SUMMARY
Many bacterial species respond to the quorum sensing signal autoinducer-2 (AI-2) by regulating different niche specific genes. Here, we show that Sinorhizobium meliloti, a plant symbiont lacking the gene for the AI-2 synthase, while not capable of producing AI-2 can nonetheless respond to AI-2 produced by other species. We demonstrate that S. meliloti has a periplasmic binding protein that binds AI-2. The crystal structure of this protein (here named SmLsrB) with its ligand reveals that it binds (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (R-THMF), the identical AI-2 isomer recognized by LsrB of Salmonella typhimurium. The gene encoding SmLsrB is in an operon with orthologs of the lsr genes required for AI-2 internalization in enteric bacteria. Accordingly, S. meliloti internalizes exogenous AI-2, and mutants in this operon are defective in AI-2 internalization. S. meliloti does not gain a metabolic benefit from internalizing AI-2, suggesting that AI-2 functions as a signal in S. meliloti. Furthermore, S. meliloti can completely eliminate the AI-2 secreted by Erwinia carotovora, a plant pathogen shown to use AI-2 to regulate virulence. Our findings suggest that S. meliloti is capable of ‘eavesdropping’ on the AI-2 signaling of other species and interfering with AI-2-regulated behaviors such as virulence.
Keywords: Sinorhizobium meliloti, Erwinia carotovora, AI-2, luxS, quorum sensing
INTRODUCTION
Quorum sensing is a cell-cell signaling process that enables bacteria to regulate gene expression as a function of population density. It is becoming increasingly apparent that quorum sensing signals, called autoinducers, can provide bacteria more information than simply the number of cells in the vicinity. By sensing combinations of various autoinducer signals in the environment, bacteria can determine, for instance, the species composition of the population or if they are inside or outside their host. Bacteria translate the information provided by the different autoinducers into specific gene expression responses leading to the promotion or inhibition of group behaviors such as bioluminescence, biofilm formation, and production of virulence factors. Additionally, some bacteria have mechanisms that enable them to interfere with other species’ ability to correctly sense and respond to autoinducer signals. It is likely that this interference with quorum sensing provides a benefit during competition for colonization of a common niche.
Most autoinducers are species specific; however, one autoinducer, autoinducer-2 (AI-2), and its synthase, LuxS, have been identified in many bacteria including both gram-negative and gram-positive species. Likewise, bacterial species have been shown to respond to AI-2 with behaviors such as bioluminescence in Vibrio harveyi (Mok et al., 2003; Schauder et al., 2001; Waters and Bassler, 2006), motility in Helicobacter pylori (Rader et al., 2007), interference with AI-2 regulated quorum sensing in Escherichia coli (Xavier and Bassler, 2005a), cell division and stress response in Streptococcus mutans (Sztajer et al., 2008), virulence and formation of biofilms in Vibrio cholerae (Hammer and Bassler, 2003, 2007; Miller et al., 2002; Xavier and Bassler, 2005a) and Staphylococcus epidermis (Li et al., 2008), and mutualistic biofilm growth in co-cultures of Actinomyces naslundii and Streptococcus oralis (Rickard et al., 2006). AI-2 is hypothesized to play an important role in enabling cross-species communication by allowing bacteria to regulate gene expression in response to the density and species composition of the bacterial populations they encounter. In some species (such as the examples given above), AI-2 has been shown to be the chemical signal responsible for inducing regulation of those specific phenotypes; in other species, the role played by AI-2 might be more complex and further studies are needed to distinguish between the metabolic effect of disrupting the AI-2 synthase and the responses caused by the signal itself (Turovskiy et al., 2007; Vendeville et al., 2005).
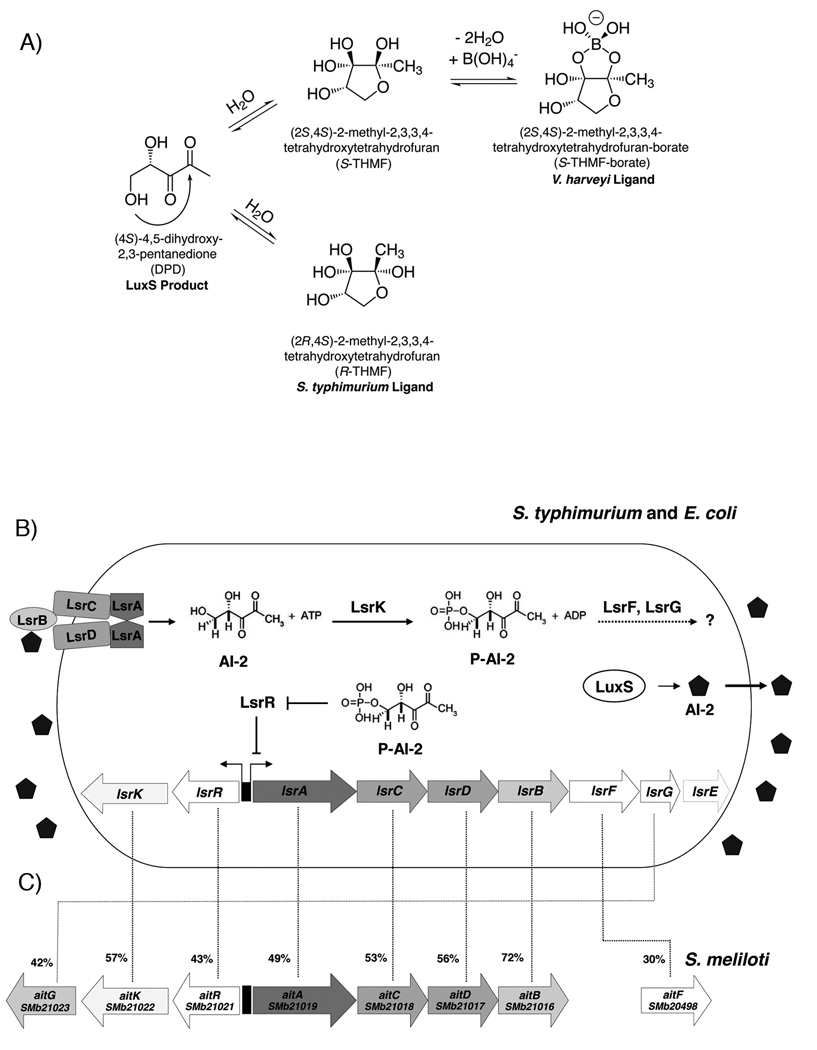
Despite the large number of studies identifying AI-2 regulated phenotypes (reviewed in in (Hardie and Heurlier, 2008; Xavier and Bassler, 2003)), the mechanisms of AI-2 detection and signal transduction have only been determined in two Vibrio species (V. harveyi and V. cholerae) (Chen et al., 2002; Lenz et al., 2004; Miller et al., 2002; Mok et al., 2003; Waters and Bassler, 2006) and the enteric bacteria Salmonella typhimurium and E. coli (Taga et al., 2001; Taga et al., 2003; Xavier and Bassler, 2005b; Xavier et al., 2007). LuxS catalyzes the production of 4,5-dihydroxy-2,3-pentanedione (DPD) from S-ribosylhomocystine, however, DPD is not directly recognized by these species as AI-2 (Chen et al., 2002; Miller et al., 2004; Schauder et al., 2001). Rather, crystal structures of the AI-2 receptor/ligand complexes revealed that these bacterial species recognize different adducts of DPD as AI-2 signals. In V. harveyi, the AI-2 signal is formed by cyclization of DPD, followed by hydration and addition of borate. In enteric bacteria, the LsrB protein recognizes an AI-2 moiety that lacks boron and is a different stereoisomer than the signal recognized by V. harveyi (Fig. 1A). Importantly, although different bacterial species recognize chemically distinct molecules as AI-2, these molecules interconvert spontaneously in solution, allowing different bacterial species to respond to one another (Miller et al., 2004; Xavier and Bassler, 2005a).
