Abstract
Biosynthesis of the DNA base thymine depends on activity of the enzyme thymidylate synthase (TS) to catalyze the methylation of the uracil moiety of 2’-deoxyuridine-5’-monophosphate (dUMP). All known thymidylate synthases (TSs) rely on an active site residue of the enzyme to activate dUMP1, 2. This functionality has been demonstrated for classical TSs, including human TS, and is instrumental in mechanism-based inhibition of these enzymes. Here we report the first example of thymidylate biosynthesis that occurs without an enzymatic nucleophile. This unusual biosynthetic pathway occurs in organisms containing the thyX gene, which codes for a flavin-dependent thymidylate synthase (FDTS), and is present in several human pathogens3–5. Our findings indicate that the putative active site nucleophile is not required for FDTS catalysis, and no alternative nucleophilic residues capable of serving this function can be identified. Instead, our findings suggest that a hydride equivalent (i.e. a proton and two electrons) is transferred from the reduced flavin cofactor directly to the uracil ring, followed by an isomerization of the intermediate to form the product, 2’-deoxythymidine-5’-monophosphate (dTMP). These observations indicate a very different chemical cascade than that of classical TSs or any other known biological methylation. The findings and chemical mechanism proposed here, together with available structural data, suggest that selective inhibition of FDTSs, with little effect on human thymine biosynthesis, should be feasible. Since several human pathogens depend on FDTS for DNA biosynthesis, its unique mechanism makes it an attractive target for antibiotic drugs.
Classical thymidylate synthases (TSs), encoded by the thyA gene, are present in most eukaryotes, including humans, and are frequently targeted by chemotherapeutic and antibiotic drugs. A recently discovered class of TSs, the flavin-dependent thymidylate synthases (FDTSs)3, 6, 7, is encoded by the thyX gene and has been found primarily in prokaryotes and viruses3, 8, including several pathogens and bio-warfare agents9. Several organisms, including human pathogens, rely solely on thyX for thymidylate synthesis (e.g. all Rickettsia lack genes coding for DHFR, TS, and thymine kinase). It has recently been suggested that thyX limits chromosomal replication in these organisms10. FDTSs share no structure or sequence homology with classical TSs, and thus present a promising new frontier for antibacterial/antiviral drug development5–7.
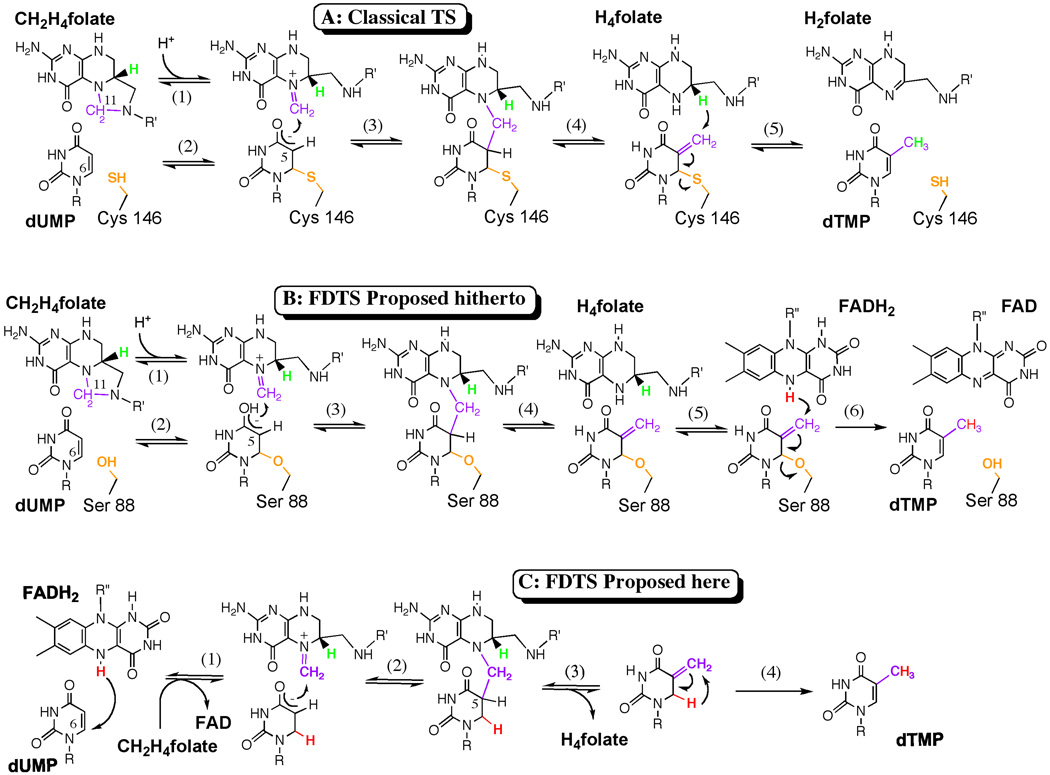
The catalytic mechanism of classical TSs is presented in Figure 1A1, 2. A strictly conserved active site cysteine covalently activates the uracil ring by nucleophilic Michael-addition at the C6 position of dUMP (Figure 1A, step 2). The resulting enolate then attacks (step 3) the iminium form of N5,N10-methylene-5,6,7,8-tetrahydrofolate (CH2H4folate), followed by elimination of tetrahydrofolate (H4folate, step 4). Finally, a hydride transfer from C6 of the H4folate yields the products dTMP and dihydrofolate (H2folate, step 5). Common drugs that target classical TS either covalently bind to the catalytic nucleophile (e.g., 5F-dUMP) or noncovalently bind the folate binding pocket (e.g. Tomudex).
