, its target genes mdm2 and p21/waf1, and apoptosis are typical cellular responses to DNA damage. Their striking induction in dnmt1 mutants suggests that loss of Dnmt1 catalytic activity results in genomic changes that may be sensed as DNA damage.
The increased de novo beta cell neogenesis observed in dnmt1 mutants and morphants after beta cell ablation suggests that surviving pancreatic cells have an increased capacity to differentiate into beta cells. There is burgeoning evidence that regulation of genomic methylation patterns can be manipulated to control the reprogramming of cells and alter their potency. For instance, reprogramming of promoter methylation at pluripotency genes may be a key mechanism by which epigenetic regulation of pluripotency is effected (Farthing et al., 2008; Mikkelsen et al., 2008). Also, the methylation status of donor nuclei can strongly influence the efficiency of deriving totipotent ES cell lines by nuclear transfer from differentiated cells (Blelloch et al., 2006). Furthermore, inhibition of Dnmt1 activity with 5-aza-2-deoxycytidine may facilitate the reprogramming of mouse embryonic fibroblasts into pluripotent stem cells (Huangfu et al., 2008). Our data suggest that genomic hypomethylation caused by disrupted Dnmt1 activity is correlated with a greater capability to form de novo beta cells in response to ablation. Further studies are needed to determine the biological mechanism of this enhanced ability, as well as the source of the new beta cells. Increased beta cell regeneration in Dnmt1-depleted zebrafish may result from reprogramming of terminally differentiated pancreatic cells, the facilitation of beta cell production from multipotent progenitors, or simply an increased capacity for endocrine cell differentiation in the absence of exocrine tissue. Our findings could have implications for the therapeutic regeneration of beta cells, if coaxing of endogenous lineage-committed progenitor cells as well as terminally differentiated cells into new fates could be aided by manipulation of DNA methylation levels either globally, or at specific loci.
Materials and Methods
Zebrafish strains and lines
Zebrafish were raised under standard laboratory conditions at 28°C. We used the following lines: dnmt1s872, dnmt1s904, Tg(ins:CFP-NTR)s892 (Curado et al., 2007), Tg(XlEef1a1:GFP)s854 (Field et al., 2003b), Tg(fabp10:dsRed, ela3l:GFP)gz12 (Farooq et al., 2008), Tg(ins:dsRed)m1081 (gift of W. Driever), Tg(−4.0ins:GFP)zf5 (Huang et al., 2001), Tg(kdrl:EGFP)s843 (Beis et al., 2005), and Tg(ptf1a:EGFP)jh1 (Godinho et al., 2007), and the WIK strain for mapping.
Genetic screening
Mutagenesis of Tg(XlEef1a1:GFP)s854 was previously described (Ober et al., 2006; Shin et al., 2008). For the 2CLIP screen, 36 males received six 1 hour exposures to 3.5 mM ENU at weekly intervals. From the number and extent of F2 families screened, we calculate that we surveyed 662 genomes.
Genetic mapping
We mapped the ddns872 mutation to chromosome 3 using standard SSLP markers. For fine mapping, 466 mutant embryos were tested with SSLP markers in the critical region. We isolated, sequenced, and analyzed dnmt1 cDNA from WT, and mutant embryos. Additionally, genomic DNA was isolated and sequenced from WT and s904 mutant embryos to confirm the splice acceptor mutation.
Methylation analyses
At 7 dpf, larvae were sorted as WT or dnmt1s872 based on phenotype. Genomic DNA was isolated from pools of 10 phenotypically WT or mutant larvae. DNA (100ng) was digested with the restriction enzyme MspI, HpaII or mock digested in buffer alone. Following digestion, samples were electrophoresed through a 0.9% agarose gel then transfered to a nylon membrane. Membranes were probed with radiolabeled DANA sequence amplified from the genome using primers 5’GGCGACRCAGTGGCGCAGTRGG and 5’TTTTCTTTTTGGCTTAGTCCC (Izsvak et al., 1996).
