Abstract
In a series of cyclic peptides based on GS10, an analog of gramicidin S (GS), the ring size was varied from 10 to 16 amino acids. Alternative addition of basic and hydrophobic amino acids to the original GS10 construct generated a variety of even-numbered rings, i.e. GS10 [cyclo-(VKLdYPVKLdYP)], GS12 [cyclo-(VKLKdYPKVKLdYP)], GS14 [cyclo-(VKLKVdYPLKVKLdYP), and GS16 [cyclo-(VKLKVKdYPKLKVKLdYP)] (d stands for D-enantiomers). The odd-numbered analogs (11, 13 and 15-mers) were derived from these four peptides either by addition or deletion of single basic (Lys) or hydrophobic (Leu or Val) amino acids. The resulting peptides, divided into three groups on the basis of peptide ring size (10- to 12-meric, 13- and 14-meric, and 15- and 16-meric) illustrated a diverse spectrum of biological activity correlated to their ring size, degree of β-structure disruption, charge, hydrophobicity, amphipathicity and affinity for lipid membranes. Two of these peptides with potent antimicrobial activities and high therapeutic indexes (4.5–10 folds compared with GS) are promising candidates for development of broad-spectrum antibiotics.
INTRODUCTION
Resurgence of once extinguished microbes, their resistance against existing drugs and reappearance of lethal diseases caused by these “modern microbes” has caused serious concern worldwide.1 Development of potent broad-spectrum antimicrobial peptides with low toxicity against mammalian cells has been a challenge and an important component of novel drug design.2,3 The main barrier in development of novel antimicrobial peptide drugs is the complex mechanism of their biological activity. Mammalian and bacterial cell membranes have been considered as the primary, if not the only, target of antimicrobial peptides. Strong and destructive interaction of peptides with mammalian membranes, such as erythrocytes, is toxic and therefore should be avoided in the design of effective antimicrobial peptides.
In our previous studies on cyclic cationic antimicrobial peptides, we have chosen a naturally found decameric peptide, gramicidin S (GS), as the principal peptide construct in the systematic design of antimicrobial peptides with enhanced antimicrobial and reduced hemolytic activities.4–8 GS, a cyclic cationic peptide, is biosynthesized in the Bacillus brevis and has been widely used as a topical antibiotic.9 GS structure is amphipathic and composed of a double-stranded antiparallel β-sheet connected by a pair of type II' β-turns.10–12 Despite its potent activity against a broad range of microbes, which include Gram-positive and Gram-negative bacteria and fungi, GS is highly hemolytic against mammalian erythrocytes.13 It has been shown that GS and GS-like cationic peptide antibiotics interact with both mammalian and microbial cell membranes to change or destroy the membrane integrity causing lysis, which could lead to cell death. Interaction with the microbial membranes may not be the only mechanism for the biological activity of GS or GS-like antimicrobial peptides. Generally, the mechanism of biological activity of antimicrobial peptides can include inhibitory interactions with the components of metabolic pathways and immune systems of organisms.14 Specifically, it has been reported that GS interacts with other biomolecules, such as carbohydrates15 and enzyme components of metabolic pathways.16
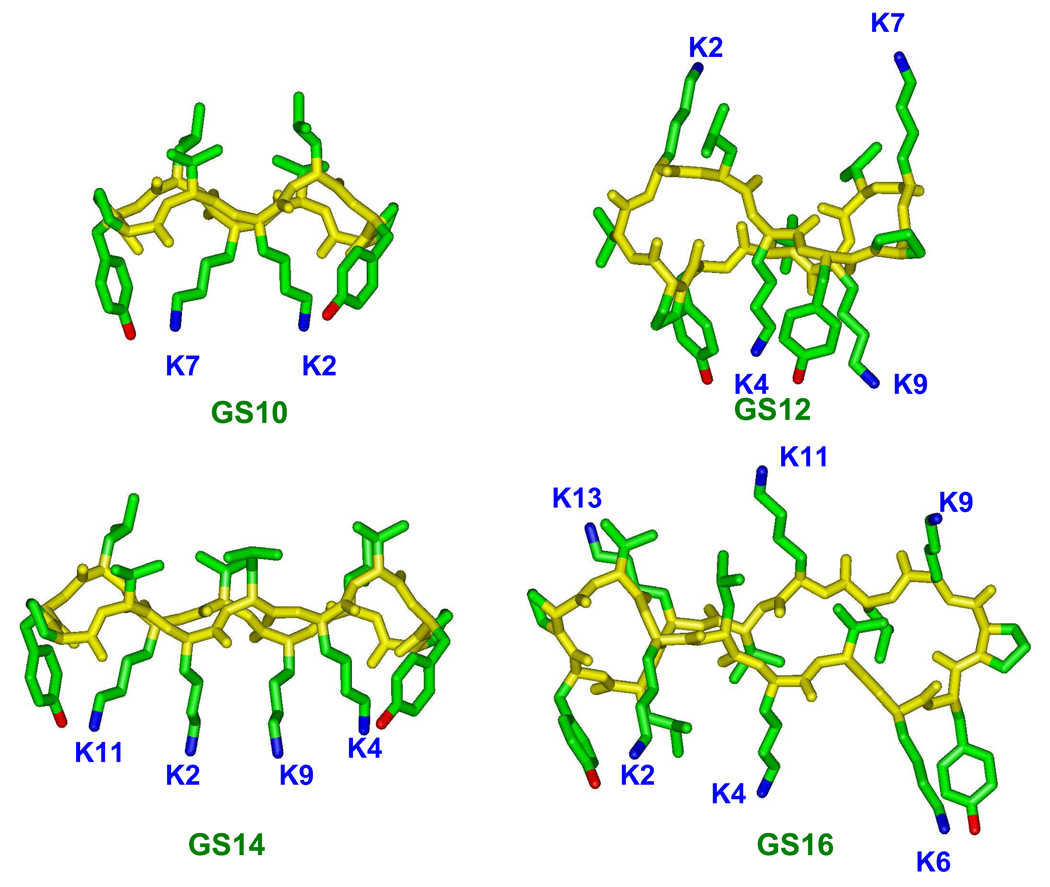
We have previously reported that amphipaticity of cyclic cationic peptides could be utilized in dissociating their antimicrobial and hemolytic activities5 and have also studied the detailed biophysical properties and mechanism of interaction of these peptides with biological membranes.17–20 In this comprehensive study, we investigate the effect of ring size, amphipathicity and interaction with mammalian and bacterial model membranes on the biological activity of 10- to16-meric cyclic cationic peptides (13 peptides in total, Table 1). The original construct for the peptide series in Table 1 is GS10, a D-Tyr- and Lys-containing analog of GS (Figure 1 and Table 1). Among these peptides, GS10 and GS14 have positively-charged residues on one face (2 and 4 charges, respectively) and hydrophobic residues on the opposite face of the molecule (amphipathic structure) and possess the highest β-sheet-β-turn (β-structure) content (Figure 1). On the other hand, GS12 and GS16 have evenly-distributed charged residues on both faces of the molecule (4 and 6 charges in total, with 2 and 3 charges on each side, respectively) (non-amphipathic structure) and possess the lowest β-structure content (Figure 1). Odd-numbered cyclic peptides were derived from these four peptides either by addition or deletion of single basic (Lys) or hydrophobic (Leu or Val) amino acids, and had a variety of conformations, positive charges and amphipathicities.
Table 1.
Peptide sequencesa, molecular massb, retention timec, relative hydrophobicityd, and hemolytic activitye
| Peptide (# ofAA) | Ch/Hypf | Sequence | MW(Da): c/m | RT (min) | [ΔGWO] | [ΔGWIF] | [ΔΔGIFO] | Hemolytic activity % |
|---|---|---|---|---|---|---|---|---|
| GS (10) | 2/4 | Cyclo-(VOLFP5VOLFP10) | 1140.7/1140.8 | 33.5 | −0.096 | −0.036 | −0.060 | 100 (1) |
| GS10 (10) | 2/4 | Cyclo-(VKLYP5VKLYP10) | 1200.7/1200.8 | 30.3 | 0.104 | 0.002 | 0.102 | 100 (1) |
| GS12 (12) | 4/4 | Cyclo-(VKLKYP6KVKLYP12) | 1456.9/1457.2 | 22.9 | 0.55 | 0.17 | 0.39 | 5.1(19.6) |
| GS12-K7(11) | 3/4 | Cyclo-(VKLKYP6*VKLYP11) | 1328.8/1330.0 | 24.4 | 0.35 | 0.092 | 0.26 | 4.9 (20.4) |
| GS12-K4(11) | 3/4 | Cyclo-(VKL*YP5KVKLYP11) | 1328.8/1328.9 | 25.7 | 0.35 | 0.092 | 0.26 | 8.6(11.4) |
| GS12-L3(11) | 4/3 | Cyclo-(VK*KYP5KVKLYP11) | 1343.8/1345.0 | 19.1 | 0.72 | 0.23 | 0.48 | 3.1(32.2) |
| GS14 (14) | 4/6 | Cyclo-(VKLKVYP7LKVKLYP14) | 1669.1/1670.3 | 30.9 | 0.35 | 0.11 | 0.24 | 100 (1) |
| GS14-V5 (13) | 4/5 | Cyclo-(VKLK*YP6LKVKLYP13) | 1570.0/1570.3 | 25.8 | 0.41 | 0.11 | 0.30 | 20.2 (4.9) |
| GS14-K4 (13) | 3/6 | Cyclo-(VKL*VYP6LKVKLYP13) | 1541.0/1540.3 | 28.7 | 0.16 | 0.04 | 0.12 | 78.2(1.3) |
| GS14-L3 (13) | 4/5 | Cyclo-(VK*KVYP6LKVKLYP14) | 1556.0/1556.2 | 26.1 | 0.48 | 0.16 | 0.32 | 15.7 (6.4) |
| GS16 (16) | 6/6 | Cyclo-(VKLKVKYP8KLKVKLYP16) | 1925.3/1925.6 | 22.8 | 0.66 | 0.22 | 0.44 | 12.7 (7.9) |
| GS16-K6 (15) | 5/6 | Cyclo-(VKLKV*YP7KLKVKLYP15) | 1797.2/1797.5 | 25.9 | 0.52 | 0.17 | 0.35 | 82 (1.2) |
| GS16-V5 (15) | 6/5 | Cyclo-(VKLK*KYP7KLKVKLYP15) | 1826.2/1826.5 | 21.6 | 0.73 | 0.23 | 0.50 | 7.3 (13.7) |
| GS16-K4 (15) | 5/6 | Cyclo-(VKL*VKYP7KLKVKLYP15) | 1797.2/1797.5 | 22.7 | 0.52 | 0.17 | 0.35 | 9.6(10.4) |
O represents ornithine; underlined amino acids (AAs) represent the D-enantiomers; the “*” symbol shows the missing amino acids compared with the parent peptide in bold letters.