Figure 1. TSase mechanisms.
A. The chemical mechanism for the classical TS catalyzed reaction 1, 2. B. The chemical mechanism for the FDTS proposed hitherto 12. C. The newly proposed mechanism for the FDTS that does not rely on an enzymatic nucleophile. The conserved enzymatic nucleophile is orange, the methylene is purple, the reducing hydride from H4folate is green, and the hydride from FADH2 is red. R= 2’-deoxyribose-5’-phosphate and R’= (p-aminobenzoyl)-glutamate. R”= adenosine-5’-pyroposphate-ribityl.
It has previously been proposed that FDTSs have a chemical mechanism analogous to the classical TS mechanism, but with a serine residue acting as a nucleophile (Figure 1B, step 2), and a flavin cofactor providing a hydride to terminate the reaction (Figure 1B, step 6). This results in the production of H4folate rather than Hsfolate11, 12. The suggestion that a serine serves as the active site nucleophile in FDTS was originally based on sequence alignments of thyX genes that indicated no conserved cysteine but a strictly conserved serine. Crystal structures of FDTSs from three very different organisms7, 13, 14 placed this conserved serine about 4 Å from the C6 position of dUMP (e.g., Figure 2A). However, without a neighboring general base this serine will not be deprotonated, decreasing its potential reactivity, casting doubt on this serine’s putative role as a nucleophile.
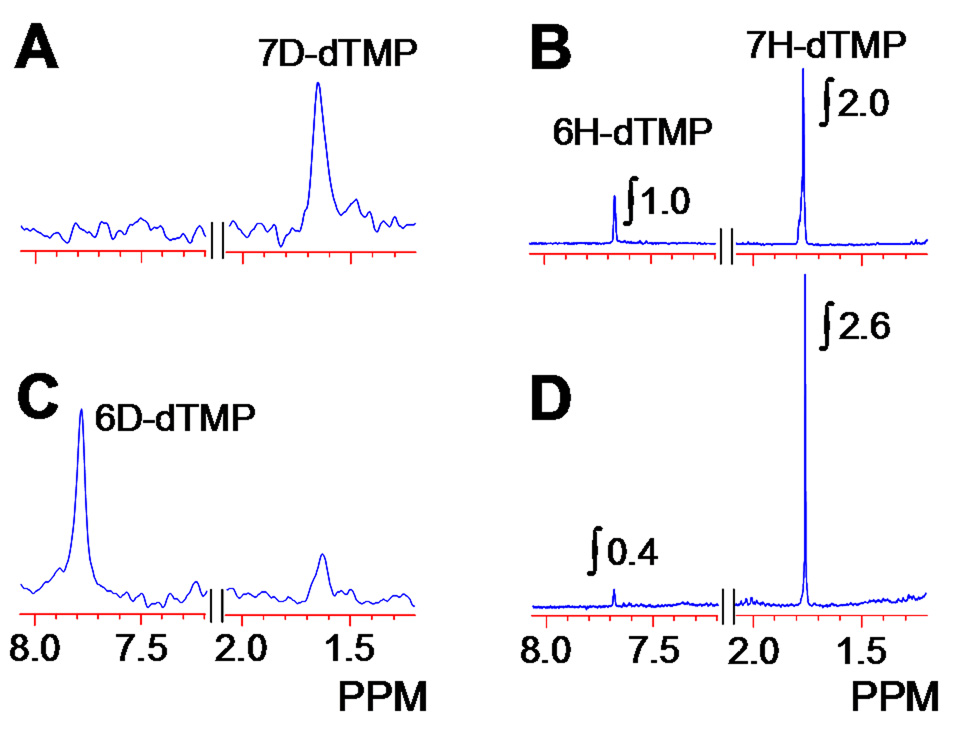
Figure 3. 2H-NMR (A & C) and 1H-NMR (B & D).
spectra of dTMP produced in the FDTS catalyzed reaction of dUMP in D2O (Experiment A in Figure 4). Spectra A and B were from the reaction at 65 °C, and spectra C and D from the same reaction at 37 °C. The latter clearly indicate the presence of 6D-dTMP (~60 %).