Fig. 1. The interconversion of DPD into the known AI-2 ligands and the AI-2-dependent internalization system.
A. Proposed equilibrium between the currently known forms of AI-2 and their common precursor, DPD. The V. harveyi and S. typhimurium ligands have previously been shown to interconvert in solution.
B. S. typhimurium and E. coli Lsr-mediated transport and processing of AI-2. In S. typhimurium and E. coli, AI-2 is produced within the cell by LuxS and is secreted to the medium. As the concentration of extracellular AI-2 increases, AI-2 binds to the periplasmic binding protein LsrB and is internalized by the Lsr system, an ABC-type transport system. Once in the cytoplasm, AI-2 is phosphorylated (P-AI-2) by LsrK. P-AI-2 binds to the repressor of the lsr operon, LsrR, inactivating LsrR, relieving repression, and inducing transcription of lsr. This causes a rapid increase in the production of the Lsr transporter and, consequently removal of AI-2 from the environment. P-AI-2 is further processed by a mechanism not fully understood involving LsrG and LsrF. The lsrE gene is also present in the operon of S. typhimurium but not in E. coli and its function is not known.
C. The lsr orthologues in S. meliloti operon. S. meliloti has orthologs to all the genes of the lsr operon except lsrE. We named the S. meliloti lsr-like operon ait (for autoinducer transporter). The percent identity to the Lsr proteins from S. typhimurium is shown.
In S. typhimurium and E. coli, AI-2 induces the production of a transport apparatus responsible for internalizing, phosphorylating, and processing of the AI-2 signal (Fig. 1B). The genes encoding this transport system are in the operon lsr (for LuxS Regulated), along with other genes involved in AI-2 processing and response (Taga et al., 2001; Taga et al., 2003; Xavier and Bassler, 2005b). The Lsr transport system internalizes endogenously produced AI-2 as well as AI-2 produced by other bacterial species, eliminating the signal from the environment. Thus, in cultures composed of different species, these enteric bacteria are capable of interfering with the AI-2-mediated signaling of other species by disrupting their ability to regulate group behaviours (Xavier and Bassler, 2005a). Recently, Demuth and co-workers have studied the function of the Lsr homologue in the oral pathogen Aggregatibacter (Actinobacillus) actinomycetemcomitans and showed that this organism is also capable of internalizing AI-2 from the environment via the Lsr system. Further, they demonstrated that the LsrB homologue is required to mediate the complete, AI-2-dependent activation of biofilm formation in this organism (Shao et al., 2007a; Shao et al., 2007b).
Of the bacteria that have a complete genome sequence in the KEGG database as of this writing, there are 16 different species with protein sequences that have greater than 60% sequence identity to the LsrB protein from S. typhimurium. Interestingly, two of these bacterial species, Sinorhizobium meliloti and Rhodobacter sphaeroides, do not have orthologs to the LuxS protein from S. typhimurium. This leads us to hypothesize that although these bacteria do not make their own AI-2 they could use their LsrB homologues to recognize AI-2 produced by other species. Additionally, sequence analysis revealed that S. meliloti also has orthologs to all the proteins of the Lsr systems from the enteric bacteria (Fig. 1B and 1C), except LsrE (which is present in S. typhimurium but not in E. coli and has no known function), suggesting that this operon could be involved in AI-2 internalization in S. meliloti. It should be noted that the LsrF orthologue (SMb20498) has the lowest sequence homology to its S. typhimurium counterpart and is not located in the same operon, raising the doubt as to whether it plays the same role as E. coli and S. typhimurium LsrF.
S. meliloti is a soil bacterium well known for its capacity to establish a symbiotic relationship with legume plants from the genera Medicago, Melilotus and Trigonella. S. meliloti symbiosis is initiated under nitrogen-limiting conditions by the exchange and recognition of specific signals between the plant and the bacteria. S. meliloti has at least two, and in some strains three, quorum sensing systems dependent on homoserine lactone-type autoinducers (Gonzalez and Marketon, 2003). While it has been shown that these species-specific quorum sensing systems regulate functions crucial for symbiosis between this bacterium and its host, they are not expected to facilitate bacterial inter-species quorum sensing. Here, we show that S. meliloti does not produce the inter-species signal AI-2 but does contain a functional AI-2 receptor protein, and furthermore that S. meliloti recognizes the same form of AI-2 previously described for S. typhimurium: (2R,4S )-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (R-THMF). S. meliloti is able to internalize exogenously supplied AI-2 from its environment, and it responds to the AI-2 signal by up-regulating transcription of its Lsr-like operon. By importing and responding to a signal it does not produce, S. meliloti is apparently employing a different strategy for AI-2-based signaling than that employed by previously characterized species possessing the Lsr system, in effect eavesdropping on other bacteria rather than participating in the conversation.
RESULTS
S. meliloti contains an AI-2 Binding Protein
The SMb21016 hypothetical protein from S. meliloti is 72% identical to the LsrB protein from S. typhimurium and, significantly, the gene SMb21016 is located in an operon that includes orthologs to all the genes necessary for AI-2 internalization in S. typhimurium (Fig. 1B and 1C). To demonstrate that the protein encoded by SMb21016, the putative S. meliloti AI-2 receptor gene, is capable of binding AI-2, we cloned and overexpressed the protein in an E. coli strain producing AI-2. The candidate protein was purified and tested for AI-2 binding. Ligand was released from the receptor by thermal denaturation and the denatured protein removed from solution by pelleting. The resulting supernatant containing released ligand was subsequently tested for its ability to induce bioluminescence in a reporter strain of V. harveyi, MM32, which produces light only in the presence of exogenous AI-2.