Calculated (c), and measured (m) molecular masses (MW) of peptides, using electrospray mass spectrometry. Molecular masses were measured as [M+H]+.
Retention times (RT) on the RP-HPLC Zorbax 300 SD C8 (150 × 2.1 mm ID, 5 µm particle size, 300 Å pore size, Rockland Technologies, Wilmington, DE) column. Solvents A (water+0.05%TFA) and B (acetonitrile+0.05%TFA) were used at a flow rate of 0.2 mL/min with a gradient from 100%A ⟶100%B in 40 minutes.
Relative free energy of transfer of the peptides from water to octanol (ΔGWO), water to POPC bilayer interface (ΔGWIF), and the difference between the two (ΔΔGIFO) were calculated on the basis of whole-residue hydrophobicity scales.26 The free energy of transfer of peptide bonds to octanol and the effect of the cyclic peptide backbone structure were not considered in these calculations. “W” stands for water; “O” stands for octanol; “IF” stands for interfacial; “IFO” stands for interfacial to octanol; All reported values in brackets are total free energy of transfer per residue. Free energies are in kcal/(mol.nr), where “nr” is the “number of residues” in peptides.
Percentage of the hemolysis of human red blood cells at 100 µg/ml peptide concentration in 24 h at 37 °C. Numbers in parentheses represent the fold reduction (improvement) of peptide hemolytic activity relative to 100% hemolysis (1 in parenthesis). These numbers are calculated as 100 / % hemolytic activity.
“Ch/Hyp” is the ratio of the number of charged residues (Ch) (Lys or Orn) to the number of hydrophobic residues (Hyp) (Leu and Vol) in peptides.
Figure 1.
Molecular models for even-numbered ring cyclic cationic peptides. GS10 and GS14 are β-sheet-β-turn amphipathic peptides and GS12 and GS16 are non-amphipathic peptides with unordered disrupted β-sheet-β-turn structures.
EXPERIMENTAL
Materials
Gramicidin S was purchased from Sigma (St. Louis, MO, U.S.A.) and further purified by reversed-phase high performance liquid chromatography (RP-HPLC). As previously described, all other peptides were synthesized by solid-phase procedures using Boc-Chemistry and Boc-Pro-PAM resin to initiate the synthesis.5,6 Briefly, HBTU-HOBT coupling reagents were used to activate C-terminus of the amino acids. Lys side-chains were formyl-protected throughout the syntheses. Linear peptides were cleaved from the resin and purified by RP-HPLC. Purified linear peptides were then cyclized in a head-to-tail manner (Pro at the C-terminus) using BOP, HOBt and DIEA in DMF. Finally, Lys residues of the cyclized peptides were deprotected in dilute methanolic HCl at 40 °C. Peptides were purified by RP-HPLC and their molecular masses were determined by electrospray mass spectrometry (Table 1). All peptides were of ≥95% purity as verified by analytical RP-HPLC. Concentrations of the peptides in aqueous stock solutions were determined by amino acid analysis (± 5% error). Phospholipids, 1-palmitoyl-2-oleoyol-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyol-sn-glycero-3-phosphoethanolamine (POPE) and 1-palmitoyl-2-oleoyol-sn-glycero-3-[phosphor-rac-1-glycerol] (sodium salt) (POPG) were from Avanti Polar (Alabaster, AL, U.S.A.). All other chemicals were of reagent grade and used as received without further purification.
Preparation and characteristics of large unilamellar vesicles
Large unilamellar vesicles (LUVs) were prepared as reported previously.6,18,19 In brief, chloroform solutions of the lipids, POPC and POPE/POPG (7/3 molar ratio) were dried under a mild nitrogen flush to form a thin film, which was further dried overnight under reduced pressure. The lipid film was then hydrated with a buffer solution containing 10 mM Tris, 150 mM NaF and 0.1 mM EDTA, at pH 7.4. Obtained multilamellar vesicle dispersions were then freeze-thawed a few times and extruded through a 100 nm filter in a LiposoFast apparatus (Avestin Inc., Ottawa, Canada).21 Prepared unilamellar vesicles (average diameter of ~ 80 nm) were stable in dark at room temperature and for several days at 4 °C. Lipid concentrations were measured as described previously.6
Two phospholipid vesicle systems were utilized for this study. The first system consisted of POPC vesicles that can be considered as a model for mammalian red blood cell membranes. The second system was composed of POPE and POPG phospholipids ([PE]/[PG]= 7/3) to model the negatively-charged bacterial membranes. These two lipid bilayer systems were in the liquid crystalline state, which is compatible with biological membranes.
Instrumentation
Peptides were analyzed on a Hewlett Packard 1100 Chromatograph (Agilent Technologies, Santa Clara, CA, U.S.A.) at 70 °C. Retention times were measured on a RP-HPLC Zorbax 300 SD C8 (150 × 2.1 mm ID, 5 µm particle size, 300 Å pore size, Rockland Technologies, Wilmington, DE) column. Solvents A (water+0.05%TFA) and B (acetonitrile+0.05%TFA) were used at a flow rate of 0.2 mL/min with a gradient from 100%A →100%B in 40 minutes.
Far-UV CD spectra were measured on a Jasco 720 spectropolarimeter (Tokyo, Japan). Ellipticities are reported as mean residue ellipticity, [θ]MRE. Spectra were measured in buffer solutions and phospholipid vesicle dispersions in buffer. The buffer solution was composed of Tris (10 mM), NaF (150 mM) and EDTA (0.1 mM), at pH 7.4. All measurements were in quartz cells with 0.1 cm pathlength, at 25 °C. Sodium fluoride was used in buffer to reduce the high noise-to-signal levels of chloride ions at wavelengths below 200 nm.
Molecular modelling of peptides was based on the global minimization of peptide structures, using the Insight II software (Accelrys, San Diego, CA, U.S.A.) on a Silicon Graphics workstation. 5,6
Antimicrobial and hemolytic activity assays
Microbial strains, both bacteria and fungi, were prepared as described previously. 5–7 Briefly, minimal inhibitory concentrations (MICs) were measured using a standard microtitre dilution method in Luria Broth no salt medium utilizing the same bacterial strains. MICs were determined as the lowest peptide concentration that inhibited growth after 24 h at 37° C.
Hemolytic activity of peptides was measured in saline in the presence human erythrocytes as described,5–7 and reported as the percentage of hemolysis at 100 µg/mL peptide concentrations in 24 h at 37°C.
RESULTS
For a systematic evaluation of the relation between structure and biological activity, the 10- to 16-meric cyclic peptides exhibited in Table 1 are divided into three groups on the basis of their ring size and amino acid composition (number of charged and hydrophobic residues). The first group includes GS10, GS12 and its three 11-meric analogs (GS {10–12} group). GS is also included in this group for comparison. The second group includes GS14 and its three 13-meric analogs (GS {13,14} group). The third group is composed of GS16 and its three 15-meric analogs (GS {15,16} group). Structures of the peptides with even-numbered rings are shown in Figure 1. These structures are based on an NMR study of the periodicity of the β-sheet content of cyclic gramicidin S analogs.22 Stereochemical considerations23 and experimental results22 both confirm that cyclic peptides with 2(2n+1) residues (n=1,2,3,…)(GS, GS10 and GS14 in Table 1 and Figure 1) can form amphipathic β-sheet ring structures, with hydrophobic residues (Val, Leu) on one face and hydrophilic residues (Lys or Orn) on the other face of the ring. On the other hand, cyclic peptides composed of 4n residues (n=2,3,…) (GS12 and GS16 in Table and Figure 1) can only form unordered non-amphipathic conformations (distorted β-sheets), with a distribution of hydrophobic and hydrophilic residues on both faces of the ring. Odd-numbered cyclic peptides of Table 1 (GS11, GS13 and GS15 peptides) contain intermediate conformations, between β-sheet and unordered, with different degrees of β-structure (β-sheet-β-turn) content and amphipathicity. A detailed study of structure-function relationships considering the β-structure content, amphipathicity and biological activity in the 10- to 16-meric cyclic peptides will be described in the following experimental results.
Biological Activities and Conformation of GS {10–12} Group
Peptides of this group, their retention times (RTs) and their charged-to-hydrophobic residue ratios (ch/hyp) are listed in Table 1. Odd-numbered ring 11-meric peptides of this group were generated by deleting charged (GS12-K4 and GS12-K7) or hydrophobic (GS12-L3) residues from GS12.
