Abstract
In all known organisms, amino acids are predominantly thought to be synthesized and used as their L-enantiomers. Here, we found that bacteria produce diverse D-amino acids as well, which accumulate at millimolar concentrations in supernatants of stationary phase cultures. In Vibrio cholerae, a dedicated racemase produced D-Met and D-Leu, while Bacillus subtilis generated D-Tyr and D-Phe. These unusual D-amino acids appear to modulate synthesis of peptidoglycan, a strong and elastic polymer that serves as the stress-bearing component of the bacterial cell wall. D-amino acids influenced peptidoglycan composition, amount, and strength, both via their incorporation into the polymer and by regulating enzymes that synthesize and modify it. Thus, synthesis of D-amino acids may be a common strategy for bacteria to adapt to changing environmental conditions.
In all kingdoms of life, cells predominantly use L-amino acids. In most bacteria, the only D-amino acids (D-aa) produced in significant quantities are D-Ala and D-Glu, which are incorporated into peptidoglycan (PG)(1). PG is a strong and elastic polymer of the bacterial cell wall that is synthesized and modified by penicillin binding proteins (PBPs). PG counteracts the cell’s osmotic pressure, maintains cell shape, and anchors components of the cell envelope. The chemistry and structure of PG is dynamic (1, 2) but the factors that regulate alterations in the composition and architecture of PG are not well understood.
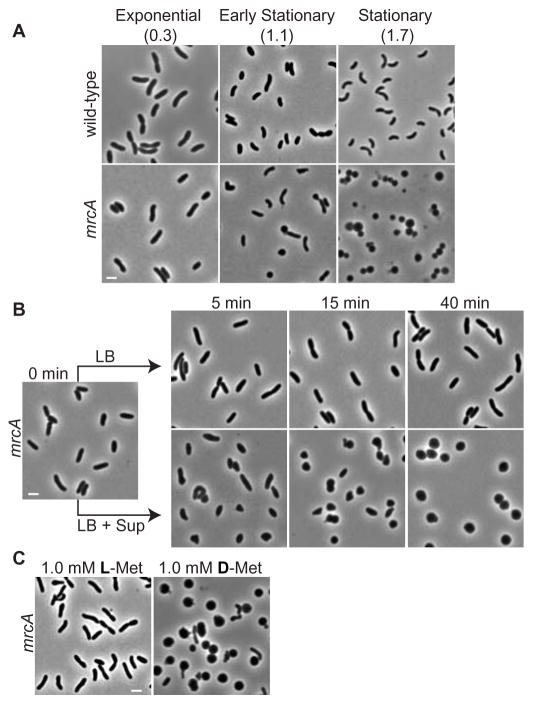
Here we found that a mutant in Vibrio cholerae mrcA, which encodes a PBP1A homologue (1, 2), retained its normal rod-shape during exponential growth but became spherical in stationary phase (Fig. 1A). The mutant’s shape change was due to factor(s) that accumulated in the culture media, because exponentially growing rod-shaped mrcA cells started to become spherical within 5 min after addition of cell-free supernatants from stationary phase cultures (Fig. 1B). We used the rod to sphere shape transition of the mrcA mutant as an indicator for such factors, and examined their role in the development of PG from wild-type cells.
Figure 1.
D-aa induce rod-shaped mrcA V. cholerae cells to become spheres. (A) Growth phase-dependent morphology of wild-type and mrcA V. cholerae; culture densities (OD600nm). (B) Kinetics of the change in mrcA cell morphology after addition of stationary phase supernatant (Sup) + LB or LB alone. (C) Morphologic effects 45 min after addition of 1 mM L-Met or D-Met to rod-shaped mrcA cells. Bar size = 2 μm.
Stationary phase supernatants were fractionated and assayed for their sphere-inducing activity. The active fractions consisted of four amino acids: Met, Leu, Val and Ile. The D-forms, and not L-forms, of these amino acids stimulated the conversion of mrcA rods to spheres (Fig. 1C). A 1.0 mM mixture of these D-aa had sphere-inducing activity nearly equivalent to that of unfractionated stationary phase supernatant. Thus, the absolute stereochemistry at the alpha carbon of these amino acids determines their sphere-inducing activity.