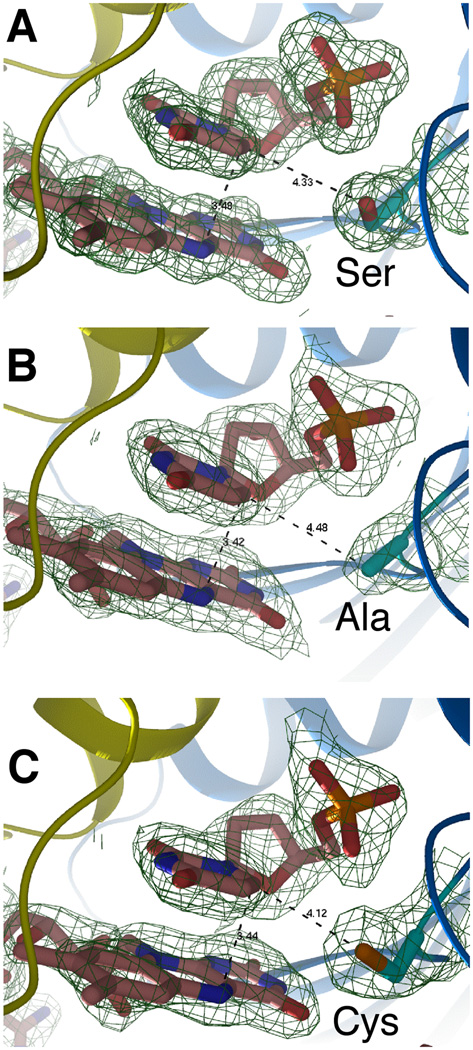
Figure 2. Crystal structures of the FDTS-FAD-dUMP complex for: (A) Wild type tmFDTS, (B) S88A mutant, and (C) S88C mutant.
The distance between the C6 carbon of dUMP and the reducing center of the flavin (N5 of FAD) is 3.4 Å for all three enzymes. The distances of the side-chain of residue 88 to C6 are 4.3, 4.5, and 4.1 Å, for wtFDTS, S88A, and S88C, respectively. The electron density maps are 2Fo-Fc with a contour level of 1.0 sigma.
Point mutation studies were performed with FDTS from H. pylori (hpFDTS), where the conserved serine residue was mutated to either alanine or cysteine (S84A and S84C). While both mutations were found to retain activity4, it was assumed that an adjacent serine (Ser85) could have rescued the activity of S84A. Abolished enzyme activity of a double mutant (S84A/S85A) supported this hypothesis. Furthermore, MALDI-TOF MS analysis showed that the S84C mutant forms a covalent adduct with dTMP. These results were used to propose that Ser84 activates dUMP (Figure 1B)4, 12.
To further test this hypothesis, we performed similar mutation studies using FDTS from T. maritima (tmFDTS). The active site of this enzyme contains a strictly conserved serine, Ser88, and no alternative nucleophilic residues (see refs 6, 7 and the Supplementary Information - SI). Activity tests for S88A and S88C (for details see ref 12 and SI) indicated that both mutants were still active. Possible contamination by classical ecTS was ruled out by a series of control experiments to ensure that mutant FDTS was the sole source of the observed activity (See SI). The FDTS activity of these mutants and the lack of classical TS activity of S88C demonstrate that the conserved active site serine does not serve as a catalytic nucleophile in the FDTS reaction, in stark contrast to the mechanism proposed hitherto (Figure 1B).
Crystal structures of the S88A and S88C mutants were obtained at 1.95 and 2.05 Å resolution, respectively (see data collection/refinement statistics in the SI), and their electron density is compared to that of the wild-type tmFDTS (Figure 2). The electron densities for both mutants indicate minimal changes in folding and active site configuration. It is apparent from Figure 2C that there is no covalent bond between the cysteine and dUMP in the crystal. Nevertheless, a MALDI-TOF analysis of the trypsin-digested S88C tmFDTS indicated that Cys88 is bound to dUMP (see SI), as previously reported for hpFDTS4. Even in solution though, cysteine covalently binds to the C6 position of uracil15. These facts together with the low activity of S88C, suggest that the observed Cys88-dUMP complex is not part of the FDTS catalytic cascade, but rather an inhibitory dead-end complex.
A critical piece of evidence for the covalent bond between the active site cysteine in classical TS and dUMP is the crystal structure of a covalently bound 5-flouro-dUMP (5F-dUMP) in complex with CH2H4folate (PDB 1tls16). In contrast to classical TS, FDTS does not covalently bind 5F-dUMP upon incubation with CH2H4folate, as confirmed by both MALDI-TOF analysis (see SI), and crystal structure analysis (PDB 1o287) obtained under similar conditions. Another test for similar Michael Addition to the C6 of dUMP is the dehalogenation of 5Br-dUMP2, 17. This test resulted in no reactivity with FDTS (see SI). We also solved the crystal structures of tmFDTS with FAD and both 5-halogenated-dUMPs (PDB 1o27 and 1o28) and Hol and coworkers solved the 5Br-dUMP-FAD structure for FDTS from M. tuberculosis (PDB 2af6)14. These structures are nearly the same as the complex with dUMP (Figure 2A) and do not support a nucleophilic attack of any enzyme residue on the C6 of dUMP. These observations emphasize the distinctions between the mechanisms of classical TS and FDTS, and in light of the activity of the S88A mutant, support a mechanism in which FDTS does not involve a Michael-addition of an enzymatic nucleophile.