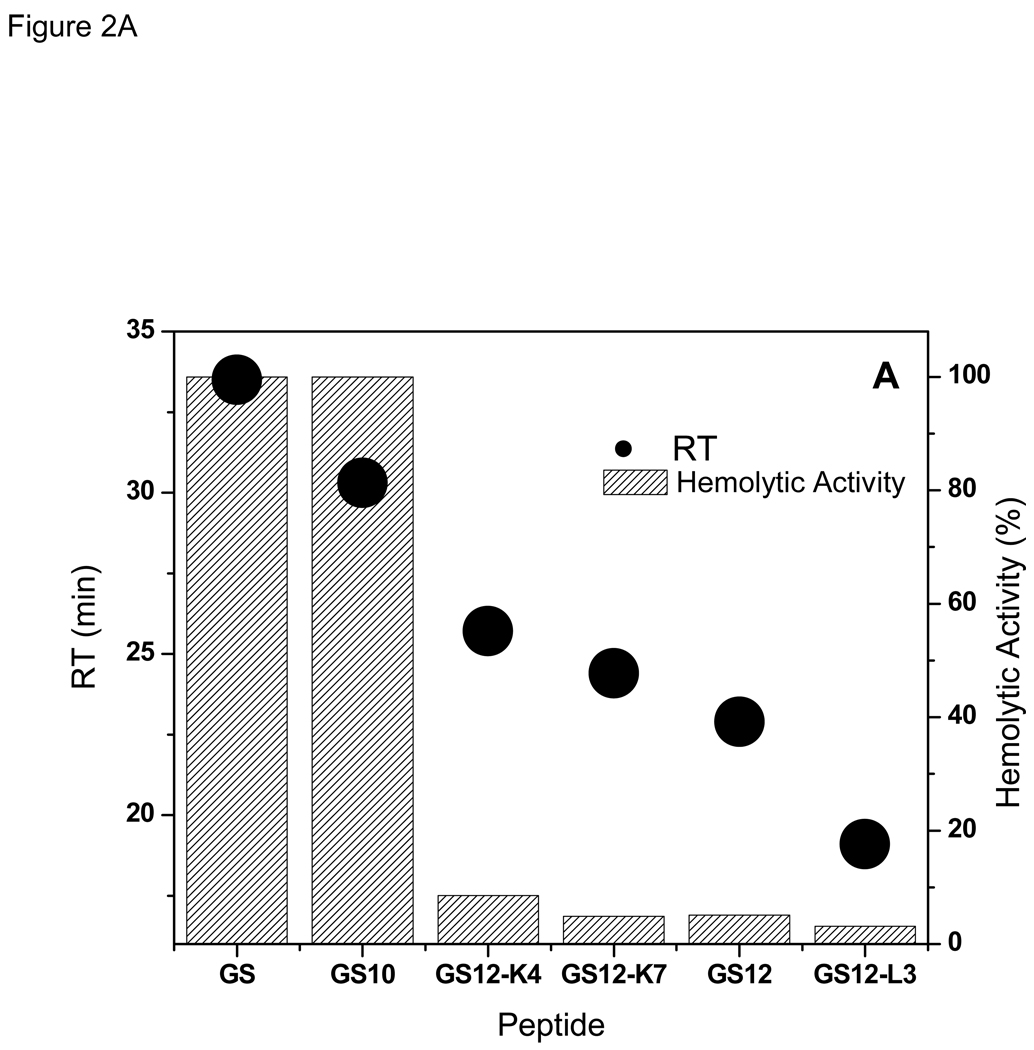
The direct relation between the hemolytic activity and RTs for GS {10–12} peptides is exhibited in Figure 2A and Table 1. GS data is also shown for comparison. Retention times represent the degree of interaction of the hydrophobic surface of the peptides and the stationary phase of the reversed-phase HPLC column: the longer the RT, the higher the affinity of the peptide for the stationary phase.24 As seen in this Figure, decrease in RTs is accompanied by decrease in hemolytic activity. The β-sheet containing peptide GS10 (Figure 1), as well as GS, had the strongest hemolytic activity and highest RT, and the less- or non-amphipathic peptides of this group had lower retention times and weaker hemolytic activites.
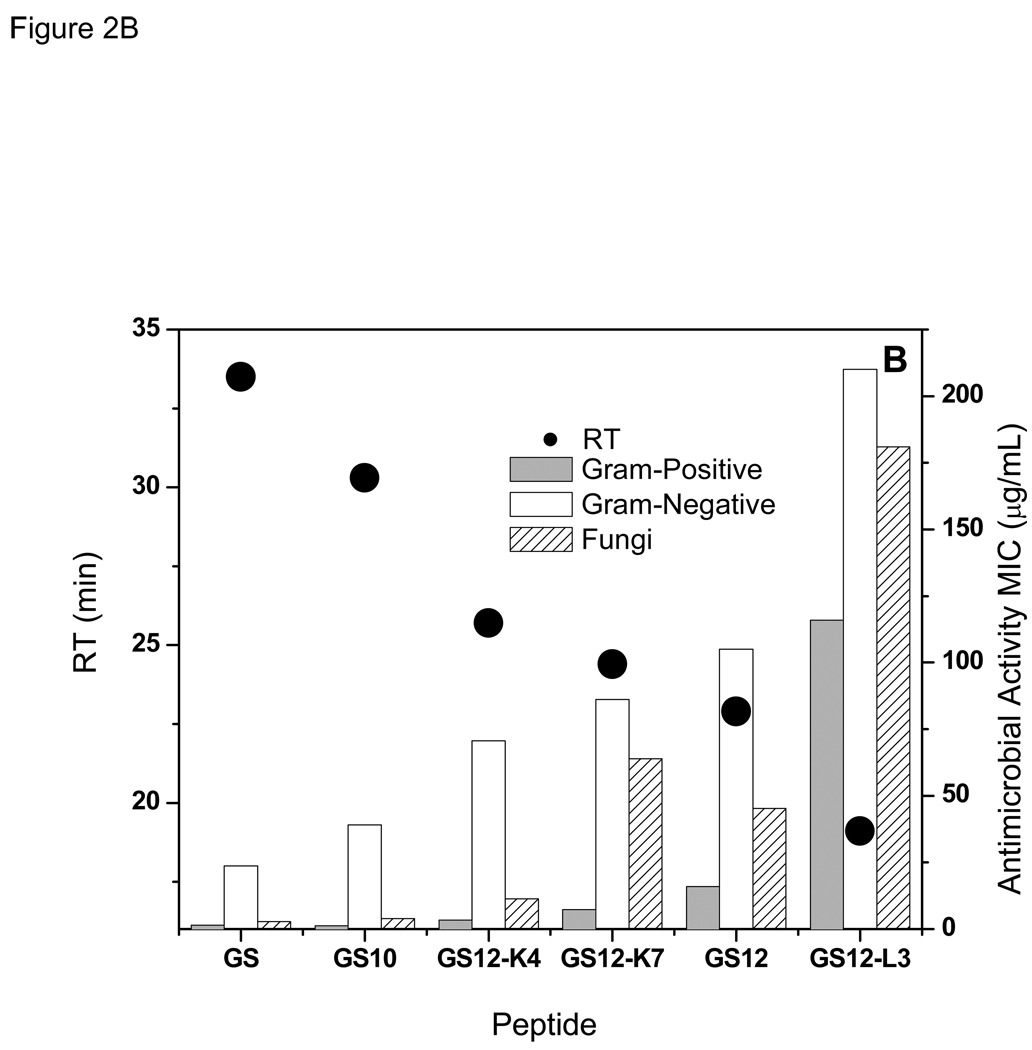
Figure 2.
Relation between retention times with hemolytic (A), and antimicrobial (B) activities of the peptides in GS {10–12} group. GS data is shown for comparison.
As can be observed in Figure 2B and Table 2 and Table 3, there is also a direct relation between the antibacterial activities of the peptides in GS12 {10–12} group and their RTs- the higher the RT the stronger the activity (lower MIC values) against both Gram-positive and Gram-negative bacteria. On average, activities against Gram-positive bacteria were much higher than Gram-negative bacteria. Similar to antibacterial activities, antifungal activities of the peptides of this group decreased with decrease in their retention times (Figure 2B and Table 4) -an exception was GS12, which had a lower RT but slightly stronger antifungal activity compared with GS12-K7.
Table 2.
Activity of peptides against Gram-positive bacteria a
| Peptide | E. faecalis (µg/mL) |
S. aureus (µg/mL) |
S. epidermis (µg/mL) |
S. mitis (µg/mL) |
S. pneumoniae (µg/mL) |
S. pyrogenes (µg/mL) |
C. jeikeium (µg/mL) |
Mean activity b (µg/mL) |
|---|---|---|---|---|---|---|---|---|
| GS | 2 (50.0) | 2 (50.0) | 2 (50.0) | 0.5 (200) | 1 (100) | 0.5 (200) | 4 (25.0) | 1.4 (74.0) |
| GS10 | 2 (50.0) | 2 (50.0) | 2 (50.0) | 1 (100) | 1 (100) | 0.5 (200) | 1 (100) | 1.2 (82.0) |
| GS12 | 64(31.0) | 64(31.0) | 64(31.0) | 2 (980) | 16 (122) | 2 (980) | 16 (122) | 16.0 (122) |
| GS12-K7 | 32 (64.0) | 64 (32.0) | 16 (128) | 2 (1020) | 8 (255) | 1 (2040) | 2 (1020) | 7.2 (281) |
| GS12-K4 | 8(145) | 16 (73.0) | 16 (73.0) | 1(1160) | 2(581) | 0.5 (2330) | 2(581) | 3.3 (354) |
| GS12-L3 | 256 (12.0) | 256 (12.0) | 256 (12.0) | 64 (50.0) | 256 (12.0) | 8 (403) | 128 (25.0) | 115.9(28.0) |
| GS14 | 32 (3.0) | 32 (3.0) | 16 (6.0) | 2 (50.0) | 4 (25.0) | 1(10) | 8 (12.0) | 7.2(14.0) |
| GS14-V5 | 4 (124) | 4 (124) | 4 (124) | 0.5 (990) | 0.5 (990) | 0.125(3960) | 2 (248) | 1.2 (406) |
| GS14-K4 | 4 (32.0) | 4 (32.0) | 4 (32.0) | 1 (128) | 0.5 (256) | 0.5 (256) | 4 (32.0) | 1.8(71.0) |
| GS14-L3 | 8 (80.0) | 4 (159) | 2(318) | 0.5 (1270) | 0.5 (1270) | 0.5 (1270) | 2(318) | 1.5 (429) |
| GS16 | 4(197) | 8 (98.0) | 2 (394) | 2 (394) | 1 (787) | 0.5 (1570) | 1 (787) | 1.8 (435) |
| GS16-K6 | 2 (61.0) | 2 (61.0) | 2 (61.0) | 0.5 (244) | 0.5 (244) | 0.5 (244) | 2 (61.0) | 1.1(110) |
| GS16-V5 | 8(171) | 64 (21.0) | 8(171) | 8(171) | 2 (685) | 0.5 (2740) | 8(171) | 5.9 (230) |
| GS16-K4 | 4 (260) | 32 (32.0) | 16 (65.0) | 4 (260) | 1 (1040) | 0.25 (4170) | 1 (1040) | 3.0 (350) |
Numbers in parentheses are the “therapeutic indices (TIs)”, calculated as 100×(100 /% hemolytic activity) / (antimicrobial activity). Best TI values for different bacterial strains and the peptide with best overall TI are highlighted.