Immunofluorescence
Antibody staining was performed as described (Shin et al., 2008) using the following antibodies: Dnmt1 (Santa Cruz SC-20701), Islet-1, Nkx6.1, and Alcam (Developmental Studies Hybridoma Bank clones 39.4D5, F55A10, and zn-8), Somatostatin (Serotec 8330−0154), Glucagon (Sigma G2654), phospho-histone (Abcam ab14955), and Caspase-3 (Cell Signaling 9661S).
mRNA expression
In situ hybridization was performed as described (Shin et al., 2007). The DANA probe was based upon GenBank accession # L42295.1; template was PCR amplified from cDNA using 5’GGTTA GTGGC GCAGT GGGAA GT and 5’GGTTA TTTAG GTGAC ACTAT AGTTA ATCCA GGGTC ACCAC AG, and transcribed with SP6 polymerase. For real-time RT-PCR, mRNA was extracted from 20 pooled 5 dpf WT or s872 mutant embryos using Trizol (Invitrogen #15596018) and then reverse-transcribed using a Superscript III kit (Invitrogen #11732020). Optimized primers targeting each gene and β-actin were designed using the Plexor Primer Design System (Promega): β-actin (5’GTGGT CTCGT GGATA CCGCA A and 5’CTATG AGCTG CCTGA CGGTC A); p53 (5’CACCA TCAGC TTCTT TCCCT and 5’TAAGT GATGT GGTGC CTGCC); mdm2 (5’CCCTC ACTGC GAGAT ATACC and 5’GAGGC ATAAG TCGGA CAGCT); and p21/waf1 (5’TAGAC GCTTC TTGGC TTGGT and 5’ATCCC GAAAA CACCA GAACG). An aliquot of each cDNA sample and the appropriate primers were added to Power SYBR Green master mix (Applied Biosystems). The 7900HT Real-Time PCR System (Applied Biosystems) was used to obtain Ct values. The relative expression of each sample was determined after normalization to β-actin using the relative standard curve method (Larionov et al., 2005).
Proliferation and apoptosis analyses
Click-IT (Invitrogen C10085) was adapted for S-phase labeling. Larvae were injected pericardially with 5nl of EdU (200 uM+2% DMSO+0.1% phenol red), incubated 60 min at 28°C, and then fixed in 3% formaldehyde. Before detection, samples were washed with PBS+0.3% Triton X-100 + 0.1% Tween-20 for 20 min. For whole mount samples > 5 optical sections at 4 μm intervals were analyzed for each sample. For cell area measurements, and manual counting of EdU or PH3+ cells, images were analyzed with ImageJ software. Significance was assessed with unpaired t-test. TUNEL was performed as described (Curado et al., 2007). For H2BRFP labeling, embryos were injected with 100 pg of H2BRFP mRNA (Megason and Fraser, 2003).
Chemical inhibitors
Larvae were exposed to 10 uM of SB-203580 p38 inhibitor (A.G. Scientific) or 2-Morpholin-4-yl-6-thianthren-1-yl-pyran-4-one ATM inhibitor (Calbiochem) from 48−108 hpf.
Morpholino injections
p53 was knocked down using 8 ng of a previously described splice-inhibiting morpholino (Chen et al., 2005), while dnmt1, was knocked down using 2 or 4 ng of a previously described translation-inhibiting morpholino (Rai et al., 2006).
Beta cell ablation and recovery
WT and dnmt1s872 mutant larvae bearing Tg(ins:CFP-NTR)s892 were treated with metronidazole from 96−120 hpf as described (Curado et al., 2008), followed by 48 hr recovery. The beta cell mass was visualized at 168 hpf with Insulin staining. Statistical significance was assessed with unpaired t-test.