To expose the nature of the FDTS catalyzed reaction we followed the flow of hydrogens along the catalytic pathway by isotopic substitution of a specific hydrogen. We have previously found that when conducting the FDTS reaction in D2O (50 % D), deuteration of the reduced flavin leads to deuterated dTMP (using ESI-MS analysis), and that reaction with tritiated 6T-CH2H4folate yields 6T-H4folate12. These results contrast the same experiments with classical TSs, where reactions performed in D2O do not incorporate deuterium into the dTMP and the labeled hydride from CH2H4folate always transfers to the dTMP18. In the past, we and others4, 12 suggested that these findings support the mechanism illustrated in Figure 1B, but the current findings however, contradict that mechanism and required further tests. By repeating the experiment in D2O (this time > 99.6 % D), and analyzing the product using ESI-MS, 1H-NMR, and 2H-NMR, we found that at 65 °C (close to the physiological temperature of T. maritima) the product was indeed deuterated at the C7 position (Figure 3 A and B). However, when we performed the same experiment at 37 °C, NMR analyses indicated the formation of both 6D-dTMP (60%) and 7D-dTMP (40%) (Figure 3 C and D). This result is quite intriguing, as no mechanism previously proposed for FDTS predicts formation of 6D-dTMP, and such phenomenon has never been reported for any TS reaction.
The lack of an obvious enzymatic nucleophile and the ability to trap deuterium from D2O at C6 of the product demonstrate that the chemical mechanisms of FDTS and classical TS differ substantially. Without an enzymatic nucleophile, the FDTS catalyzed reaction could proceed via Michael-addition of a hydroxide ion or though participation of the flavin prosthetic group. For hydroxide to serve as a nucleophile, a water molecule must be activated by a general base in the active site (e.g. the catalytic triad in hydrolytic enzymes). All crystal structures of FDTSs indicate that there is no such basic system available in the active site. Additional experiments using reduced 5-carba-5-deaza-FAD resulted in dTMP formation, excluding the possibility that the reduced N5 of FADH2 is the nucleophile. Importantly, the FDTS mechanism requires a hydrogen transfer to the C6 of the uracil moiety to explain the formation 6D-dTMP from reactions performed in D2O, which is inconsistent with either hydroxyl or flavin as Micheal nucleophiles.
In Figure 1C we propose a new chemical mechanism consistent with current data and previous findings19, wherein a hydride equivalent from the N5 of FADH2 is transferred to C6 of dUMP (Figure 1C, step 1). The resulting enolate anion nucleophilically attacks the iminium methylene of CH2H4folate, and an elimination of H5 from dUMP and H4folate results in a C5=C7 double bond (steps 2 and 3). This exocyclic-methylene intermediate then isomerizes to form the product, dTMP (step 4). The intermediate proposed here is unique in nucleotide biochemistry, but this isomer of the thymine moiety is chemically feasible and quite stable in solution20. This mechanism is compatible with our previously findings19 on the oxidative half-reaction if the equilibrium constant for the first step lies to the left. Since we have no experimental data regarding the methylene transfer and the initial activation (if not H-transfer), steps 2 and 3 are proposed here as a logical path toward the product and step 1 might be preceded by other activation steps.
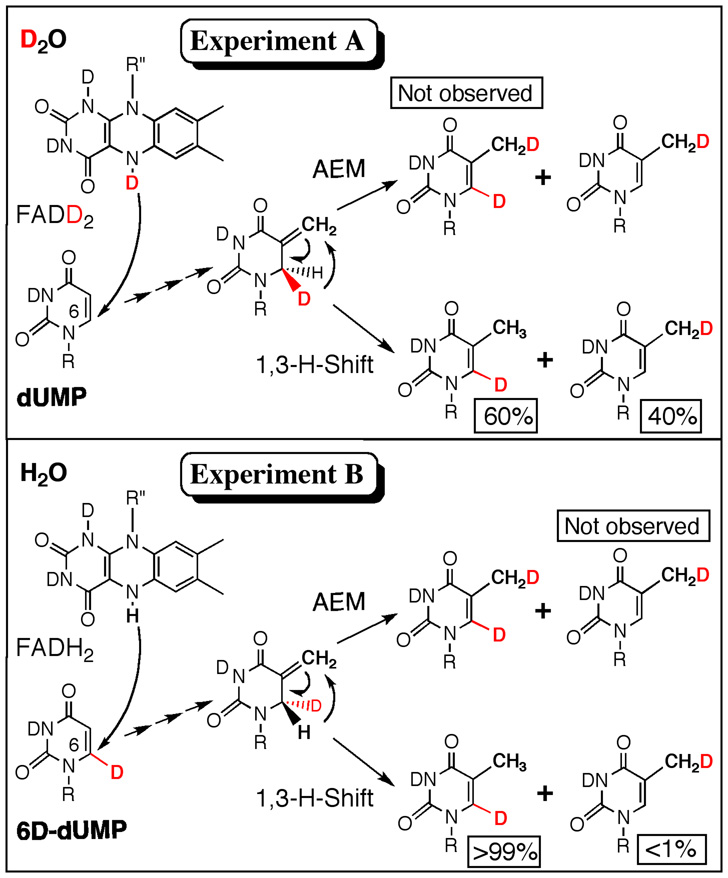
Since the isomerization of the putative intermediate (Figure 1C, step 4) does not occur rapidly in solution20, the enzyme could catalyze this transformation by the two mechanisms illustrated in Figure 4. An enzymatic acid could catalyze this step via an addition-elimination mechanism (AEM), in which a proton is added to the C5=C7 double bond and the intermediate cation loses a proton from C6 to form the product. Alternatively, the thermodynamic driving force (>6 kcal/mol as estimated from semiempirical QM calculations) could favor a 1,3-sigmatropic rearrangement (1,3-hydride shift)21. When conducting the reaction in D2O, an AEM would lead to 6D,7D-dTMP, but ESI-MS analysis (Figure S4) did not indicate such product. Therefore, a suitable explanation is an enzyme-catalyzed isomerization via a 1,3-H-shift (Figure 4, lower path).