Geometric Mean of MIC values (after 24 h) against Gram-positive bacteria. Geometric mean for “n” numbers: .
Table 3.
Activity of peptides against Gram-negative bacteria a
| Peptide | A. caloaceticus (µg/mL) | E. cloacae (µg/mL) | E. coli (µg/mL) | K. pneumoniae (µg/mL) | P. aeruginosa (µg/mL) | S. maltophilia (µg/mL) | S. marcescens (µg/mL) | Mean Activity b (µg/mL) |
|---|---|---|---|---|---|---|---|---|
| GS | 8 (12.0) | 32 (3.0) | 16 (6.0) | 16 (6.0) | 32 (3.0) | 8 (12.0) | 256 (0.3) | 23.8 (4.0) |
| GS10 | 4 (25.0) | 64 (2.0) | 32 (3.0) | 32 (3.0) | 128(1.0) | 16 (6.0) | 256 (0.3) | 39.0 (2.0) |
| GS12 | 16 (122) | 256 (8.0) | 64(31.0) | 64(31.0) | 256 (8.0) | 128 (15.0) | 256 (8.0) | 105.0(19.0) |
| GS12-K7 | 8 (255) | 256 (8.0) | 64 (32.0) | 64 (32.0) | 256 (8.0) | 64(31.0) | 256 (8.0) | 86.1 (24.0) |
| GS12-K4 | 8(145) | 256 (4.0) | 64 (18.0) | 32 (36.0) | 256 (5.0) | 32 (36.0) | 256 (4.0) | 70.7 (16.0) |
| GS12-L3 | 64 (50.0) | 256 (13.0) | 256(13.0) | 256(13.0) | 256(13.0) | 256 (13.0) | 256(13.0) | 210.0(15.0) |
| GS14 | 16 (6.0) | 256 (0.4) | 128(1.0) | 128(1.0) | 64 (2.0) | 64 (2.0) | 256 (0.4) | 95.1(1.0) |
| GS14-V5 | 2 (248) | 128 (4.0) | 16(31.0) | 8 (62.0) | 64 (8.0) | 16(31.0) | 256 (2.0) | 26.3 (19.0) |
| GS14-K4 | 4 (32.0) | 256 (0.5) | 64 (2.0) | 64 (2.0) | 256 (0.5) | 32 (4.0) | 256 (0.5) | 70.7 (2.0) |
| GS14-L3 | 2(318) | 128 (5.0) | 16 (4.0) | 8 (80.0) | 128 (5.0) | 16 (40.0) | 256 (2.5) | 29.0 (22.0) |
| GS16 | 2 (394) | 64 (12.0) | 4(197) | 4(197) | 64 (12.0) | 32 (25.0) | 256 (3.0) | 19.5 (40.00) |
| GS16-K6 | 2 (61.0) | 64 (2.0) | 8 (15.0) | 8 (15.0) | 128(1.0) | 4 (30.0) | 256 (0.5) | 19.5 (6.0) |
| GS16-V5 | 2 (685) | 256 (5.0) | 32 (43.0) | 8(171) | 128(11.0) | 64 (21.0) | 256 (5.0) | 43.1 (32.0) |
| GS16-K4 | 2(521) | 256 (4.0) | 16 (65.0) | 8(130) | 256 (4.0) | 8(130) | 256 (4.0) | 32.0 (32.0) |
Numbers in parentheses are the “therapeutic indices (TIs)”, calculated as 100×(l00 /% hemolytic activity) / (antimicrobial activity). Best TI values for different bacterial strains and the peptide with best overall TI are highlighted.
Geometric Mean of MIC values (after 24 h). For the definition of geometric mean, please see the footnote “b” inTable 2.
Table 4.
Activity of peptides against fungi a
| Peptide | C. albicans (µg/mL) | C. neoformans (µg/mL) | Mean activityb (µg/mL) |
|---|---|---|---|
| GS | 8 (12.0) | 1 (100) | 2.8(35.0) |
| GS10 | 16 (6.0) | 1 (100) | 4.0 (25.0) |
| GS12 | 128(15.0) | 16 (122) | 45.2 (43.0) |
| GS12-K7 | 128 (16.0) | 32 (12.0) | 64.0 (32.0) |
| GS12-K4 | 64 (18.0) | 2(581) | 11.3(103) |
| GS12-L3 | 256(13.0) | 128 (25.0) | 181.0(18.0) |
| GS14 | 64 (2.0) | 2 (50.0) | 11.3(9.0) |
| GS14-V5 | 32(15.0) | 1 (500) | 5.7 (88.0) |
| GS14-K4 | 32 (4.0) | 1 (128) | 5.7 (23.0) |
| GS14-L3 | 16 (40.0) | 1 (637) | 4.0(159) |
| GS16 | 64 (12.0) | 64 (12.0) | 64.0 (12.0) |
| GS16-K6 | 32 (4.0) | 1 (122) | 5.7 (22.0) |
| GS16-V5 | 64 (21.0) | 64 (21.0) | 64.0 (21.0) |
| GS16-K4 | 64 (16.0) | 8(130) | 22.6 (46.0) |
Numbers in parentheses are the “therapeutic indices (TIs)”, calculated as 100×(100 / % hemolytic activity) / (antimicrobial activity). Best TI values for different fungal strains and the peptide with best overall TI are highlighted.
Geometric Mean of MIC values (after 24 h). For the definition of geometric mean, please see the footnote “b” in Table 2
To investigate possible relations between the biological activity of peptides and their conformation, CD spectra of the peptides were measured in three different milieus –buffer, PC vesicles (erythrocyte model), and PE/PG vesicles (bacterial membrane model).
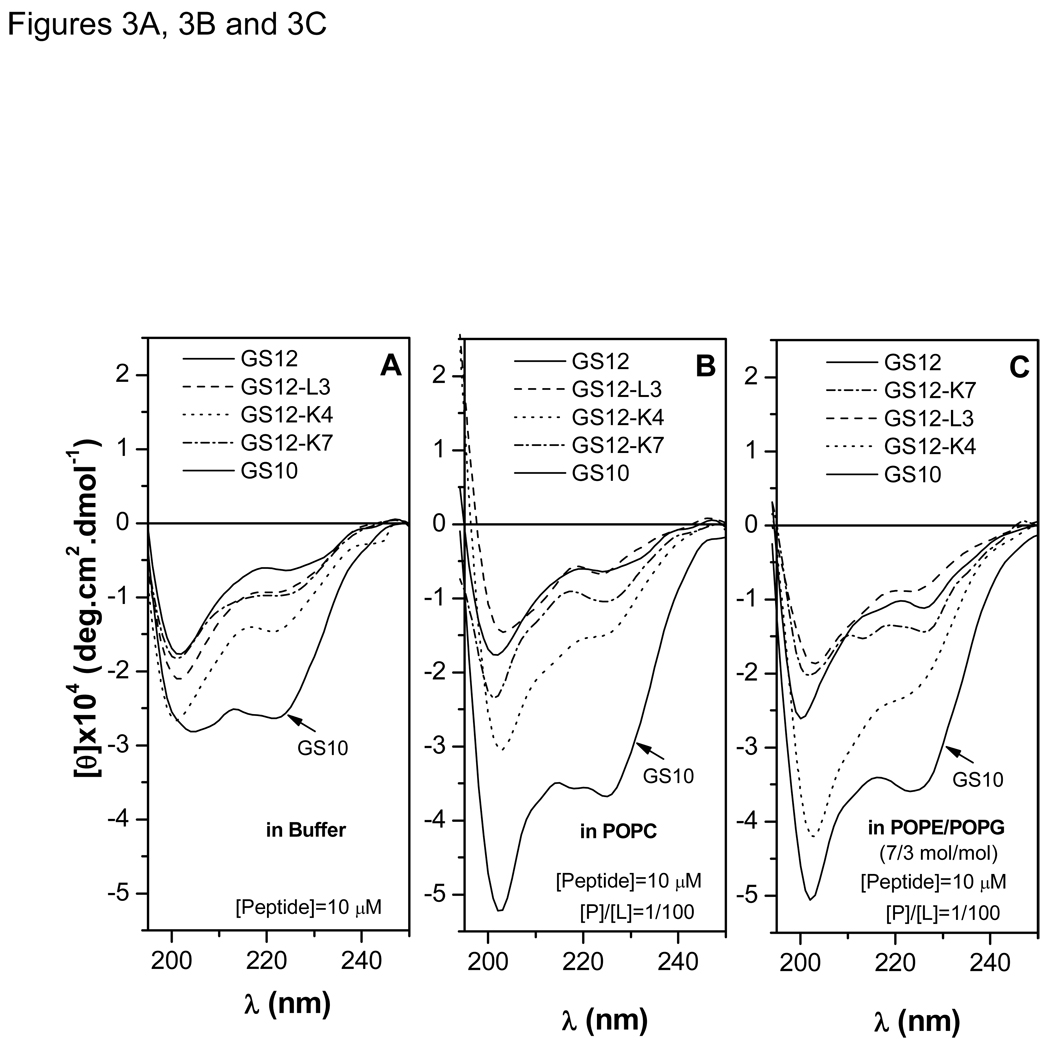
CD spectra for the GS12 {10–12} group are exhibited in Figure 3A to 3C. Figure 3A shows a β-sheet-β-turn spectrum for GS10, with two negative maxima at 205 and 222 nm. The positive maximum of the spectra (185–188 nm) is not shown. In comparison with GS10 spectrum, substantial decrease in the negative maxima at 222 nm (45–75%) and blue shift of the lower wavelength negative maxima are indicative of conformational change and decrease of the β-structure (β-sheet-β-turn structure) content of GS12 and 11-meric peptides in buffer. Negative ellipticities of peptides were generally enhanced in both zwitterionic PC (Figure 3B) and negatively-charged PE/PG (Figure 3C) lipid systems, which imply that these positively-charged cyclic peptides interact with lipid membranes with different affinities and change or enhance their conformation. Detailed mechanistic studies on interaction of cyclic cationic peptides with lipid membrane systems have been reported previously.18,19
Figure 3.
