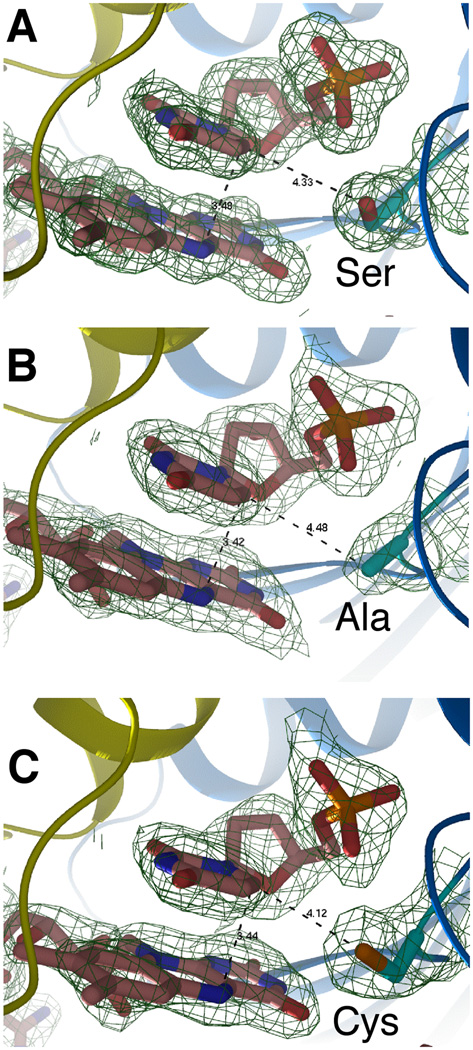
Figure 2. Crystal structures of the FDTS-FAD-dUMP complex for: (A) Wild type tmFDTS, (B) S88A mutant, and (C) S88C mutant.
The distance between the C6 carbon of dUMP and the reducing center of the flavin (N5 of FAD) is 3.4 Å for all three enzymes. The distances of the side-chain of residue 88 to C6 are 4.3, 4.5, and 4.1 Å, for wtFDTS, S88A, and S88C, respectively. The electron density maps are 2Fo-Fc with a contour level of 1.0 sigma.