Figure 4. Hydride flow.
An illustration of two experimental approaches to examine the hydride flow in the reaction catalyzed by the thermophilic tmFDTS at reduced temperature (37 °C). Experiment A was performed in a D2O buffer using 6H-dUMP (i.e., unlabeled, dUMP). Experiment B was performed in an H2O buffer using 6D-dUMP (see SI). Percentages below each species represent the relative quantities of product formation as indicated by 1H and 2H NMR.
The finding that two different isotopically labeled products are formed at 37 °C was intriguing and warranted further investigation. Thus, we performed the FDTS reactions using dUMP, with D at its C6, in an H2O buffer at 37 °C (Figure 4, Experiment B). NMR analysis of the product showed the formation of 6D-dTMP (>99%). If any 6H-dTMP was formed, it was below our detection limits (<1%). The observations for both experiments (6H-dUMP in D2O and 6D-dUMP in H2O) can be explained by the combination of normal kinetic isotope effect (KIE: H reacts faster than the heavier D) and reduced stereoselectivity at reduced temperature (37 °C). Lack of stereoselectivity at reduced temperature has already been observed during the reductive-half reaction of FDTS, which transfers both 4-(R) and 4-(S) hydride of NADPH12. A KIE of 10 and stereoselectivity of 87 %, for example, would result in production of 60:40 C6:C7-dTMP when 6H-dUMP reacts with FDTS in D2O, and more than 99:1 C6:C7-dTMP produced when 6D-dUMP reacted in H2O (see SI for a more detailed discussion).
The proposed hydrogen transfer to the C6 of dUMP by the flavin cofactor is further supported by the crystal structures, which show a short distance (3.4 Å) between N5 of the flavin ring and C6 of dUMP (Figure 2). Such hydride transfer from FADH2 to a uracil ring is an atypical chemistry for thymidylate synthases and nucleotide methylation in general, but is not unprecedented in enzymology. For example, dihydrooratate dehydrogenase (DHOD) and old yellow enzyme (OYE) are other flavo-proteins that catalyze similar chemistry22, 23.
To the best of our knowledge, neither hydride transfer to the uracil ring, nor an isomerization of such an intermediate has been reported for any thymine biosynthetic pathway or other nucleotide methylations. Importantly, such a chemical mechanism is very different from that of classical TSs, and along with structural differences, may help explain why classical TS inhibitors have a reduced effect on FDTSs5. These findings suggest that selective inhibition of FDTS should be feasible, and may further alleviate the constraint of an enzymatic nucleophile from structure-based rational drug design efforts. Rationally designed compounds could mimic the non-covalently bound intermediate or the transition states for its formation and isomerization. Such compounds may inhibit FDTS with little effect on classical TSs and thus may serve as leads to selective antibiotics that would not interfere with human thymine biosynthesis.
Methods summary
Purification and activity of FDTS enzymes
The FDTS from T. maritima (TM0449, GeneBank accession number NP228259), and its mutants S88A and S88C were expressed and purified as previously described6. The activities of these enzymes were determined using a [2-14C]dUMP assay which is a modification of the procedure developed and described in ref 12. Mutant reactivity was also determined by oxidation of chemically reduced enzyme by CH2H4folate and dUMP under an atmosphere of purified Ar.
Halogenated substrate derivatives
The 5Br-dUMP assay was adopted from ref 17. A TS inhibitor, 5F-dUMP, was assessed as a covalent inhibitor of FDTS by incubating it with the enzyme in the presence of CH2H4folate and sodium dithionite, followed by removal of small molecules by ultra filtration. Activities of the incubated enzymes were determined prior to MALDI-TOF analyses.
Mass spectroscopy and NMR analyses
All MALDI-TOF and ESI-MS analyses were conducted at the High Resolution Mass Spectrometry Facility (HRMSF) of the University of Iowa. Enzymes were analyzed following trypsin digestion. NMR analyses were performed at the University of Iowa NMR Central Research Facility using Bruker model Av-300 (for 1H-NMR measurements), and Av-800 (for 2H-NMR measurements) spectrometers.