CD spectra of the GS {10–12} group peptides in buffer (A), POPC (B), and POPE/POPG (C) vesicles. Relation between ellipticities sensitive to changes in β-structure (205, 215 and 225 nm) and retention times are shown in D.
Ellipticities at three wavelengths (205, 215 and 225 nm) can indicate the changes in the β-sheet (212–218 nm) and β-turn (205–225 nm) content of the peptide conformations.25 As mentioned above, since there is also a direct correspondence between RTs and the biological activity (hemolytic and antibacterial) of the peptides in GS {10–12} series, the β-structure content in PC (erythrocyte membrane) and PE/PG (bacterial membrane) lipid systems can be directly related to the biological activity –the higher the β-structure content of the peptides (higher negative ellipticities) the more potent their biological activities. Among the peptides of the GS {10–12} group, GS10 and GS12-L3 had the highest and lowest overall biological activities, RT and β-sheet content, respectively. As observed in Table 2–Table 4, the therapeutic index (TI) parameters, calculated as the ratio of hemolytic to antimicrobial activities, is used to define the degree of dissociation of hemolytic (toxic, lethal) activity from antimicrobial activities. In the GS {10–12} group, GS12-K4 had the highest overall TI against Gram-positive bacteria and fungi, however, despite better average antibacterial activity against Gram-negative bacteria, its TI was slightly lower than GS12-K7 (Table 2–Table 4).
Biological Activities and Conformation of GS {13,14} Group
Odd-numbered ring 13-meric peptides of this group were generated by deleting charged (GS14-K4) or hydrophobic (GS14-L3 and GS14-V5) residues from GS14, and their sequences, RTs and ch/hyp ratios are listed in Table 1.
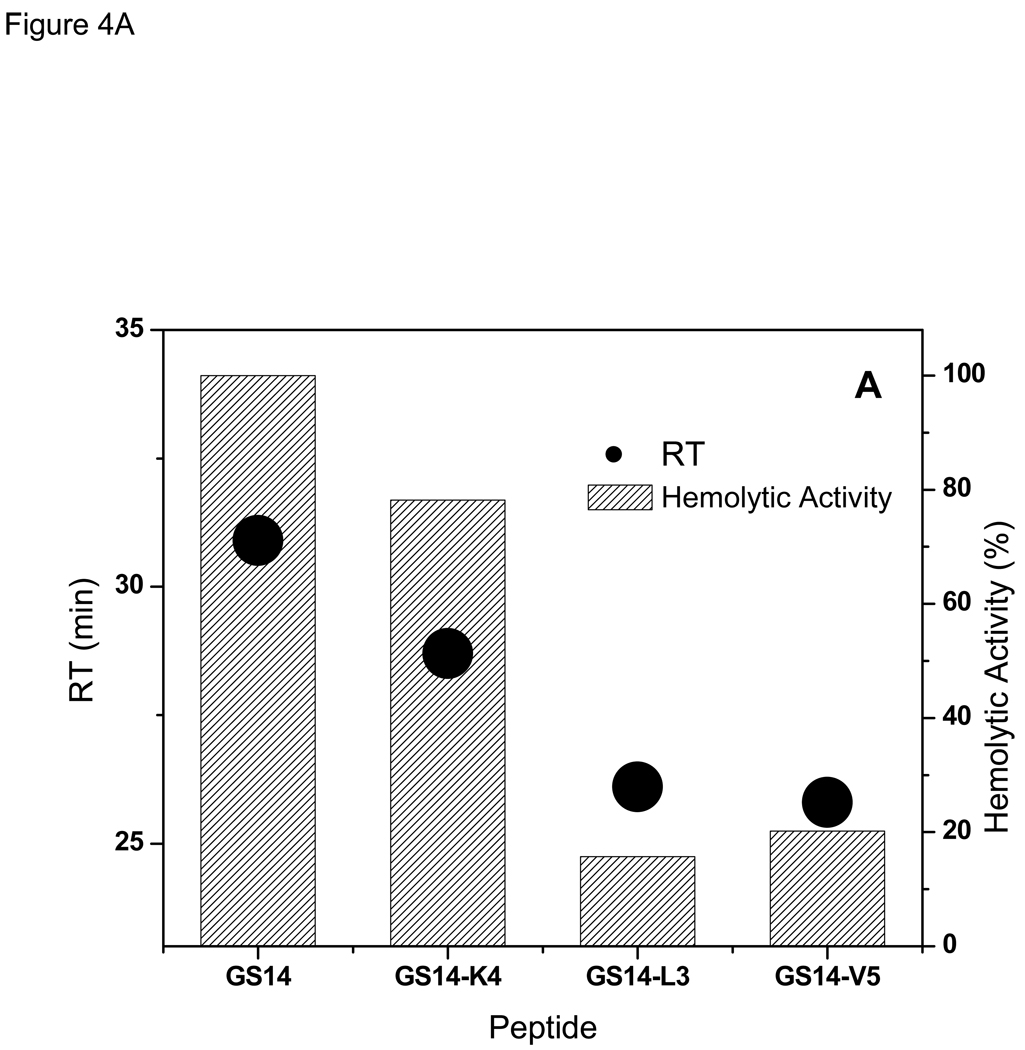
Hemolytic activity and RTs for GS {13,14} peptides are directly related (Figure 4A and Table 1). As observed in Figure 4A, decrease in RTs is accompanied by decrease in hemolytic activity. The amphipathic β-sheet containing peptide GS14 (Figure 1) had the most potent hemolytic activity and highest RT, and the less-amphipathic peptides of this group had lower RTs and weaker hemolytic activities. GS14-L3 and GS14-V5 peptides had comparable RTs, hemolytic activities and ch/hyp ratios (4/5) (Table 1).
Figure 4.
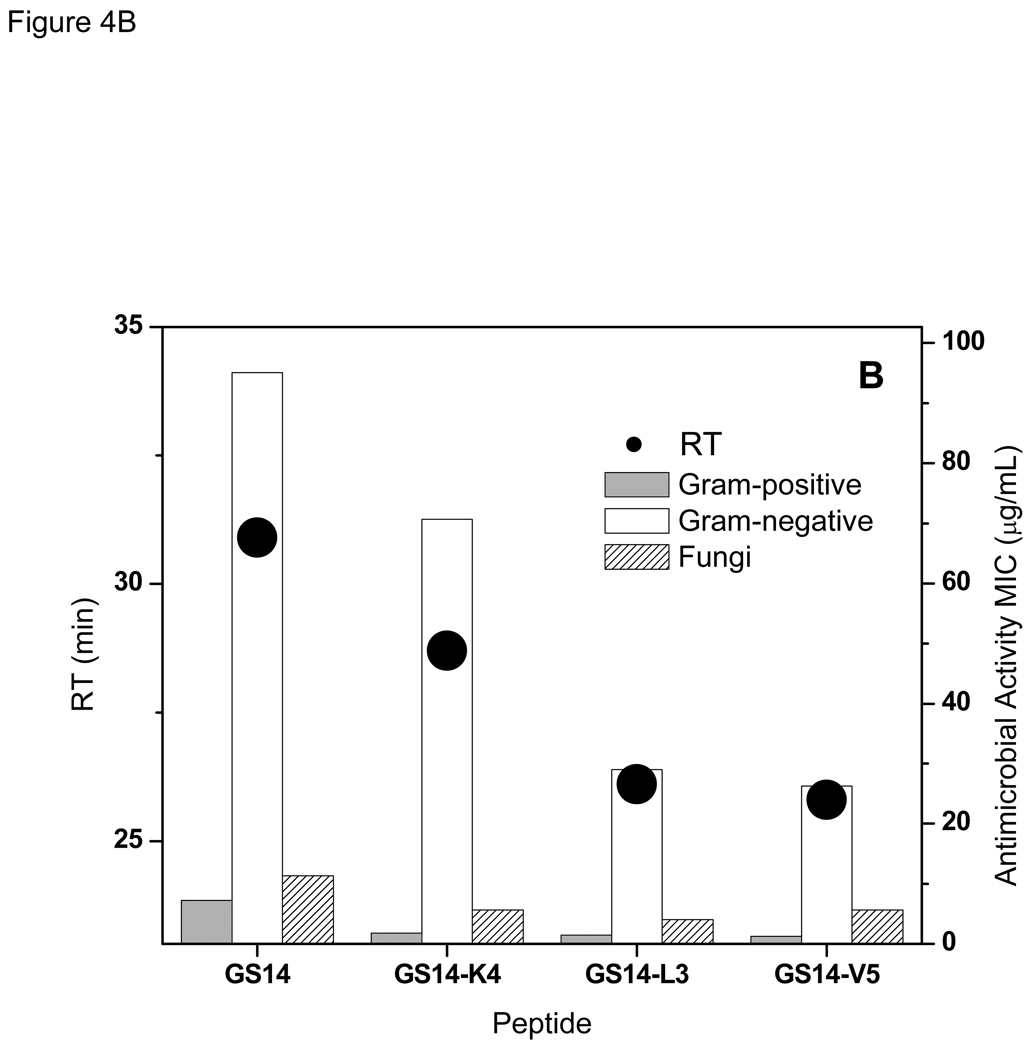
Relation between retention times with hemolytic (A), and antimicrobial (B) activities of the peptides in GS {13,14} group.
In contrast to the 10- to 12-meric peptides, there is a inverse relation between the antibacterial activities of the peptides in GS {13,14} group and their RTs- the higher the RT the lower the activity against both Gram-positive and Gram-negative bacteria (Table 2 and Table 3, and Figure 4B). Similar to their hemolytic activities, GS14-L3 and GS14-V5 had comparable antibacterial activities. Antifungal activities of the peptides of this group also decreased with increase in their retention times (Figure 4B and Table 4).