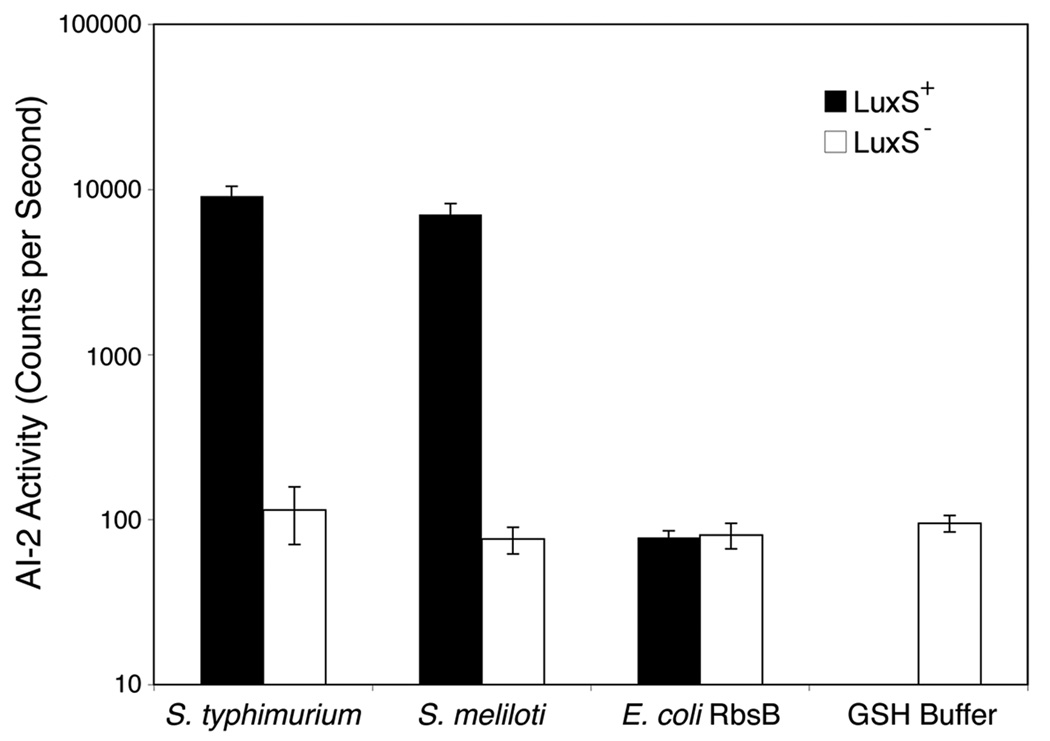
Ligand released from the S. meliloti AI-2 receptor induced a light response similar to that of ligand released from S. typhimurium LsrB (Fig. 2, black bars). To confirm that the light response was specifically induced by AI-2, receptor proteins from S. meliloti and S. typhimurium were overexpressed in FED101, an E. coli strain lacking LuxS and thus unable to produce AI-2. As expected, denaturation of these proteins released no ligand capable of inducing light production in MM32 (Fig. 2, white bars). As a further control, ribose binding protein from E. coli was overexpressed as a GST-fusion and tested for AI-2 binding ability; the protein showed no ability to bind AI-2 in this bioassay (Fig. 2). Thus, the periplasmic binding protein (PBP) encoded by the S. meliloti SMb21016 has the capacity to bind AI-2, and we named it SmLsrB (for S. meliloti LsrB).
Fig. 2. Binding of AI-2 to potential receptor proteins.
Light produced by V. harveyi strain MM32 (LuxN−, LuxS−) was assayed following the addition of ligand released from purified protein expressed in either LuxS+ (black bars) or LuxS− (white bars) E. coli (strains BL21 and FED101, respectively). The E. coli ribose binding protein RbsB and protein-free GSH-buffer were included as negative controls. AI-2 activity is reported as c.p.s. of MM32 bioluminescence. Error bars represent the standard deviations for three independent cultures.
Structure of the S. meliloti AI-2-receptor complex
To identify the form of AI-2 recognized by S. meliloti, we determined the crystal structure of the SmLsrB/AI-2 complex. SmLsrB crystallized with ligand bound, and the structure was solved at 1.8Å resolution by molecular replacement (bound ligand was omitted from the molecular replacement model). The S. meliloti AI-2 receptor has a classic PBP fold, with two α/β domains connected via a three-stranded hinge. The structure is very similar to that of LsrB from S. typhimurium, with an RMSD of 0.6Å. As with other PBPs, the receptor binds the ligand in a cleft between the two domains (Fig. 3A). The structure has been deposited in the PDB with ID code 3EJW.
Fig. 3. Structure of S. meliloti LsrB and its ligand.
A. Ribbon diagram of SmLsrB. The ribbon diagram is colored in rainbow order from N- to C- terminus. The bound ligand and the corresponding electron density are shown.
B. Stereoview of 2Fo-Fc ligand electron density. The DPD isomer R-THMF is shown modeled into non-protein electron density in the receptor binding site.
C. Comparison of the of the S. meliloti and S. typhimurium LsrB AI-2 binding sites. Overlay of the SmLsrB (green) and S. typhimurium LsrB (yellow) binding sites based on overall alignment of the protein structures as calculated by PyMOL. Residue numbers are from the S. meliloti sequence. Dashed red lines indicate potential hydrogen bonds and the interacting residues are labeled in red.
After the structure of the protein was modeled and refined, electron density corresponding to the ligand was easily interpretable. This density was modeled as R-THMF (Fig. 3B), the same form of AI-2 recognized by S. typhimurium (Fig. 1A). The electron density does not make it possible to unequivocally rule out the enantiomer S-THMF as the ligand, but the assignment of R-THMF is consistent with the chemical environment of the binding site. If the R- form of AI-2 is bound, the methyl group on the cyclized ligand is positioned in a hydrophobic pocket, surrounded by residues Phe43, Ala224, Leu268, and Trp269 (Fig. 3C). In contrast, modeling the S- form of the ligand into the binding site leads to the less energetically favorable positioning of a hydrophilic hydroxyl group in this hydrophobic pocket. Further, in the S- form, the nearest potential hydrogen bonding partner for this hydroxyl group is the backbone nitrogen from Ala224, but the interatomic distance of 4.12Å and poor geometry rule out a significant hydrogen bond interaction. When the R- form is modeled into the binding site electron density, this hydroxyl group is 3.35Å from the backbone oxygen of Pro222 (within hydrogen bonding distance) and 3.85Å from the side chain of Gln169 and thus reasonably positioned for a beneficial electrostatic interaction. Thus, we expect that these interactions selectively stabilize the receptor-ligand complex with bound R-THMF. Furthermore, these interactions are consistent with those observed previously in the S. typhimurium LsrB/AI-2 complex; in that case, higher resolution electron density further supported the assignment of R-THMF as the bound form of AI-2. Thus, all the evidence supports the conclusion that R-THMF is the predominant species recognized by S. meliloti as AI-2.
S. meliloti removes exogenously supplied AI-2 from extracellular medium
S. meliloti does not have an ortholog to the luxS gene from S. typhimurium, and thus we expected that this bacterium does not produce AI-2. Accordingly, cell-free culture fluids collected from S. meliloti cultures grown on LBMC do not possess AI-2 activity, as measured by induction of bioluminescence in a V. harveyi BB170 AI-2-reporter assay (Fig. 4A, squares).
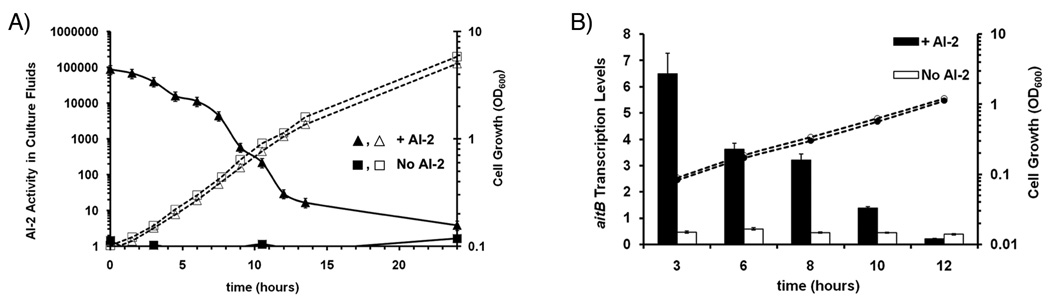
Fig. 4. S. meliloti internalization of exogenously supplied AI-2.
A. Extracellular AI-2 activity in S. meliloti cultures. Wild-type S. meliloti Rm1021 was cultured in LBMC in the presence (triangles) and absence (squares) of in vitro synthesized AI-2 and aliquots were taken at the specified times. AI-2 activity in cell-free culture fluids is reported as fold induction of light production by V. harveyi BB170 (solid lines). Samples with fold inductions above 1000 were diluted in LBMC and values shown were calculated taking account the dilution factor. Cell growth was monitored by optical density (dashed lines).