Following the flow of deuterium during the FDTS reaction in D2O
Experiments were performed at 65 and 37 °C using dUMP or 6D-dUMP, NADPH, CH2H4folate, and enzyme in D2O or H2O under anaerobic conditions. The product dTMP was then purified using semi-preparative reverse phase HPLC.
Methods
Materials
All materials were reagent grade and used without further purification unless specified. 2’-deoxyuridine-5’-monophosphate (dUMP), reduced nicotinamide adenosine dinucleotide phosphate (NADPH), 5-bromo-2’-deoxyuridine (5Br-dU), 5-fluoro-2’-deoxyuridine-5’-monophosphate (5F-dUMP), trypsin protease, ammonium bicarbonate, tris(hydroxymethyl)aminomethane, D2, D2O, phosphocreatine, and creatine kinase were purchased from Sigma. Radiolabed [2-14C]dUMP was obtained from Moravek Biochemicals. N5,N10-methylene-5,6,7,8-tetrahydrofolate (CH2H4folate) was a generous gift from Eprova Inc, Switzerland. The FDTS from T. maritima (TM0449, GeneBank accession number NP228259), and its mutants S88A and S88C were expressed and purified as previously described6. The thymidine kinase plasmid was obtained from Dr. Robert Stroud’s lab at UCSF and expressed and purified as described in ref 24.
Synthesis of 5-bromo-2’-deoxyuridine-5’-monophosphate (5Br-dUMP)
5Br-dUMP was synthesized by phosphorylation of 5Br-dU at 37°C in a 100 mM Tris, 10 mM MgCl2 buffer at pH = 7.5. The reaction mixture contained 1.5 mM 5Br-dU, 5 mM ATP, 50 mg/mL phosphocreatine, 2 mg/mL creatine kinase and ~1 µM of thymidine kinase. The 5Br-dUMP product was purified by HPLC-UV/Vis (following 280 nm absorbance) and analyzed by ESI-MS.
Synthesis of dUMP with deuterium at C6
This procedure was adapted from ref 25. dUMP (225 mg) was dissolved twice in 5 mL of D2O (>99.96 D-atom) under Ar gas, and evaporated under vacuum (< 50 mTorr) to dryness to reduce proton contamination. The dUMP was then dissolved in 5 mL of D2O and stirred in the presence of Pt(IV) oxide under 1 atm D2 gas (>99.96 D-atom) for 3 hours. Vacuum filtration removed the catalyst and the remaining solution was lyophilized to dryness. NMR analysis confirmed >99.5 % D-atom substitution of both the 5 and 6 positions of the uracil ring. A method to substitute the 5D into 5H without affecting 6D has been developed15, however, such substitution was not used in the current preparation because the TS reaction is a substitution reaction where a methyl group replaces the 5H of dUMP to form dTMP. Since the 5H position is always replaced during the synthesis of dTMP1, 2, 4 its isotopic labeling can be disregarded.
Methods
Analytical methods
All analytical separations were performed on an Agilent series HPLC model 1100, equipped with online degasser, UV/Vis diode array detector and 500TR series Packard flow scintillation analyzer (FSA). Supelco reverse phase column (Discover series 250 mm × 4.6 mm) was used starting with 100 mM KH2PO4 (pH = 6.0) followed by a methanol gradient as described elsewhere12. The enzyme active site concentration was determined by the 454 nm absorbance of bound FAD (ε = 11,300 cm−1M−1).
Purification methods
Separation by HPLC was performed using a semi-preparative reverse phase Supelco Column (Discovery series 250 mm × 10 mm). Mobile phase used for separation was a gradient of 100 mM KH2PO4 and methanol. Eluent was collected according to the UV spectral absorbance of the purified species and then lyophilized to dryness.
Protein crystallization
The protein-FAD-dUMP complex was prepared by treating 15 mg/mL of the enzyme with around 10 molar excess of dUMP. The well solution for crystallization contained 35 – 45% PEG200 and 0.1M Tris-HCl (pH 8.0) buffer.
Activity assays
The activity assay ([2-14C]dUMP assay) was a modification of the procedure developed and described in ref 12. All experiments were performed in 200 mM tris buffer (exchanged with Ar) pH = 8.0 at 65 °C with standard reaction conditions of: 100 µM dUMP (including 0.5 Mdpm [2-14C]dUMP), 200 µM CH2H4folate, 5 mM CH2O (to stabilize CH2H4folate) and 5 mM sodium dithionite. Reactions were initiated by addition of 0.1 – 2 µM (final concentration) of enzyme, quenched with HCl (to a final pH = 1) and stored at −80 °C until analysis. HPLC-FSA analysis was used to determine the conversion of [2-14C]dUMP to [2-14C]dTMP.