As with the peptides in group GS {10–12}, biological activities of GS {13,14}peptides were related to their conformation. CD spectra of the peptides in buffer and the two lipid vesicle systems are exhibited in the supporting information (Figure I). Similar to GS10, GS14 has a typical β-sheet-β-turn conformation.18 Compared with GS14, the CD spectra of 13-meric peptides imply a decrease in the β-structure content of these peptides in buffer and lipid vesicles. Enhancement of negative ellipticities of peptides in both PC and PE/PG lipid systems indicate interaction of peptides with lipid membranes as well as their conformational changes (see above).18,19 Similar to the peptides of the GS {10–12} group, there is a direct relation between the β-structure content of the peptides in lipid vesicles and their RTs and hemolytic activities –the lower the content of β-structure the lower the RTs and the weaker the hemolytic activities. However, antibacterial activities of the peptides of the GS {13,14} group are inversely related to their β-structure content in the PE/PG lipid system-the lower the β-structure content of the peptides the more potent their antimicrobial activities (Figure 4 and supporting information). Among the peptides of the GS {13,14} group, GS14-L3 and GS14 had the highest and lowest overall TIs against Gram-positive and Gram-negative bacteria as well as fungi, respectively (Table 2 to Table 4).
Biological Activities and Conformation of GS {15,16} Group
Odd-numbered ring 15-meric peptides of this group were generated by deleting charged (GS16-K4 and GS16-K6) or hydrophobic (GS16-V5) residues from GS16. The sequences, RTs and ch/hyp ratios of the GS {15,16} group are listed in Table 1.
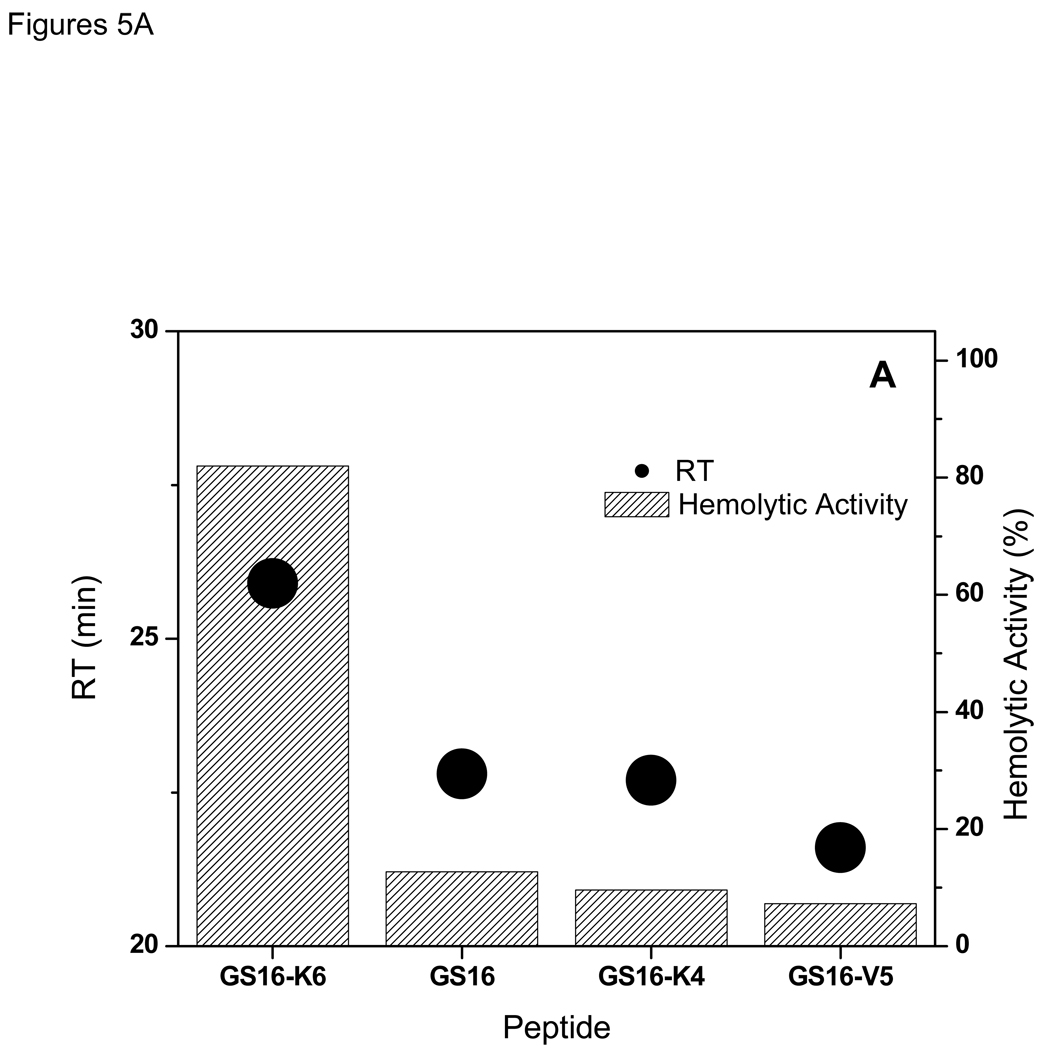
Similar to GS {10–12} and GS {13,14} peptides, the hemolytic activities and RT values for GS {15,16}peptides are directly related (Figure 5A and Table 1). As observed in Figure 5A, decrease in RTs is accompanied by decrease in hemolytic activity. The non-amphipathic peptide of this group, GS16 (Figure 1) had a weak hemolytic activity and second highest RT, and the most amphipathic peptide of this group, GS16-K6, had the highest RTs and strongest hemolytic activities.
Figure 5.
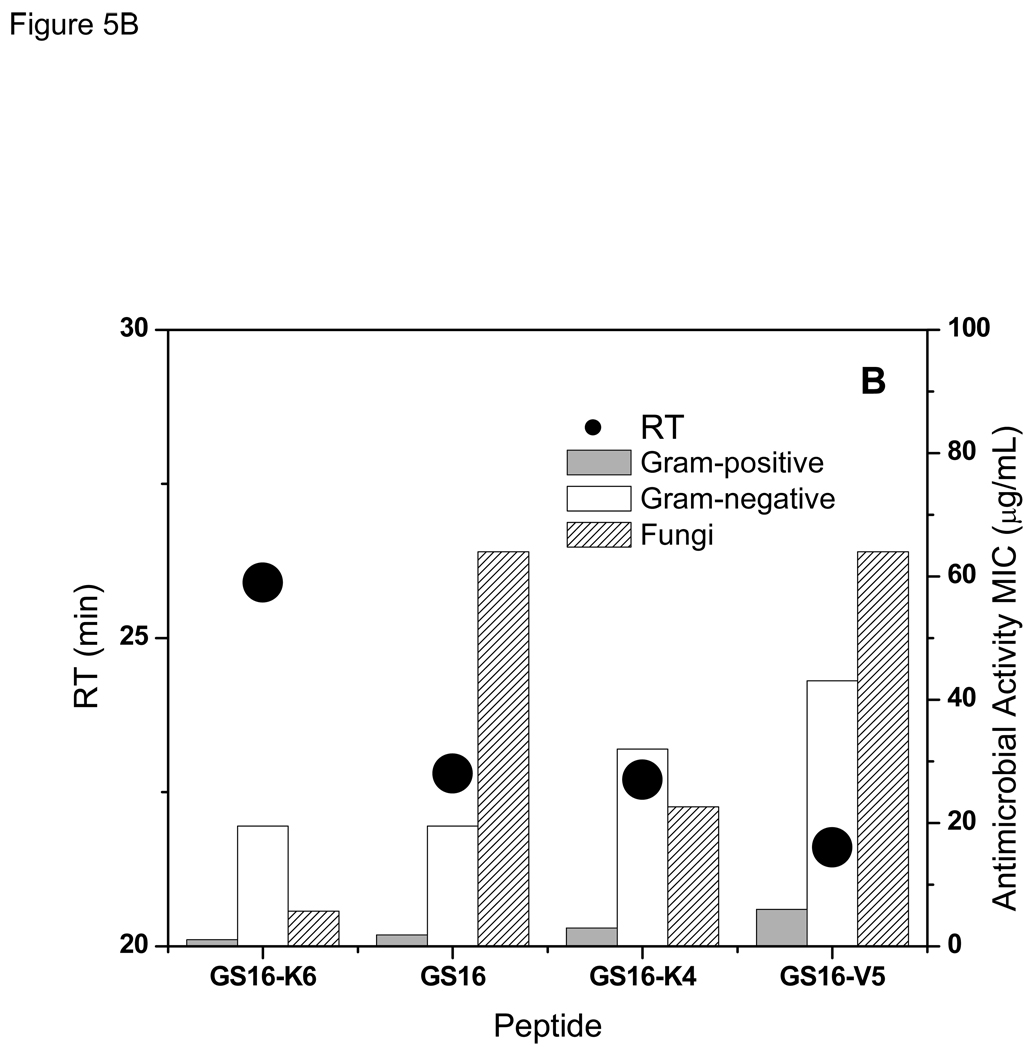
Relation between retention times with hemolytic (A), and antimicrobial (B) activities of the peptides in GS {15,16} group.
Similar to the 10- to 12-meric peptides, and in contrast to GS {13,14} series, there is a direct relation between the antibacterial activities of the peptides in GS {15,16} group and their RTs- the higher the RT the higher the activity against both Gram-positive and Gram-negative bacteria (Table 2 and Table 3, and Figure 5B). Like the two other groups, activities of the 15- and 16-meric peptides against Gram-positive bacteria were much higher than Gram-negative bacteria. Antifungal activities of the peptides of this group also increased with increase in their retention times (Figure 5B and Table 4) -an exception was GS16, which had a higher RT but lower antifungal activity compared with GS16-K4.
As with the peptides of the two previous groups, biological activities of GS {15,16} peptides were related their conformation. CD spectra of the peptides in buffer and the two lipid vesicle systems are exhibited in the supporting information (Figure II). Compared with GS10 and GS14 (β-sheet-β-turn peptides), all peptides of this group have low β-structure contents. The spectra in the PC lipid system were different from those in PE/PG system, which imply different conformations and peptide-lipid interactions in this milieu. Overall the CD spectra in lipid systems, especially PE/PG, indicate that the cyclic peptides interact with lipid membranes and change their conformation. As in the cases of GS {10–12} and GS {13,14} groups, there is a direct relation between the β-structure content of the peptides and their RTs and hemolytic activities–the lower the content of β-structure in lipid vesicles the lower the RTs and the weaker the hemolytic activities. However, the relation between antimicrobial activity and the β-structure content in PE/PG lipid system is not as straightforward. GS16 with the second best overall antibacterial activity had the lowest β-structure content in the GS {15,16} group. In this group of peptides, GS16 had the highest overall TI against both Gram-positive and Gram-negative bacteria, whereas GS16-K4 had the highest TI against fungi (Table 2–Table 4).