B. Expression of the S. meliloti aitB transcript in the presence and absence of AI-2. RNA levels in cultures of wild-type S. Meliloti RM1021 grown in the presence (black bars) and absence (white bars) of in vitro synthesized AI-2 were measured using quantitative real-time PCR. aitB transcript levels are reported as fold increase of aitB transcript in relation to the rpsL transcript. Cell growth in the presence (black circles) and in the absence (white circles) of AI-2 was measured by optical density.
We cultured S. meliloti in medium supplemented with in vitro synthesized AI-2 and followed the AI-2 activity in the cell-free culture fluids over time. As shown in Fig. 4A, the initial AI-2 activity due to the addition of synthetic AI-2 results in 100,000-fold induction of bioluminescence in the V. harveyi assay (Fig. 4A, black triangles). However, after 12 hours of growth, the AI-2 activity measured in the cell-free culture fluids decreases drastically to only 20-fold induction. This decrease in extracellular AI-2 activity is consistent with S. meliloti having a functional Lsr operon and, thus, the ability to remove exogenously supplied AI-2 from its environment.
The decrease in apparent AI-2 levels in S. meliloti cultures supplemented with AI-2 could also be due to accumulation of an inhibitor of the V. harveyi AI-2 response or to the degradation of AI-2 by some extracellular factor. To eliminate the first possibility, we added AI-2 to S. meliloti cell-free culture fluids collected after 24 hours of growth (OD=6.0). These samples of cell-free culture fluids were subsequently tested in the V. harveyi bioluminescence assay and found to give high-level light induction (Fig. S1). Thus, no inhibitor was present. To control for the possibility of AI-2 degradation, we collected cell-free culture fluids from S. meliloti grown in the presence of AI-2 for 18 hours (OD=3.6). As shown in Fig. S2A, no AI-2 activity can be detected in these cell-free culture fluids. We then supplemented this cell-free culture fluid with synthetic AI-2 and incubated this sample for 24 hours at 30°C to determine the stability of AI-2 in this cell-free sample. Results from a BB170 bioluminescence assay show that AI-2 activity did not decrease during this incubation period (Fig. S2B). Therefore, we conclude that the decrease of extracellular AI-2 activity observed during growth of S. meliloti results from AI-2 internalization by the cells and not from inhibition of AI-2 detection or degradation of AI-2 by an extracellular factor.
Increased transcription of the lsr-like operon ait occurs in response to AI-2
In S. typhimurium and E. coli, the lsr-operon is induced by the presence of AI-2. Induction of lsr in these bacteria leads to increased production of the AI-2 transport proteins and, thus, a positive feedback loop resulting in increased removal of AI-2 from the extracellular medium. As shown in Fig. 1B and 1C, the S. meliloti orthologs to the proteins of the S. typhimurium Lsr transport system (LsrA, LsrC, LsrD, and LsrB) have a sequence identity above 49% to their orthologs. Thus, we predicted that the S. meliloti lsr-like operon was involved in the removal of AI-2 from culture fluids reported above and we named the S. meliloti lsr-like operon ait (for autoinducer transporter). This name was chosen because the name lsr has already been given to another gene in S. meliloti with an unrelated function (Luo et al., 2005).
We anticipated that S. meliloti would respond to exogenously supplied AI-2 with a similar up-regulation of its ait operon. To test this premise, we used RT-PCR to determine whether the addition of AI-2 to S. meliloti cultures would induce transcription of this operon. As predicted, levels of aitB (SMb21016) mRNA were higher in the culture that was supplemented with synthetic AI-2 than in AI-2-free cultures (Fig. 4B, black and white bars respectively). Moreover, induction of aitB transcription decreases overtime, an observation in accordance with the decrease of extracellular AI-2 during growth shown in Fig. 4A (triangles).
S. meliloti aitK and aitA mutants are impaired in the ability to remove AI-2 from the medium
To verify if the S. meliloti Lsr-like system (Ait) was capable of internalizing AI-2, we constructed a mutant in aitA (previously SMb21019), the S. meliloti gene homologous to lsrA from S. typhimurium (Fig. 1B). In S. typhimurium and E. coli, this gene is predicted to encode the ATP-binding subunit of the Lsr transporter. Comparison of AI-2 activity in cell-free culture fluids of S. meliloti WT cultures with cultures of the aitA mutant shows that the mutant is defective in AI-2 internalization (Fig. 5, diamonds and squares respectively), supporting our prediction that this operon encodes a functional AI-2 transporter that accounts, at least partially, for the observed AI-2 internalization.
Fig. 5. Extracellular AI-2 activity in S. meliloti ait mutants.
Cultures of the following S. meliloti strains, RM1021 (WT, diamonds), MET2000 (aitA, squares) and MET2002 (aitK, triangles), were grown in LBMC with in vitro synthesized AI-2. AI-2 activity in the cell-free culture fluids is reported as fold induction of light production by V. harveyi BB170 (solid lines) and cell growth was monitored by optical density (dashed lines). The average optical density of the three cultures is shown (circles) with the corresponding standard deviation. Fold inductions above 1000 correspond to AI-2 concentrations that saturate the V. harveyi bioassay and are highlighted by the grey shadow
In S. typhimurium and E. coli, the phenotype of the lsrK mutant with respect to AI-2 internalization is even more pronounced than a transport mutant because phosphorylation of AI-2 is required for trapping the signal inside the cell (Taga et al., 2001; Taga et al., 2003; Xavier and Bassler, 2005b). As shown in Fig. 5, this is also true for the S. meliloti mutant in aitK (previously SMb21022) the lsrK orthologue (Fig. 5, triangles). In cultures of the aitK mutant, AI-2 activity persists in the extracellular medium for much longer than in wild type S. meliloti. The fact that aitA transport mutant is less defective than the aitK mutant in AI-2 removal suggests that at least one more transport mechanism for AI-2 exists, a finding consistent with previous results in enteric bacteria (Taga et al., 2001; Taga et al., 2003; Xavier and Bassler, 2005b). Nonetheless, our data indicate that in S. typhimurium, E. coli, and S. meliloti the alternate system(s) for AI-2 uptake is less efficient in AI-2 transport than the Lsr and Ait systems.
We have also tested the ability of the S. meliloti aitA mutant to colonize its host, the plant Medicago sativa, and found that plants inoculated with the aitA mutant or the aitK mutant were indistinguishable from WT-inoculated plants. Additionally, the aitA mutant did not have a competitive defect when co-inoculated with the WT (data not shown). We interpret this result to mean that the S. meliloti Ait system is not essential in this single-species symbiosis process. This result is not surprising since, as we demonstrate in Fig. 4B, the Ait system in S. meliloti is only induced in the presence of exogenously supplied AI-2.