5Br-dUMP assay
Reactions with either ecTS or tmFDTS were performed in a 200 mM tris buffer at pH = 8.0, containing 100 µM 5Br-dUMP, 5 mM sodium dithionite, and 5 mM β-mercaptoethanol. Enzyme was added to the reaction mixture which was then incubated at 37 °C for 3 hours. Product conversion was determined by HPLC-UV/Vis analysis of the reaction mixtures by following both 256 and 280 nm absorptions for dUMP and 5Br-dUMP, respectively.
Oxidative Half-Reaction of S88A
A solution of oxidized S88A (20 µM) and dUMP (1 mM) was made anaerobic in a sealed cuvette by successive cycles of evacuation and equilibration with an atmosphere of purified Ar. The oxidized enzyme was titrated spectrophotometrically to complete reduction with a solution of dithionite. CH2H4folate was added anaerobically from a side-arm to initiate the reaction (25°C). The absorbance spectrum of oxidized enzyme returned before the first scan, indicating a rapid reaction of the mutant enzyme.
Protein digestion
All enzyme digestion reactions were performed in 100 mM ammonium bicarbonate buffer adjusted to pH = 8.0 at 37°C. Enzyme solutions were diluted to 1 µM protein concentration followed by addition of trypsin to a final concentration of 10 ng/µL. All digestions were allowed to incubate for 3 hours at 37°C and stored at −20 °C prior to MALDI-TOF MS analysis.
Assessment of 5F-dUMP as a covalent inhibitor of FDTS
These experiments were performed at 37°C in 200 mM tris buffer at pH = 8.00. Both wt-FDTS and S88A (11 µM active site concentration) were incubated for 30 minutes in the presence of 50 µM 5F-dUMP, 200 µM CH2H4folate, and 5 mM sodium dithionite. After incubation, activities of the enzymes were determined using the standard activity assay conditions (except with a residual 5 µM 5F-dUMP). Samples of native and trypsin-digested enzymes were prepared for MALDI-TOF MS analysis. The remaining enzyme solutions were exchanged with 40 mL of Tris buffer at 4°C, and concentrated by centrifugal filtration (using a Millipore 10,000 MWCO filtration device) to 11 µM active site concentration. Once concentrated, the activity of both FDTS and S88A were determined using the standard activity assay conditions. As described in the report, no covalent adduct of 5F-dUMP to enzyme was identified and both enzymes recovered 100 % activity after the removal of the 5F-dUMP from the reaction mixture.
Following the flow of deuterium during the FDTS reaction
For studies in D2O, all substrates in 100 mM tris buffer were exchanged twice by dissolving in D2O (>99.96 % D-atom) and lyophilizing to dryness prior to use. Experiments were performed in 100 mM tris buffer (99.96 % D2O or H2O) pH = 8.0 at 65 and 37 °C using 4 mM dUMP or 6D-dUMP, 8 mM NADPH, and 8mM CH2H4folate, under Ar. To maintain anaerobic conditions 10 mM glucose and 100 units of glucose oxidase were added to the reaction mixture. Reactions were initiated by adding enzyme (previously lyophilized and resuspended in D2O or H2O) to a final concentration of 1 – 10 µM. The reaction mixtures were incubated (at 65 or 37 °C) for 20 hours under Ar and stored at −20 °C. The product dTMP was then purified using semi-preparative HPLC, lyophilized, triturated into methanol, and dried under vacuum. The dTMP was dissolved in D2O or H2O for 1H and 2H NMR analysis, respectively.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgements
This work was supported by NIH R01 GM065368 and NSF CHE 0715448 to AK, the Iowa Center for Biocatalysis and Bioprocessing to EMK, NIH R01 GM61087 to BAP, NIH training grant GM08270 to JAC, and JCSG grant U54GM074898 to SAL. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory (SSRL), a national user facility operated by Stanford University on behalf of DOE, OBER. The SSRL Structural Molecular Biology Program is supported by DOE, OBER, and by NIH, NCRR, Biomedical Technology Program, and NIGMS.
Footnotes
Author Information: Atomic coordinates and structure factor files have been deposited with the Protein Data Bank under the accession codes 3G4A and 3G4C. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions.
References
- 1.Carreras CW, Santi DV. The catalytic mechanism and structure of thymidylate synthase. Annu. Rev. Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 2.Finer-Moore JS, Santi DV, Stroud RM. Lessons and conclusions from dissecting the mechanism of a bisubstrate enzyme: thymidylate synthase mutagenesis, function and structure. Biochemistry. 2003;42:248–256. doi: 10.1021/bi020599a. and many cited therein. [DOI] [PubMed] [Google Scholar]
- 3.Myllykallio H, et al. An alternative flavin-dependent mechanism of thymidylate synthesis. Science. 2002;297:105–107. doi: 10.1126/science.1072113. [DOI] [PubMed] [Google Scholar]
- 4.Leduc D, et al. Functional evidence for active site location of tetrameric thymidylate synthase X at the interphase of three monomers. Proc. Nat. Acad. Sci. U.S.A. 2004;101:7252–7257. doi: 10.1073/pnas.0401365101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chernyshev A, Fleischmann T, Kohen A. Thymidyl biosynthesis enzymes as antibiotic targets. Appl. Microbiol. Biotech. 2007;74:282–289. doi: 10.1007/s00253-006-0763-1. [DOI] [PubMed] [Google Scholar]
- 6.Lesley SA, et al. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11664–11669. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathews II, et al. Functional analysis of substrate and cofactor complex structures of a thymidylate synthase-complementing protein. Structure. 2003;11:677–690. doi: 10.1016/s0969-2126(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 8.Leduc D, et al. Two distinct pathways for thymidylate (dTMP) synthesis in (hyper)thermophilic Bacteria and Archaea. Biochem. Soc. Trans. 2004;32:231–235. doi: 10.1042/bst0320231. [DOI] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. http://www.cdc.gov.