Retention Time, Hydrophobicity, Amphipathicity and Conformation
Table 1 exhibits the relation between the apparent amphipathicity, as measured by RTs, and hydrophobicity of the cyclic peptides. Hydrophobicities of peptides were calculated on the basis of whole-residue hydrophobicity scale by calculating the sum of the free energy of transfer of individual amino acids from water to octanol (ΔGWO), representing the lipid bilayer hydrophobic core.26 To compare the hydrophobicity of peptides with different ring sizes, free energy of transfer values are shown as [ΔGWO]= ΔGWO per residue. As seen in this Table, peptides with comparable RT values can have different overall hydrophobicity (for example, GS14-L3, GS16-K6, GS14-V5 and GS12-K4) and vice versa (for example GS12-K4 and GS12-K7). For a relation between the amphipaticity (RTs) and β-sheet content of all three peptide groups in buffer and lipid membranes see Figure III in the supporting information.
DISCUSSION
Systematic disruption of the β-sheet-β-turn structure of amphipathic β-sheet peptides GS10 and GS14 by single enantiomeric substitution of the amino acid residues of the peptide rings (D-enantiomer in place of L-amino acids and L-enantiomer in place of D-Tyr) resulted in significant changes in the biological activities of these peptides5–7,27 These results showed that by systematic disruption of the β-structure, the larger more flexible rings can adopt a diverse array of conformations with different degrees of amphipathicity, and this diversity in conformation and amphipathicity could be the key factor in design of novel peptides with higher TI and antimicrobial specificity. Another method to disrupt the structure of cyclic β-sheet peptides and their amphipathicity is systematic addition and deletion of amino acids in the peptide ring. By adopting the latter method in this study, odd- and even-numbered rings were generated and divided into three groups on the basis of their ring size: rings with limited conformational flexibility, GS {10–12} group; rings with moderate conformational flexibility, GS {13,14}; and conformationally flexible rings, GS {15,16}. Interestingly, variation in the ring size and charge of the cationic cyclic peptides resulted in diversity and specificity of biological function, dissociation between hemolytic and antimicrobial activities as well as improvement in TIs.
Among the peptides of Table 1, less- or non-amphipathic peptides of group GS {10–12}, with disrupted β-sheet had the weakest hemolytic activities. After GS10, GS12-K4 (+3) (more amphipathic than GS12 and less amphipathic than GS10 (+2)) had the second best overall biological activity as well as the best overall TI in this group (Table 2–Table 4). Since there is a direct correlation between RTs, β-structure content, hemolytic and average antibacterial activities in this group, an optimal β-structure content and amphipathicity can be achieved for the best dissociation between the hemolytic and antimicrobial activities (best TI). GS12-K4, the lead peptide of this group, had an RT of 25.7 min, as well as ~45% less β-structure content and weaker overall antimicrobial activity compared with GS10.
In contrast to the GS {10–12} group, in GS {13,14} group amphipathicity was reduced by deleting amino acid residues from the β-sheet-β-turn peptide GS14 (+4). Hemolytic activities were also reduced with reduction in β-structure content, but were stronger than 11-meric peptides and GS12. GS14 was the most hemolytic peptide of this group, but had the lowest TIs. The low antimicrobial activity of GS14 can be related to its strong interaction with and entrapment in the non-lipid components of the bacterial cell walls. In this group there is an inverse correlation between RTs and β-structure content, and average antibacterial activities in this group. GS14-L3, with highest TI, had an RT of 26.1 min and 55% and 45–50% less β-structure content in buffer and the two lipid systems, respectively, compared with GS14.
All peptides of the GS {15,16}group had disrupted β-sheet (CD spectra and RT values), and two of these peptides, GS16-K4 (+5) and GS16-V5 (+6), with comparable conformations in PC vesicles, had very weak hemolytic activities. GS16-K6 (+5) was the most amphipathic peptide of this group (highest RT and β-structure content) and had the strongest hemolytic and average antimicrobial activities. However, due to its potent hemolytic activity, this peptide had the lowest TIs of the GS {15,16} group. GS16, had the best overall TIs of this group for antibacterial activity, and GS16-K4 had the best TI for antifungal activity. Similar to the GS {10–12} group, there is a direct correlation between RTs, β-structure content, hemolytic and average antibacterial activities in the GS {15,16}group.
Overall, peptides with comparable apparent amphipathicity (as estimated by RT values), but with different ring size and positive charge, can have different hemolytic or antimicrobial activities (for example, GS14-L3, GS16-K6 and GS12-K4 or GS12 and GS16). On the other hand, peptides with comparable ring size, charge and RT values can have comparable biological activities (for example, GS14-L3 and GS14-V5). Hydrophobicity of peptides ([ΔGWO]), exhibited in Table 1, is correlated to the affinity of peptides for lipid bilayer cores. Interestingly, with exception of GS, all [ΔGWO] values for the peptides are positive. Apparently, based on the hydrophobicity of their amino acid sequences, none of the peptides in the three groups has a tendency to interact with the lipid bilayer hydrophobic core (negative ΔGWO) or bilayer interface (negative ΔGWIF). However, the whole-residue hydrophobicity scale does not consider the structure of peptides. By considering the overall topology, amphipaticity and hydrophobic surfaces of the peptides and based on experimental evidence,17–20 cyclic peptides in the three groups interact with the lipid membrane surface with different affinities. In a series of isothermal titration calorimetry (ITC) experiments, the binding affinity of some of the cyclic peptides (GS10, GS12, GS14, GS14-L3, GS16 and GS16-K4) for PC and PE/PG systems was measured (19 and unpublished results, L. Wheaton and M. Jelokhani-Niaraki). Based on these measurements, with the exception of GS14, interactions of peptides with PC vesicles were weak, endothermic and entropy-driven. In contrast to all other peptides, GS14 had an exceptionally strong and exothermic binding to PC vesicles. Compared with PC vesicles, all peptides (including GS14) had endothermic binding to PE/PG vesicles.
In this study, biophysical properties and biological activities of the cyclic peptides show a clear relation between their structure and function. Disruption of the β-structure in 11-, 13-, 15- and 16-meric peptides was the decisive step taken in reducing the hemolytic activity (as much as 32 fold), amphipathicity and increasing the selectivity (increase in TI). Consequently, by systematic modulation of ring size, β-structure content, amphipathicity and affinity for microbial membrane surfaces, we were able to find lead peptides with significantly improved TIs compared with GS and its decameric analog GS10. These peptides are highlighted in Table 2 to Table 5. In each of the three groups, peptide conformation and its affinity for model lipid membranes of erythrocytes and bacteria was closely related to its biological activity. This finding implies that, regardless of other possible additional mechanisms for the biological function of antimicrobial peptides, interaction with mammalian and bacterial cell membranes in these series of peptides is an essential component of their biological activity. From the results of this study it is also clear that more flexible and larger rings, higher number of positive charges and lower affinity for lipid membranes are important factors for improving the TIs, while preserving the antimicrobial potency. As exhibited in Table 5, for average activity against the Gram-positive bacteria, GS16 and GS14-L3 were as potent as GS and had the best TI (~6 fold improvement compared with GS). However, compared with GS16, GS14-L3 was more specific (higher TI) for some strains such as S. aureus, S. mitis and S. pneumoniae; and GS16-K4 was more specific against E. faecalis, S. pyrogenes and C. jeikeium (Table 2). Also, both GS14-V5 and GS12-K4 had good average activities and were specific against certain bacteria (Table 2 and Table 5). Activity against Gram-negative bacteria was generally weaker than activity against Gram-positive bacteria. Peptides were practically inactive against S. marcescens and only weakly active against E. cloacae or P. aeruginosa (Table 3). GS16 also had the best TI (10 fold improvement and stronger activity, compared with GS) for average activity against the Gram-negative bacteria (Table 5) and was most specific towards most strains (Table 3). GS16-V5 and GS16-K4 were more specific against A. caloaceticus and S. maltophilia, respectively (Table 3). For average activity against fungi, GS14-L3 had the best TI (4.5 fold improvement compared with GS) and specificity against the tested strains, followed by GS12-K4 and GS14-V5 (Table 4 and Table 5). In addition to physicochemical properties of the antimicrobial peptides, the specificity of peptide activity against different strains of bacteria could be related to the composition and structure of different bacterial cell walls and membranes. Mechanism of interaction of the cyclic cationic peptides with animal and bacterial cytoplasmic cell membranes has been the subject of our previous studies.18,19 It is noteworthy to mention that we have also examined the biological activities and conformations of linear forms of the even-numbered peptides (unpublished results, M. Jelokhani-Niaraki and L.H. Kondejewski). Compared to their cyclic versions, these linear peptides had weak biological activities and little ordered secondary structures, moreover, their conformations were dependent on the amino acids at their N- and C-termini.
Table 5.
Summary of therapeutic index, mean antimicrobial activity and fold improvement of the lead peptides
| Microbial Strain | GS | GS10 | GS12-K4 | GS14-L3 | GS14-V5 | GS16 | GS16-K4 | |
|---|---|---|---|---|---|---|---|---|
| Grant-positive | TI | 74 | 82 | 354 | 429 | 406 | 435 | 350 |
| Activity (µg/mL)a | 1.4 | 1.2 | 3.3 | 1.5 | 1.2 | 1.8 | 3.0 | |
| Fold Improvement b | - | 1.1 | 4.8 | 5.8 | 5.5 | 5.9 | 4.7 | |
| Gram-negative | TI | 4 | 2 | 16 | 22 | 19 | 40 | 32 |
| Activity (µg/mL) | 23.8 | 39.0 | 70.7 | 29.0 | 26.3 | 19.5 | 32.0 | |
| Fold Improvement | - | 0.5 | 4 | 5.5 | 4.8 | 10 | 8 | |
| Fungi | TI | 35 | 25 | 103 | 159 | 88 | 12 | 46 |
| Activity (µg/mL) | 2.8 | 4.0 | 11.3 | 4.0 | 5.7 | 64.0 | 22.6 | |
| Fold Improvement | - | 0.7 | 2.9 | 4.5 | 2.5 | 0.3 | 1.3 |
Geometric mean of antimicrobial activity. For the definition of geometric mean, please see the footnote “b” in Table 2.