S. meliloti does not grow with AI-2 as a sole carbon source
Some bacteria, such as Variovorax paradoxus, are capable of degrading specific acyl-homoserine lactone autoinducers and using these molecules as a source of nitrogen and energy when grown in minimal medium (Leadbetter and Greenberg, 2000). This led us to test whether, in conditions of low nutrient availability, S. meliloti could similarly use AI-2 as a sole carbon source. We grew cultures of S. meliloti in minimal medium (M9) using AI-2 as a carbon source and compared growth to cultures grown in M9 supplemented with a variety of sugars (glucose, sucrose, and ribose). S. meliloti was able to grow on glucose, sucrose, and ribose as a sole carbon source (at a concentration of 2 mM) but not when the same concentration of AI-2 was provided (Fig. S3). After 72 hours of incubation the OD600 of a culture of S. meliloti in M9 supplemented with 2 mM AI-2 remained as low as the control where no carbon source was added. Additionally, our results (Fig. 4A and Fig. 5) show that in rich medium (LBMC) addition of AI-2 to cultures of S. meliloti WT or ait mutants does not affect the growth rate of these strains. Thus, we have no evidence indicating that S. meliloti can gain any metabolic benefit from internalizing AI-2.
S. meliloti clears AI-2 produced by Erwinia carotovora in co-cultures of these two species
We have shown that S. meliloti is capable of removing exogenously supplied synthetic AI-2 from culture fluids and that the S. meliloti Ait system is involved in this process. Importantly, we also showed that the Ait system is induced only when exogenous AI-2 is supplied to the culture. To test if S. meliloti grown in the presence of an AI-2 producing bacterial species could use the Ait system to remove AI-2 produced by that species, we cultured S. meliloti in the presence of Erwinia carotovora (wild type strain SCC3193). E. carotovora is a Gram-negative plant pathogen that can co-exist with S. meliloti in the rhizosphere of several plants. E. carotovora has a luxS homolog and has been shown to produce AI-2 (Laasik et al., 2006). Accordingly, AI-2 activity was detected in cell-free culture fluids of cultures of E. carotovora and in co-cultures with wild type strains of both S. meliloti and E. carotovora (Fig. 6, triangles and circles respectively) but not when S. meliloti was co-cultured with an E. carotovora luxS mutant (strain SCC6023) incapable of producing AI-2 (Fig. 6, crosses). In assays of mixed cultures of the wild type strains of S. meliloti and E. carotovora, we observed that AI-2 activity in culture cell-free fluids increased for 4 hours and then began to decrease (Fig. 6, circles). After 6 hours, AI-2 activity in this culture was almost undetectable. In contrast, AI-2 activity in cell-free culture fluids remained high when E. carotovora is grown as a pure culture (Fig. 6, triangles) or in mixed cultures of E. carotovora and the S. meliloti aitK mutant (Fig. 6, squares), showing that the disappearance of AI-2 from the extracellular medium in these mixed cultures requires S. meliloti with a functional Ait system.
Fig. 6. Extracellular AI-2 activity in co-cultures of S. meliloti with E. carotovora.
Extracellular AI-2 activity was measured in a pure culture of E. carotovora WT strain SCC3193 (triangles) or co-cultures of the following combinations: S. meliloti (WT) with E. carotovora WT strain SCC3193 (circles),S. meliloti (WT) with E. carotovora luxS strain SCC6023 (crosses), and S. meliloti aitK strain MET2002 with E. carotovora WT strain SCC3193 (squares). All strains were grown in LBMC medium. AI-2 activity in the cell-free culture fluids is reported as fold induction of light production by V. harveyi BB170.
DISCUSSION
As of this writing, approximately 40% of the nearly 800 sequenced bacterial genomes contain the luxS gene (based on genomes with homologues to the luxS gene of S. typhimurium with an e-value smaller than 10−12 (http://www.genome.jp/kegg/)), suggesting that there are a large number of bacterial species capable of producing DPD, the AI-2 precursor. Additionally, luxS/AI-2 has been implicated in the regulation of a variety of niche-specific functions. For these reasons, AI-2 has been proposed to function as a universal bacterial signal. Here we provide the molecular mechanism for AI-2 detection and response in S. meliloti, an organism that lacks the AI-2 synthase and thus is incapable of producing its own AI-2. We have determined the crystal structure of the AI-2/receptor complex in S. meliloti and have shown that transcription of this receptor is dependent on exogenously supplied AI-2, produced either synthetically or by organisms capable of synthesizing AI-2.
This work demonstrates that AI-2 signaling can influence levels of gene expression in non-AI-2 producers, a fact that increases the range of species with the potential to be involved in exchange of, or response to, the AI-2 molecule beyond those that carry the luxS gene. The S. meliloti response to AI-2 emphasizes the role of AI-2 as an inter-species signal; because S. meliloti is incapable of producing AI-2, other organisms are the only source of AI-2 in the environment. Significantly, the S. meliloti case is not an isolated example; in fact, Surette and colleagues have shown (Duan and Surette, 2007) that expression of several genes in Pseudomonas aeruginosa, another organism lacking luxS, is influenced by the presence of AI-2, although the molecular mechanism involved in this process has yet to be defined. We predict that this phenomenon is not limited to these two species, and other species lacking LuxS will be shown to respond to AI-2.
Understanding the role played by AI-2 across species requires understanding of the molecular mechanisms by which different species recognize and respond to AI-2. To date, the AI-2 receptors of the marine bacterium V. harveyi and the human pathogen S. typhimurium have been characterized. Here, by studying AI-2 response in a plant symbiont, we expand our understanding of the molecular mechanism of AI-2 detection into a new environmental niche, the soil. Previous work has shown that S. typhimurium and V. harveyi recognize chemically distinct forms of the AI-2 molecule and that levels of the various forms of AI-2 present in a particular environment are dictated by the chemistry of that environment (Chen et al., 2002; Miller et al., 2004). In this work, we demonstrate that S. meliloti recognizes the same form of AI-2 as the enteric bacterium S. typhimurium despite the fact that these bacteria are usually isolated from chemically different niches (the soil and the human gut).
Previous work has shown that S. typhimurium and E. coli have the ability to internalize AI-2 via their Lsr system, thus removing the molecule from the environment. These species can use this ability to interfere with AI-2 based signaling of other species (Xavier and Bassler, 2005a). Here we show that S. meliloti also has a functional AI-2-inducible Lsr-like system (Ait) capable of removing AI-2 from the environment. S. meliloti colonizes the rhizosphere of several legume plants and therefore it shares its habitat with many AI-2-producing bacterial species. Our results show that S. meliloti can use the Ait system to clear the AI-2 signal produced by E. carotovora, a plant pathogen that can co-exist with S. meliloti in the rhizosphere and that has been reported to regulate virulence by AI-2 quorum sensing (Laasik et al., 2006). Thus, it is reasonable to presume that, like the enteric bacterium, S. meliloti can use the AI-2 internalization system for interference. However, this strategy of interference likely functions somewhat differently for S. meliloti than for the enteric species, since in S. meliloti the ait operon can only be induced in the presence of AI-2 produced by other bacterial species. Thus, unlike other previously characterized bacterial species, a population of S. meliloti cannot up-regulate AI-2 internalization in response to fluctuations in its own population density. Instead, a population of S. meliloti could sense the AI-2 produced by its neighbors, leading to induction of its ait operon and thus interference with the AI-2 mediated behaviors of other species in the vicinity. Moreover, S. meliloti presumably does so without allowing the other species to detect its presence via AI-2 mediated quorum sensing, effectively eavesdropping on its neighbors. It is tempting to speculate that the ability of S. meliloti to interfere with the quorum sensing of plant pathogens that use AI-2 to regulate virulence could be beneficial to the plant, decreasing the virulence of pathogens like E. carotovora. The identification and characterization of the S. meliloti AI-2 dependent Ait system has provided us an excellent tool to begin studying the influence of inter-species bacterial signaling on bacteria-plant interactions, both symbiotic and pathogenic.