ACKNOWLEDGEMENTS
We thank all members of the Stainier laboratory for support during the screen and A. Ayala, S. Waldron, and N. Zvenigorodsky for expert help with the fish. We thank E. Ober, H. Field, H. Verkade, W. Driever, and Z. Gong for providing unpublished zebrafish lines. This work was supported in part by fellowships from the JDRF (R.M.A., P.D.S.D.), the Damon Runyon Cancer Research Foundation (M.G.G.), the NIH (N.C.C., D.S., C.H.S., A.S.), the Hillblom Foundation (D.H., P.D.S.D.), the GSK Research & Education Foundation (N.C.C.), the AHA (N.C.C.) as well as grants from the NIH (M.H., D.Y.R.S.) and the Packard Foundation (D.Y.R.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Beis D, Bartman T, Jin SW, Scott IC, D'Amico LA, Ober EA, Verkade H, Frantsve J, Field HA, Wehman A, Baier H, Tallafuss A, Bally-Cuif L, Chen JN, Stainier DY, Jungblut B. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132:4193–204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- Biemar F, Argenton F, Schmidtke R, Epperlein S, Peers B, Driever W. Pancreas development in zebrafish: early dispersed appearance of endocrine hormone expressing cells and their convergence to form the definitive islet. Dev Biol. 2001;230:189–203. doi: 10.1006/dbio.2000.0103. [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–4. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Blelloch R, Wang Z, Meissner A, Pollard S, Smith A, Jaenisch R. Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem Cells. 2006;24:2007–13. doi: 10.1634/stemcells.2006-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–4. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Brennand K, Huangfu D, Melton D. All beta Cells Contribute Equally to Islet Growth and Maintenance. PLoS Biol. 2007;5:e163. doi: 10.1371/journal.pbio.0050163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KD, Robertson KD. DNMT1 knockout delivers a strong blow to genome stability and cell viability. Nat Genet. 2007;39:289–90. doi: 10.1038/ng0307-289. [DOI] [PubMed] [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988;53:3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- Chen J, Ruan H, Ng SM, Gao C, Soo HM, Wu W, Zhang Z, Wen Z, Lane DP, Peng J. Loss of function of def selectively up-regulates Delta113p53 expression to arrest expansion growth of digestive organs in zebrafish. Genes Dev. 2005;19:2900–11. doi: 10.1101/gad.1366405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, Li E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet. 2007;39:391–6. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, Stainier DY. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236:1025–35. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- Curado S, Stainier DY, Anderson RM. Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat Protoc. 2008;3:948–54. doi: 10.1038/nprot.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delporte FM, Pasque V, Devos N, Manfroid I, Voz ML, Motte P, Biemar F, Martial JA, Peers B. Expression of zebrafish pax6b in pancreas is regulated by two enhancers containing highly conserved cis-elements bound by PDX1, PBX and PREP factors. BMC Dev Biol. 2008;8:53. doi: 10.1186/1471-213X-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong PD, Munson CA, Norton W, Crosnier C, Pan X, Gong Z, Neumann CJ, Stainier DY. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat Genet. 2007;39:397–402. doi: 10.1038/ng1961. [DOI] [PubMed] [Google Scholar]
- Dunican DS, Ruzov A, Hackett JA, Meehan RR. xDnmt1 regulates transcriptional silencing in pre-MBT Xenopus embryos independently of its catalytic function. Development. 2008;135:1295–302. doi: 10.1242/dev.016402. [DOI] [PubMed] [Google Scholar]
- Easwaran HP, Schermelleh L, Leonhardt H, Cardoso MC. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep. 2004;5:1181–6. doi: 10.1038/sj.embor.7400295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq M, Sulochana KN, Pan X, To J, Sheng D, Gong Z, Ge R. Histone deacetylase 3 (hdac3) is specifically required for liver development in zebrafish. Dev Biol. 2008;317:336–53. doi: 10.1016/j.ydbio.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, Dean W, Hemberger M, Reik W. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field HA, Dong PD, Beis D, Stainier DY. Formation of the digestive system in zebrafish. II. Pancreas morphogenesis. Dev Biol. 2003a;261:197–208. doi: 10.1016/s0012-1606(03)00308-7. [DOI] [PubMed] [Google Scholar]