- 10.Escartin F, Skouloubris S, Liebl U, Myllykallio H. Flavin-dependent thymidylate synthase X limits chromosomal DNA replication. Proc. Nat. Acad. Sci. U.S.A. 2008;105:9948–9952. doi: 10.1073/pnas.0801356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myllykallio H, Leduc D, Filee J, Liebl U. Life without dihydrofolate reductase FolA. Trends Microbiol. 2003;11:220–223. doi: 10.1016/s0966-842x(03)00101-x. [DOI] [PubMed] [Google Scholar]
- 12.Agrawal N, Lesley SA, Kuhn P, Kohen A. Mechanistic studies of a flavin-dependent thymidylate synthase. Biochemistry. 2004;43:10295–10301. doi: 10.1021/bi0490439. [DOI] [PubMed] [Google Scholar]
- 13.Graziani S, et al. Catalytic mechanism and structure of viral flavin-dependent thymidylate synthase ThyX. J. Biol. Chem. 2006;281:24048–24057. doi: 10.1074/jbc.M600745200. [DOI] [PubMed] [Google Scholar]
- 14.Sampathkumar P, et al. Structure of the Mycobacterium tuberculosis flavin dependent thymidylate synthase (MtbThyX) at 2.0 resolution. J. Mol. Biol. 2005;352:1091–1104. doi: 10.1016/j.jmb.2005.07.071. [DOI] [PubMed] [Google Scholar]
- 15.Hong B, Maley F, Kohen A. The role of Y94 in proton and hydride transfers catalyzed by thymidylate synthase. Biochemistry. 2007;46:14188–14197. doi: 10.1021/bi701363s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyatt DC, Maley F, Montfort WR. Use of strain in a stereospecific catalytic mechanism: crystal structures of Escherichia coli thymidylate synthase bound to FdUMP and methylenetetrahydrofolate. Biochemistry. 1997;36:4585–4594. doi: 10.1021/bi962936j. [DOI] [PubMed] [Google Scholar]
- 17.Wataya Y, Santi DV. Thymidylate synthase catalyzed dehalogenation of 5-Bromo-and 5-iodo-deoxyuridylate. Biochem. Biophys. Res. Comm. 1975;67:818–823. doi: 10.1016/0006-291x(75)90886-4. [DOI] [PubMed] [Google Scholar]
- 18.Hong B, Kohen A. Microscale synthesis of isotopically labeled 6R-N5, N10 methylene-5, 6, 7, 8-tetrahydrofolate. J. Label. Compd. Radiopharm. 2005;48:759–769. [Google Scholar]
- 19.Gattis SG, Palfey BA. Direct observation of the participation of flavin in product formation by thyX-encoded thymidylate synthase. J. Am. Chem. Soc. 2005;127:832–833. doi: 10.1021/ja0432214. [DOI] [PubMed] [Google Scholar]
- 20.Kloetzer W. Two isomers of thymine. Monatsh. Chem. 1973;104:415–420. [Google Scholar]
- 21.Carey FA, Sundberg RJ. Advanced Organic Chemistry, Part A. New York: Kluwer Academic/Plenum Publishers; 2000. [Google Scholar]
- 22.Brown BJ, Deng Z, Karplus PA, Massey V. On the active site of Old Yellow Enzyme. Role of histidine 191 and asparagine 194. J. Biol. Chem. 1998;273:32753–32762. doi: 10.1074/jbc.273.49.32753. [DOI] [PubMed] [Google Scholar]
- 23.Fagan RL, Nelson MN, Pagano PM, Palfey BA. Mechanism of flavin reduction in class 2 dihydroorotate dehydrogenases. Biochemistry. 2006;45:14926–14932. doi: 10.1021/bi060919g. [DOI] [PubMed] [Google Scholar]
- 24.Waldman AS, Haeusslein E, Milman G. Purification and characterization of herpes simplex virus (type 1) thymidine kinase produced in Escherichia coli by a high efficiency expression plasmid utilizing a lambda PL promoter and cI857 temperature- sensitive repressor. J. biol. Chem. 1983;258:11571–11575. [PubMed] [Google Scholar]
- 25.Burdzy A, Noyes KT, Valinluck V, Sowers LC. Synthesis of stable-isotope enriched 5-methylpyrimidines and their use as probes of base reactivity in DNA. Nucleic Acids Res. 2002;30:4068–4074. doi: 10.1093/nar/gkf520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.