It has been argued that some species gain mainly a metabolic benefit from internalization of AI-2 (Winzer et al., 2002b); if this were the case, a non-AI-2 producing species could be acting as a “free-rider” in a mixed-species environment where other species are producing AI-2. Although this remains a possibility, our results indicate that S. meliloti gains no metabolic benefit from metabolizing AI-2, at least under our growth conditions. We did not observe an increase in the growth rate of S. meliloti cultured in the presence of AI-2 in either complex medium or in minimal medium with AI-2 as sole carbon source, nor did the S. meliloti ait mutants show a growth defect in the presence or absence of AI-2.
Some bacteria are capable of degrading acyl-homoserine lactone signals produced by other species (Dong et al., 2000; Leadbetter and Greenberg, 2000; Wang et al., 2007; Zhang et al., 2004). While the producing species use these molecules for species-specific quorum sensing, at least one bacterium, Variovorax paradoxus, is able to use these signal molecules as an energy source (Leadbetter and Greenberg, 2000). Although the benefit derived by V. paradoxus from removing autoinducer signals from the environment might be only metabolic, S. meliloti does not gain a metabolic benefit from internalizing AI-2 and therefore would be expected to gain another advantage. This supports the possibility that S. meliloti is using AI-2 internalization as a means to interfere with the quorum sensing of competitive species.
An alternative hypothesis for the function of the AI-2-response in S. meliloti is that AI-2 is used to distinguish between being in the soil, a mixed-species environment where, presumably, it encounters AI-2 produced by bacteria such as Erwinia or any of several bacillus, and being in its plant host where it exists inside nodules colonized exclusively by a single-species culture of S. meliloti and, thus, in a niche where it will encounter no AI-2. Given that S. meliloti lacks the ability to produce its own AI-2, it is clear that any benefits derived from AI-2 recognition and transport must arise from inter-species interactions.
EXPERIMENTAL PROCEDURES
Protein Production
AI-2 receptor proteins from S. typhimurium and S. meliloti, and RbsB from E. coli were cloned from each species’ genomic DNA (S. typhimurium 14028, S. meliloti Rm1021, E. coli MG1655, respectively) into plasmid pGEX-4T1 for expression as glutathione-S-transferase (GST) fusion proteins. In all cases, the amino-terminal signal peptides, as determined by the program SignalP (Bendtsen et al., 2004), were omitted from the construct. The primers used to PCR amplify the genes (Stfgex1 and Stfgex2 for the S. meliloti AI-2 receptor, Styl1 and Styl2 for S. typhimurium LsrB, and Strbs1 and Strbs2 for E. coli RbsB) are shown in Table S1.
Plasmids were transformed into E. coli strains BL21 and FED101 (BL21, luxS mutant), and protein expression was induced by the addition of 0.1 mM isopropyl β-D-thiogalactopyranoside. After induction, the bacteria were grown for 6 hours at 22°C before harvesting.
Cells were resuspended in 25 mM Tris, pH 8.0, 150 mM NaCl, 1 mM DTT and lysed using a M-110Y Microfluidizer (Microfluidics). The lysates were then clarified by centrifugation and fusion proteins purified by affinity chromatography using glutathione agarose (Sigma-Aldrich). Proteins for luminescence assays were eluted from the affinity resin using 25 mM Tris, pH 8.0, 150 mM NaCl, 1 mM DTT, 10 mM reduced glutathone. Eluted protein was concentrated to approximately 10 mg/ml for the luminescence assay.
To prepare the S. meliloti receptor protein for crystallization, the GST-fusion protein was digested with thrombin for 18 hours at 4°C while still bound to the glutathione agarose. The receptor protein was then eluted from the agarose in resuspension buffer and subsequently swapped into 25 mM HEPES pH 7.0, 1mM DTT using G25 resin (GE Healthcare). The receptor protein was further purified by ion exchange chromatography using a SouceQ column (GE Healthcare) with a gradient from 0 to 350 mM NaCl. As a final purification step, the protein was subjected to size exclusion chromatography on an Superdex 75 column (GE Healthcare), eluting in 25 mM HEPES pH 7.0, 150 mM NaCl, 1mM DTT. The protein was then concentrated to approximately 10 mg/ml.
AI-2 Binding Assay
Ligands were released from purified receptor proteins by heating the protein samples (10 min, 70°C). The denatured protein was then pelleted and the supernatants used in the luminescence assay. For this assay, the V. harveyi strain MM32 (luxN∷Cm, luxS∷Tn5Kan) was used as a reporter. Because this strain has an insertion in the AI-1 receptor (LuxN) it does not respond to autoinducer-1, and since it is a luxS mutant, it does not produce AI-2; thus the strain will only produce light in response to exogenous AI-2 and is effective for discriminating between the presence and absence of AI-2. V. harveyi MM32 was grown for 16 hr in AB medium at 30°C and subsequently diluted 1:5000 into fresh AB medium containing 10% released autoinducer sample or buffer. The bacteria were then grown at 30°C and luminescence measured using a Wallac Victor2 1420 multilabel counter. Bioluminescence produced by MM32 is reported as counts per second (c. p. s.) as measured by the instrument.
Crystallization and Structure Determination
Crystals of the S. meliloti AI-2 receptor protein expressed in E. coli BL21 (LuxS+) were grown via the sitting drop method with a well solution of 0.1 M Tris-HCl pH 8.5, 0.2 M MgCl2, 30% w/v PEG 4000 and developed in approximately two weeks at room temperature. Crystals were frozen in mother liquor, and data were collected at 100K using an R-AXIS-IV image plate detector mounted on a Rigaku 200HB generator. The crystals (P21, a = 57.85, b = 71.49, c = 68.04, β = 98.69) diffracted to 1.8Å resolution. Data were processed using MOSFLM (Leslie, 1992) and CCP4 (CCP4, 1994).
The structure of S. meliloti LsrB (SmLsrB) was solved via molecular replacement with PHENIX (Adams et al., 2002), using LsrB from S. typhimurium (PDB ID: 1TJY) as a search model. The automatically generated partial structure was used as a starting point for building in Coot (Emsley and Cowtan, 2004). The structure was refined using data to 1.8Å with PHENIX. To avoid bias, the ligand was omitted from the search model and not built until the protein structure had been completed and refined. The binding site electron density was clearly interpretable and modeled as R-THMF. A prime-and-switch map was also calculated via PHENIX to confirm the identity of the bound ligand. Refinement parameters for the ligand were generated with the eLBOW module of PHENIX. The final structure contains two copies of SmLsrB, including all residues in the expressed construct (29 – 343), two copies of the bound ligand, and 1113 water molecules. The structure has good geometry (Table 1), with only two residues per chain (Asp118 and Leu268) outside the allowed regions of the Ramachandran plot. Clear density exists for both of these residues. The final Rcryst and Rfree were 0.168 and 0.214 respectively. All molecular images were generated using PyMOL (DeLano, 2002).
Table 1.
Crystallographic data and refinement statistics.
| Data | |
|---|---|
| Resolution (Å) | 1.8 (1.9–1.8) |
| Unique Reflections | 50186 (7394) |
| Rmerge (outer shell) | 0.059 (0.348) |
| Mean I/σI (outer shell) | 15.3 (2.6) |
| Completeness (%) | 98.8 (100) |
| Multiplicity | 2.6 (2.5) |
| Refinement | |
| Rcryst/Rfree | 0.168/0.214 |
| RMS deviation | |
| Bond Length (Å) | 0.007 |
| Bond Angle (°) | 1.007 |
| Dihedrals (°) | 19.074 |
| Average B factor | |
| Non-solvent | 17.20 |
| Waters | 28.74 |
| All atoms | 19.26 |
AI-2 synthesis
DPD protected with cyclohexylidene was synthesized as reported previously (Semmelhack et al., 2005). The protective group was removed with H2SO4 followed by neutralization with potassium phosphate buffer, pH 7 as described in Xavier, 2007 (Xavier et al., 2007).