- Field HA, Ober EA, Roeser T, Stainier DY. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol. 2003b;253:279–90. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- Futscher BW, Oshiro MM, Wozniak RJ, Holtan N, Hanigan CL, Duan H, Domann FE. Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet. 2002;31:175–9. doi: 10.1038/ng886. [DOI] [PubMed] [Google Scholar]
- Godinho L, Williams PR, Claassen Y, Provost E, Leach SD, Kamermans M, Wong RO. Nonapical symmetric divisions underlie horizontal cell layer formation in the developing retina in vivo. Neuron. 2007;56:597–603. doi: 10.1016/j.neuron.2007.09.036. [DOI] [PubMed] [Google Scholar]
- Goll MG, Anderson R, Stainier DY, Spradling AC, Halpern ME. Transcriptional Silencing and Reactivation in Transgenic Zebrafish. Genetics. 2009 doi: 10.1534/genetics.109.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Huang H, Liu N, Lin S. Pdx-1 knockdown reduces insulin promoter activity in zebrafish. Genesis. 2001;30:134–6. doi: 10.1002/gene.1048. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–7. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izsvak Z, Ivics Z, Garcia-Estefania D, Fahrenkrug SC, Hackett PB. DANA elements: a family of composite, tRNA-derived short interspersed DNA elements associated with mutational activities in zebrafish (Danio rerio). Proc Natl Acad Sci U S A. 1996;93:1077–81. doi: 10.1073/pnas.93.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, Jaenisch R. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–9. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Knox JD, Araujo FD, Bigey P, Slack AD, Price GB, Zannis-Hadjopoulos M, Szyf M. Inhibition of DNA methyltransferase inhibits DNA replication. J Biol Chem. 2000;275:17986–90. doi: 10.1074/jbc.C900894199. [DOI] [PubMed] [Google Scholar]
- Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC Bioinformatics. 2005;6:62. doi: 10.1186/1471-2105-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–26. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Ma Y, Jacobs SB, Jackson-Grusby L, Mastrangelo MA, Torres-Betancourt JA, Jaenisch R, Rasmussen TP. DNA CpG hypomethylation induces heterochromatin reorganization involving the histone variant macroH2A. J Cell Sci. 2005;118:1607–16. doi: 10.1242/jcs.02291. [DOI] [PubMed] [Google Scholar]
- Martin CC, Laforest L, Akimenko MA, Ekker M. A role for DNA methylation in gastrulation and somite patterning. Dev Biol. 1999;206:189–205. doi: 10.1006/dbio.1998.9105. [DOI] [PubMed] [Google Scholar]
- Megason SG, Fraser SE. Digitizing life at the level of the cell: high-performance laser-scanning microscopy and image analysis for in toto imaging of development. Mech Dev. 2003;120:1407–20. doi: 10.1016/j.mod.2003.07.005. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhanni AA, McGowan RA. Variations in DNA (cytosine-5)-methyltransferase-1 expression during oogenesis and early development of the zebrafish. Dev Genes Evol. 2002;212:530–3. doi: 10.1007/s00427-002-0275-7. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milutinovic S, Brown SE, Zhuang Q, Szyf M. DNA methyltransferase 1 knock down induces gene expression by a mechanism independent of DNA methylation and histone deacetylation. J Biol Chem. 2004;279:27915–27. doi: 10.1074/jbc.M312823200. [DOI] [PubMed] [Google Scholar]
- Mitchell AR, Jeppesen P, Nicol L, Morrison H, Kipling D. Epigenetic control of mammalian centromere protein binding: does DNA methylation have a role? J Cell Sci. 1996;109(Pt 9):2199–206. doi: 10.1242/jcs.109.9.2199. [DOI] [PubMed] [Google Scholar]
- Nishino K, Hattori N, Tanaka S, Shiota K. DNA methylation-mediated control of Sry gene expression in mouse gonadal development. J Biol Chem. 2004;279:22306–13. doi: 10.1074/jbc.M309513200. [DOI] [PubMed] [Google Scholar]
- Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006;442:688–91. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–20. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- Pauls S, Zecchin E, Tiso N, Bortolussi M, Argenton F. Function and regulation of zebrafish nkx2.2a during development of pancreatic islet and ducts. Dev Biol. 2007;304:875–90. doi: 10.1016/j.ydbio.2007.01.024. [DOI] [PubMed] [Google Scholar]
- Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech Dev. 2007;124:218–29. doi: 10.1016/j.mod.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Nadauld LD, Chidester S, Manos EJ, James SR, Karpf AR, Cairns BR, Jones DA. Zebra fish Dnmt1 and Suv39h1 regulate organ-specific terminal differentiation during development. Mol Cell Biol. 2006;26:7077–85. doi: 10.1128/MCB.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461–7. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- Sakaguchi TF, Sadler KC, Crosnier C, Stainier DY. Endothelial signals modulate hepatocyte apicobasal polarization in zebrafish. Curr Biol. 2008;18:1565–71. doi: 10.1016/j.cub.2008.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanosaka T, Namihira M, Asano H, Kohyama J, Aisaki K, Igarashi K, Kanno J, Nakashima K. Identification of genes that restrict astrocyte differentiation of midgestational neural precursor cells. Neuroscience. 2008;155:780–8. doi: 10.1016/j.neuroscience.2008.06.039. [DOI] [PubMed] [Google Scholar]
- Shemer R, Kafri T, O'Connell A, Eisenberg S, Breslow JL, Razin A. Methylation changes in the apolipoprotein AI gene during embryonic development of the mouse. Proc Natl Acad Sci U S A. 1991;88:11300–4. doi: 10.1073/pnas.88.24.11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin CH, Chung WS, Hong SK, Ober EA, Verkade H, Field HA, Huisken J, Stainier DY. Multiple roles for Med12 in vertebrate endoderm development. Dev Biol. 2008;317:467–79. doi: 10.1016/j.ydbio.2008.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Shin CH, Tucker J, Ober EA, Rentzsch F, Poss KD, Hammerschmidt M, Mullins MC, Stainier DY. Bmp and Fgf signaling are essential for liver specification in zebrafish. Development. 2007;134:2041–50. doi: 10.1242/dev.000281. [DOI] [PubMed] [Google Scholar]
- Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, Nagase H, Held WA. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci U S A. 2005;102:3336–41. doi: 10.1073/pnas.0408436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancheva I, Hensey C, Meehan RR. Loss of the maintenance methyltransferase, xDnmt1, induces apoptosis in Xenopus embryos. EMBO J. 2001;20:1963–73. doi: 10.1093/emboj/20.8.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Hopkins N. vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes Dev. 2001;15:3217–29. doi: 10.1101/gad946701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi S, Tamura T, Matsuoka C, Okano M. Major and essential role for the DNA methylation mark in mouse embryogenesis and stable association of DNMT1 with newly replicated regions. Mol Cell Biol. 2007;27:8243–58. doi: 10.1128/MCB.00899-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka K, Moriguchi T, Nishida E. Activation of the protein kinase p38 in the spindle assembly checkpoint and mitotic arrest. Science. 1998;280:599–602. doi: 10.1126/science.280.5363.599. [DOI] [PubMed] [Google Scholar]
- Unterberger A, Andrews SD, Weaver IC, Szyf M. DNA methyltransferase 1 knockdown activates a replication stress checkpoint. Mol Cell Biol. 2006;26:7575–86. doi: 10.1128/MCB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee NS, Yusuff S, Pack M. Zebrafish pdx1 morphant displays defects in pancreas development and digestive organ chirality, and potentially identifies a multipotent pancreas progenitor cell. Genesis. 2001;30:137–40. doi: 10.1002/gene.1049. [DOI] [PubMed] [Google Scholar]
- Zecchin E, Mavropoulos A, Devos N, Filippi A, Tiso N, Meyer D, Peers B, Bortolussi M, Argenton F. Evolutionary conserved role of ptf1a in the specification of exocrine pancreatic fates. Dev Biol. 2004;268:174–84. doi: 10.1016/j.ydbio.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Zhang W, Glockner SC, Guo M, Machida EO, Wang DH, Easwaran H, Van Neste L, Herman JG, Schuebel KE, Watkins DN, Ahuja N, Baylin SB. Epigenetic inactivation of the canonical Wnt antagonist SRY-box containing gene 17 in colorectal cancer. Cancer Res. 2008;68:2764–72. doi: 10.1158/0008-5472.CAN-07-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]