Fold improvement of the TI values of peptide analogs relative to GS. Highlighted parameters denote the analogs with best improvement in their TI relative to GS. Average antimicrobial activities of these analogs are comparable or better than GS.
CONCLUSION
Among the peptides in three groups, the 16-meric peptide GS16, with the largest ring, most flexible and non-amphipathic structure and highest number of positive charges, was the lead compound with best TI for average activity against both Gram-positive and Gram-negative bacteria (compared with GS, 5.9 and10 fold increase, respectively). This peptide also had low affinity to interact with PC vesicles, but had a higher tendency to interact with the negatively-charged PE/PG vesicles. GS14-L3, a moderately amphipathic peptide with a relatively less flexible13-meric ring, was the lead peptide with the best TI for average activity against Gram-positive bacteria and fungi (compared with GS, 5.8 and 4.5 fold increase, respectively). With their low toxicity against erythrocytes (6–8 times less than GS), potent antimicrobial activity and specificity (comparable to GS and GS10), as well as various possibilities for optimizing their therapeutic function,7 GS16 and GS14-L3 can therefore serve as promising candidates for developing novel broad-spectrum antimicrobial peptide drugs.
Supplementary Material
SUPLEMENTARY INFORMATION on the “Conformation of Cyclic Peptides as Determined by CD Spectra” is available.
ACKNOWLEDGEMENTS
We thank Mr. Marc Genest for peptide synthesis and purification, and amino acid analysis. This study was supported by grants from the Canada Foundation for Innovation (CFI: 6786) and the Natural Sciences and Engineering Research Council of Canada (NSERC: 250119) to M.J-N., and the Protein Engineering Network of Centres of Excellence (PENCE) and National Institutes of Health (NIHR01-AI-067296) to R.S.H..
Abbreviations
- Boc
tert-butyloxycarbonyl
- BOP
benzotriazole-1-yl-oxy-tris-(dimethylamino)-phosphoniumhexafluorophosphate
- CD
circular dichroism
- DIEA
N,N-diisopropylethylamine
- DMF
N,N-dimethylformamide
- GS
gramicidin S
- HBTU
N-[(1H-benzotriazol-1yl)(dimethylamino)methylene]-N-methylmethanaminium hexafluorophosphate N-oxide
- HOBt
1-hydroxybenzotriazole
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- Tris
Tris(hydroxymethyl)aminomethane hydrochloride
REFERENCES
- 1.Wax RG, Lewis K, Salyers A, Taber H, editors. Bacterial Resistance to Antimicrobials. 2nd ed. Roca Baton, FL, U.S.A.: CRC Press; 2007. [Google Scholar]
- 2.Kleinkauf H, von Döhren H, editors. Biochemistry of peptide antibiotics. Berlin, Germany: Walter de Gruyter; 1990. [Google Scholar]
- 3.Yount NY, Yeaman MR. Immunocontinuum: perspectives in antimicrobial mechanisms of action and resistance. Protein Peptide Lett. 2005;12:49–76. doi: 10.2174/0929866053405959. [DOI] [PubMed] [Google Scholar]
- 4.Kondejewski LH, Farmer SW, Wishart DS, Kay CM, Hancock REW, Hodges RH. Modulation of structure and antibacterial and hemolytic activity by ring size in cyclic gramicidin S analogs. J. Biol. Chem. 1996;271:25261–25268. doi: 10.1074/jbc.271.41.25261. [DOI] [PubMed] [Google Scholar]
- 5.Kondejewski LH, Jelokhani-Niaraki M, Farmer SW, Lix B, Kay CM, Sykes BM, Hancock REW, Hodges RH. Dissociation of antimicrobial and hemolytic activities in cyclic peptide diasteromers by systematic alterations in amphipathicity. J. Biol. Chem. 1999;274:13181–13192. doi: 10.1074/jbc.274.19.13181. [DOI] [PubMed] [Google Scholar]
- 6.Jelokhani-Niaraki M, Kondejewski LH, Farmer SW, Hancock REW, Kay CM, Hodges RS. Diastereomeric analogues of gramicidin S: structure, biological activity and interaction with lipid bilayers. Biochem. J. 2000;349:747–756. doi: 10.1042/bj3490747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondejewski LH, Lee DL, Jelokhani-Niaraki M, Farmer SW, Hancock REW, Hodges RH. Optimization of microbial specificity in cyclic peptides by modulation of hydrophobicity within a defined structural framework. J. Biol. Chem. 2002;277:67–74. doi: 10.1074/jbc.M107825200. [DOI] [PubMed] [Google Scholar]
- 8.Lee DL, Hodges RS. Structure-activity relationships of de novo designed cyclic antimicrobial peptides based on Gramicidin S. Biopolymers (Peptide Science) 2003;71:28–48. doi: 10.1002/bip.10374. [DOI] [PubMed] [Google Scholar]
- 9.Gause GF, Brazhnikova MG. Gramicidin S and its use in the treatment of infected wounds. Nature (London) 1944;154:703b. [Google Scholar]
- 10.Schmidt GMJ, Crowfoot-Hodgkin D, Oughton BM. A crystallographic study of some derivatives of gramicidin S. Biochem J. 1957;65:744–756. doi: 10.1042/bj0650744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stern A, Gibbons WA, Craig LC. A conformational analysis of gramicidin S-A by nuclear magnetic resonance. Proc. Natl. Acad. Sci. U.S.A. 1968;61:734–741. doi: 10.1073/pnas.61.2.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hull SE, Karlsson R, Main P, Wolfson MM, Dodson EJ. The crystal structure of a hydrated gramicidin S-urea complex. Nature (London) 1978;275:206–207. [Google Scholar]
- 13.Kondejewski LH, Farmer SW, Wishart DS, Hancock REW, Hodges RH. Gramicidin S is active against both Gram-positive and Gram-negative bacteria. Int. J. Peptide Protein Res. 1996;47:460–466. doi: 10.1111/j.1399-3011.1996.tb01096.x. [DOI] [PubMed] [Google Scholar]
- 14.Brodgen KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nature Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 15.Niidome T, Murakami H, Kawazoe M, Hatakeyama T, Kobashigawa Y, Matsushita M, Kumaki Y, Demura M, Nitta K, Aoyagi H. Carbohydrate recognition of gramicidin S analogs in aqueous medium. Bioorg. Med. Chem. Lett. 2001;11:1893–1896. doi: 10.1016/s0960-894x(01)00322-5. [DOI] [PubMed] [Google Scholar]
- 16.Mogi T, Ui H, Shiomi K, Omura S, Kita K. Gramicidin S identified as a potent inhibitor for cytochrome bd-type quinol oxidase. FEBS Lett. 2008;582:2299–2302. doi: 10.1016/j.febslet.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 17.Jelokhani-Niaraki M, Prenner EJ, Kondejewski LH, Kay CM, McElhaney RN, Hodges RS. Conformation and other biophysical properties of the cyclic antimicrobial peptides in aqueous solutions. J. Peptide Res. 2001;58:293–306. doi: 10.1034/j.1399-3011.2001.00893.x. [DOI] [PubMed] [Google Scholar]
- 18.Jelokhani-Niaraki M, Prenner EJ, Kay CM, McElhaney RN, Hodges RS. Conformation and interaction of the cyclic antimicrobial peptides in lipid bilayers. J. Peptide Res. 2002;60:23–36. doi: 10.1034/j.1399-3011.2002.21003.x. [DOI] [PubMed] [Google Scholar]
- 19.Jelokhani-Niaraki M, Hodges RS, Meissner JE, Hassenstein UE, Wheaton L. Interaction of gramicidin S and its aromatic amino-acid analogs with phospholipid membranes. Biophys. J. 2008;95:3306–3321. doi: 10.1529/biophysj.108.137471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salgado J, Grage SL, Kondejewski LH, Hodges RS, McElhaney RN, Ulrich AS. Membrane-bound structure and alignment of the antimicrobial β-sheet peptide gramicidin S derived from angular and distance constraints by solid-state 19F-NMR. J. Biomol. NMR. 2001;21:191–208. doi: 10.1023/a:1012946026231. [DOI] [PubMed] [Google Scholar]
- 21.MacDonald RC, MacDonald RI, Menco BPhM, Takeshita K, Subbarao NK, Hu L-R. Small-volume extrusion apparatus for preparation of large unilamellar vesicles. Biochim. Biophys. Acta. 1991;1061:297–303. doi: 10.1016/0005-2736(91)90295-j. [DOI] [PubMed] [Google Scholar]
- 22.Gibbs AC, Kondejewski LH, Gronwald W, Nip AM, Hodges RS, Sykes BD, Wishart DS. Unusual β-sheet periodicity in small cyclic peptides. Nature Struct. Biol. 1998;5:284–288. doi: 10.1038/nsb0498-284. [DOI] [PubMed] [Google Scholar]
- 23.Shwyzer R. Chemistry of amino acids and peptides. Annu. Rev. Biochem. 1960;29:183–206. doi: 10.1146/annurev.bi.29.070160.001151. [DOI] [PubMed] [Google Scholar]
- 24.Sereda TJ, Mant CT, Hodges RS. Selectivity due to conformational differences between helical and non-helical peptides in reversed-phase chromatography. J. Chromatogr. A. 1995;695:205–221. doi: 10.1016/0021-9673(94)01147-7. [DOI] [PubMed] [Google Scholar]
- 25.Fasman GD, editor. Circular Dichroism and the Conformational Analysis of Biomolecules. New York, U.S.A.: Plenum Press; 1996. [Google Scholar]
- 26.White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu. Rev. Biochem. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 27.McInnes C, Kondejewski LH, Hodges RS, Sykes BD. Development of the structural basis for antimicrobial and hemolytic activities of peptides based on gramicidin S and design of novel analogs using NMR spectroscopy. J. Biol. Chem. 2000;275:14287–14294. doi: 10.1074/jbc.275.19.14287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPLEMENTARY INFORMATION on the “Conformation of Cyclic Peptides as Determined by CD Spectra” is available.