Bacterial strains and growth conditions
The S. meliloti strains used in this study are derived from the wild-type strain Rm1021 (Meade et al., 1982). To construct the aitA∷pJH104 mutant (MET2000), a 300 bp internal fragment of aitA was amplified by PCR from an S. meliloti Rm1021 colony with the primers P113 and P114 (Table S1), ligated into the suicide plasmid pJH104 at the SpeI and XhoI restriction sites, and transformed into E. coli DH5α. The resulting plasmid was introduced into Rm1021 by triparental mating, and integration of the plasmid was selected by growth on neomycin (0.2 mg/ml). To construct the in-frame deletion of aitK (MET2002), a 750 bp region upstream of the aitK open reading frame was amplified by PCR with primers P117 and P118 (Table S1), and a 500 bp region downstream of aitK was amplified with primers P120 and P121 (Table S1). The two PCR products were ligated in tandem into the plasmid pK18 mob sacB (Schafer et al., 1994) at the BamHI, PstI, and HindIII restriction sites, transformed into E. coli, and introduced into Rm1021 by triparental mating. Neomycin resistant exconjugants were plated on TY with 10% sucrose to select for a second recombination event. Neomycin sensitive colonies were screened by PCR for deletion of aitK. The E. carotovora ssp. carotovora strains used are the wild type SCC3193 (Pirhonen, 1991) and its isogenic luxS∷Cm mutant SCC6063 (Laasik et al., 2006). S. meliloti and E. carotovora strains were grown at 30°C with aeration in Luria–Bertani broth (LB) supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2 (LBMC).
For the studies of S. meliloti growth on different carbon sources, S. meliloti was grown overnight in LBMC at 30ºC. The culture was washed three times in M9 minimal medium (Glazebrook and Walker, 1991) with no carbon source and used to inoculate M9 media with biotin and sucrose, glucose, ribose or AI-2 to a final concentration of 2 mM. As a control, a culture was grown with M9 media and biotin but no carbon source. Cultures were grown at 30ºC with agitation for 72 hours and growth was monitored by optical density (OD600). Each carbon source was tested in duplicate.
AI-2 activity in S. meliloti and E. carotovora cultures
To monitor extracellular AI-2 activity in S. meliloti cultures during growth, overnight cultures were diluted to OD = 0.08 into LBMC medium with and without 80 µM chemically synthesized AI-2. Aliquots were collected at the times indicated and used for measurement of optical density at 600 nm (OD600) and preparation of cell-free culture fluids. The AI-2 activity in cell-free culture fluids was measured using the V. harveyi BB170 (luxN∷Tn5Kan) bioluminescence reporter assay, as described previously (Bassler et al., 1993; Bassler et al., 1994). Cell-free culture fluids were prepared by filtration of liquid cultures (Surette and Bassler, 1998, 1999). The filtered samples were analyzed in duplicate. A similar procedure was used to measure AI-2 activity in E. carotovora cultures, either in single cultures or in co-culture with S. meliloti. When S. meliloti was grown in co-culture with E. carotovora, or when single cultures of E. carotovora were tested, no synthetic AI-2 was supplied.
AI-2 activity in cell-free culture fluids is reported as fold induction of light produced by BB170 and is calculated as the ratio of light produced by BB170 supplemented with sample to light produced by BB170 supplemented with S. meliloti growth medium (LBMC). When required, serial dilutions of the cell-free fluids in LBMC were tested and the values were calculated from the dilution of each sample that resulted in half-maximal induction (approximately 500 fold). (In these experiments, BB170 was used because it allows determination of the fold induction of each sample in relation to the background luminescence when the response is not saturated (Bassler et al., 1993) and is thus appropriate for quantifying the AI-2 in the cell-free fluids. The MM32 assay, used in the AI-2 receptor binding assay, above, is more effective in discriminating between the presence and absence of AI-2.)
Quantitative real-time PCR analysis
To measure the induction of aitB transcription by AI-2 in S. meliloti culture, aliquots were collected throughout growth in the presence or absence of 80 µM AI-2. Cells were collected by centrifugation at 16,000g for 10 min. 50 µL of 5 mg/ml lysozyme was added to each sample, which were then incubated on ice for 5 minutes. Samples were frozen in liquid nitrogen and stored at −80ºC until the RNA was extracted.
S. meliloti RNA was extracted with Trizol reagent (Invitrogen) and chloroform according to the manufacturer’s protocol. RNA was precipitated using isopropanol, washed with 75% ethanol, and diluted in DEPC water. RNA samples were diluted to a concentration of 200 ng/ul and treated with DNase I (Roche). The RNeasy Mini kit from QIAGEN was used to clean the RNA. cDNA was generated in 100-µL reactions, each containing 20 µg of RNA, 5x First Strand Buffer (Invitrogen), 100 mM DTT (Invitrogen), 10 mM dNTPs (ABI), random hexamers (Roche), and SuperScript II reverse transcriptase (Invitrogen). The reverse transcriptase reactions were undertaken in a thermocycler with steps of 10 min at 25°C, 50 min at 42°C, and 15 min at 72°C. Identical reactions were performed without reverse transcriptase enzyme to ensure the absence of genomic DNA contamination. Quantitative RT-PCR reaction mixtures contained 5 µl of cDNA template, 2 µl gene-specific primers, 12.5 µl of SYBR Green Mix (Applied Biosystems), and 5.5 µl H20. For each reaction, 10 µl of reaction mixture were loaded into 384-well optical reaction plates (Applied Biosystems) using a platemate 2×2 automated liquid pipettor (Matrix), with six replicates of each sample. Real-time PCR reactions were carried out on an ABI Prism 7900HT Sequence Detector (Applied Biosystems). Real-time PCR primers were designed using Primer Express 2.0 (ABI Software) and are listed in Table S1. hfq or rpsL transcripts were used as endogenous controls for the reactions, and RNA levels were quantified using absolute quantification (standard curve analysis).
ACKNOWLEDGEMENTS
The work preformed in the laboratory of K.B.X. was supported by Marie Curie International Reintegration grant 031108 and by Fundação para a Ciência e Tecnologia (FCT) grant PTDC/BIA-BCM/73676/2006. C.S.P. was supported by FCT award SFRH/BD/28543/2006. The laboratory of S.T.M. gratefully acknowledges support from grants from the Camille and Henry Dreyfus Foundation, the Merck/AAAS Undergraduate Science Research Program, and NIH grant AI074041. We also would like to thank program FLAD/NSF Proj. 600-10/2006 for sponsoring the stay of S.T.M. in the laboratory of K. X. during his sabbatical. We are grateful to João C. Marques (IGC), Chris Maycock (ITQB), Rita Ventura (ITQB), William Brow (Princeton University), and Martin Semmelhack (Princeton University) for synthesizing and providing us with DPD necessary for this work and to Andres Mäe (Institute of Molecular and Cell Biology) for providing the E. carovotora strains used in this study. We thank Bonnie Bassler and Frederick M. Hughson (Princeton University) for critical reading the manuscript and Graham Walker (MIT) for receiving C.S.P in his laboratory to learn how to work with S. meliloti.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Wright M, Showalter RE, Silverman MR. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. 2002 [Google Scholar]
- Dong YH, Xu JL, Li XZ, Zhang LH. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci U S A. 2000;97:3526–3531. doi: 10.1073/pnas.060023897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K, Surette MG. Environmental regulation of Pseudomonas aeruginosa PAO1 Las and Rhl quorum-sensing systems. J Bacteriol. 2007;189:4827–4836. doi: 10.1128/JB.00043-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Walker GC. Genetic techniques in Rhizobium meliloti. Methods Enzymol. 1991;204:398–418. doi: 10.1016/0076-6879(91)04021-f. [DOI] [PubMed] [Google Scholar]
- Gonzalez JE, Marketon MM. Quorum sensing in nitrogen-fixing rhizobia. Microbiol Mol Biol Rev. 2003;67:574–592. doi: 10.1128/MMBR.67.4.574-592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A. 2007;104:11145–11149. doi: 10.1073/pnas.0703860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie KR, Heurlier K. Establishing bacterial communities by 'word of mouth': LuxS and autoinducer 2 in biofilm development. Nat Rev Microbiol. 2008 doi: 10.1038/nrmicro1916. [DOI] [PubMed] [Google Scholar]
- Laasik E, Andresen L, Mae A. Type II quorum sensing regulates virulence in Erwinia carotovora ssp. carotovora. FEMS Microbiol Lett. 2006;258:227–234. doi: 10.1111/j.1574-6968.2006.00222.x. [DOI] [PubMed] [Google Scholar]
- Leadbetter JR, Greenberg EP. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J Bacteriol. 2000;182:6921–6926. doi: 10.1128/jb.182.24.6921-6926.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;26 [Google Scholar]
- Li M, Villaruz AE, Vadyvaloo V, Sturdevant DE, Otto M. AI-2-dependent gene regulation in Staphylococcus epidermidis. BMC Microbiol. 2008;8:4. doi: 10.1186/1471-2180-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Yao SY, Becker A, Ruberg S, Yu GQ, Zhu JB, Cheng HP. Two new Sinorhizobium meliloti LysR-type transcriptional regulators required for nodulation. J Bacteriol. 2005;187:4562–4572. doi: 10.1128/JB.187.13.4562-4572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade HM, Long SR, Ruvkun GB, Brown SE, Ausubel FM. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- Miller ST, Xavier KB, Campagna SR, Taga ME, Semmelhack MF, Bassler BL, Hughson FM. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell. 2004;15:677–687. doi: 10.1016/j.molcel.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Mok KC, Wingreen NS, Bassler BL. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. Embo J. 2003;22:870–881. doi: 10.1093/emboj/cdg085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirhonen M, Saarilahti H, Karlsson M-B, Palva ET. Identification of pathogenicity determinants of Erwinia carotovora subspecies carotovora by transposon mutagenesis. Mol Plant-Microbe Interact. 1991;4:276–283. [Google Scholar]
- Rader BA, Campagna SR, Semmelhack MF, Bassler BL, Guillemin K. The quorum-sensing molecule autoinducer 2 regulates motility and flagellar morphogenesis in Helicobacter pylori. J Bacteriol. 2007;189:6109–6117. doi: 10.1128/JB.00246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard AH, Palmer RJ, Jr, Blehert DS, Campagna SR, Semmelhack MF, Egland PG, Bassler BL, Kolenbrander PE. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol. 2006;60:1446–1456. doi: 10.1111/j.1365-2958.2006.05202.x. [DOI] [PubMed] [Google Scholar]
- Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- Semmelhack MF, Campagna SR, Federle MJ, Bassler BL. An expeditious synthesis of DPD and boron binding studies. Org Lett. 2005;7:569–572. doi: 10.1021/ol047695j. [DOI] [PubMed] [Google Scholar]
- Shao H, James D, Lamont RJ, Demuth DR. Differential interaction of Aggregatibacter (Actinobacillus) actinomycetemcomitans LsrB and RbsB proteins with autoinducer 2. J Bacteriol. 2007a;189:5559–5565. doi: 10.1128/JB.00387-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Lamont RJ, Demuth DR. Autoinducer 2 is required for biofilm growth of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Infect Immun. 2007b;75:4211–4218. doi: 10.1128/IAI.00402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette MG, Bassler BL. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc Natl Acad Sci U S A. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette MG, Bassler BL. Regulation of autoinducer production in Salmonella typhimurium. Mol Microbiol. 1999;31:585–595. doi: 10.1046/j.1365-2958.1999.01199.x. [DOI] [PubMed] [Google Scholar]
- Sztajer H, Lemme A, Vilchez R, Schulz S, Geffers R, Yip CY, Levesque CM, Cvitkovitch DG, Wagner-Dobler I. Autoinducer-2-regulated genes in Streptococcus mutans UA159 and global metabolic effect of the luxS mutation. J Bacteriol. 2008;190:401–415. doi: 10.1128/JB.01086-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taga ME, Semmelhack JL, Bassler BL. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol Microbiol. 2001;42:777–793. doi: 10.1046/j.1365-2958.2001.02669.x. [DOI] [PubMed] [Google Scholar]
- Taga ME, Miller ST, Bassler BL. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol Microbiol. 2003;50:1411–1427. doi: 10.1046/j.1365-2958.2003.03781.x. [DOI] [PubMed] [Google Scholar]
- Turovskiy Y, Kashtanov D, Paskhover B, Chikindas ML. Quorum sensing: fact, fiction, and everything in between. Adv Appl Microbiol. 2007;62:191–234. doi: 10.1016/S0065-2164(07)62007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. Making 'sense' of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol. 2005;3:383–396. doi: 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- Wang YJ, Huang JJ, Leadbetter JR. Acyl-HSL signal decay: intrinsic to bacterial cell-cell communications. Adv Appl Microbiol. 2007;61:27–58. doi: 10.1016/S0065-2164(06)61002-2. [DOI] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev. 2006;20:2754–2767. doi: 10.1101/gad.1466506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer K, Hardie KR, Williams P. Bacterial cell-to-cell communication: sorry, can't talk now - gone to lunch! Curr Opin Microbiol. 2002b;5:216–222. doi: 10.1016/s1369-5274(02)00304-1. [DOI] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL. LuxS quorum sensing: more than just a numbers game. Curr Opin Microbiol. 2003;6:191–197. doi: 10.1016/s1369-5274(03)00028-6. [DOI] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL. Interference with AI-2-mediated bacterial cell-cell communication. Nature. 2005a;437:750–753. doi: 10.1038/nature03960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J Bacteriol. 2005b;187:238–248. doi: 10.1128/JB.187.1.238-248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier KB, Miller ST, Lu W, Kim JH, Rabinowitz J, Pelczer I, Semmelhack MF, Bassler BL. Phosphorylation and processing of the quorum-sensing molecule autoinducer-2 in enteric bacteria. ACS Chem Biol. 2007;2:128–136. doi: 10.1021/cb600444h. [DOI] [PubMed] [Google Scholar]
- Zhang HB, Wang C, Zhang LH. The quormone degradation system of Agrobacterium tumefaciens is regulated by starvation signal and stress alarmone (p)ppGpp. Mol Microbiol. 2004;52:1389–1401. doi: 10.1111/j.1365-2958.2004.04061.x. [DOI] [PubMed] [Google Scholar]