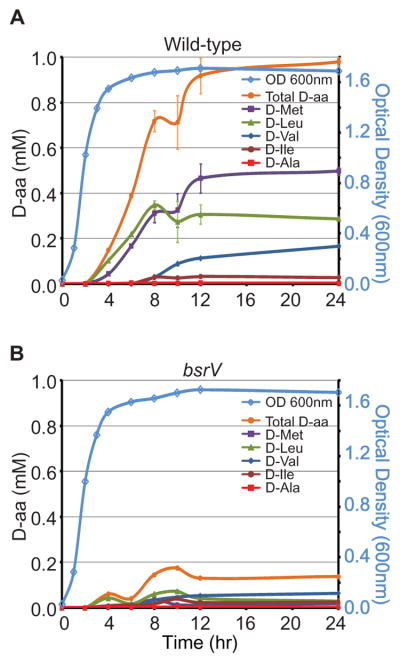
We quantified the D- and L-forms of all amino acids in V. cholerae culture supernatants. D-Met and D-Leu were the predominant D-aa detected (Fig. S1). Total D-aa concentrations were low until early stationary phase, then increased for ~8 hours and plateaued at ~1.0 mM (Fig. 2A). Thus, D-aa accumulation in stationary phase supernatants could account for sphere-inducing activity. D-Ala was not found in the supernatant, so this normal component of PG is not shed into the media. The abundance of particular D-aa, along with the ratios of the enantiomers (Fig. S1), suggests a specific mechanism for creating these D-isomers in stationary phase.
Figure 2.
BsrV is required for production of D-aa in V. cholerae supernatants. (A, B) Kinetics of the accumulation of the indicated D-aa in supernatants from wild-type (A) or bsrV (B) V. cholerae.
Bacteria generate the D-Glu and D-Ala found in PG by specific amino acid racemases, enzymes that convert the chiral carbon of these amino acids from L- to D-forms (3). The V. cholerae genome codes for a putative amino acid racemase (vc1312), in addition to Glu and Ala racemases. A strain lacking vc1312 (renamed here bsrV, broad spectrum racemase Vibrio) had normal growth and morphology but produced minimal D-Met, D-Leu, D-Val and D-Ile (Fig. 2B), suggesting that the BsrV racemase generated these four D-aa. Notably, BsrV was found in the periplasm (Fig. S2), unlike Glu and Ala racemases (3).
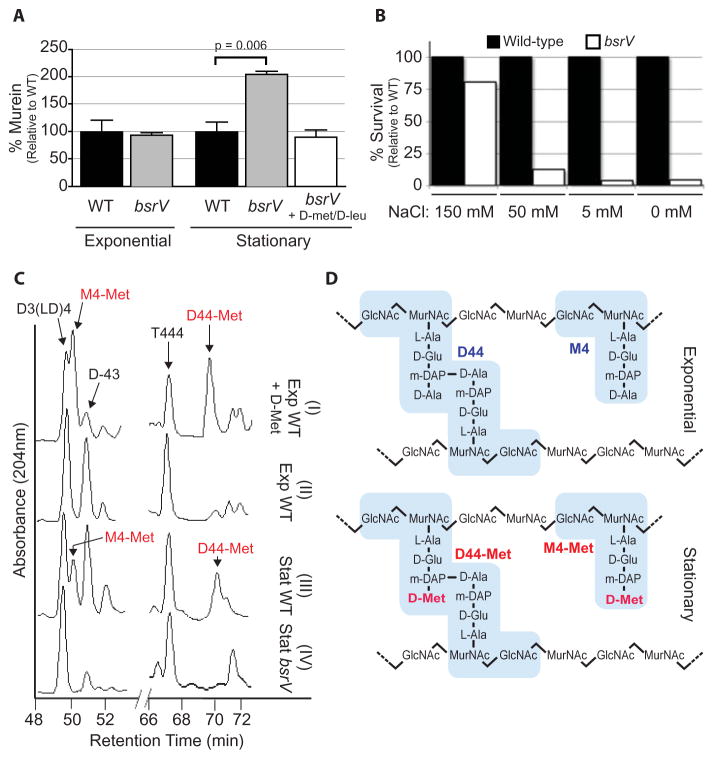
Although the morphology of wild-type cells was not altered by D-aa (Fig. S3), the metabolic expenditure of producing D-aa suggested they have important physiological function(s). Because exogenous D-aa can influence PG composition and structure (4,5), we compared PG isolated from wild-type and bsrV V. cholerae. The bsrV mutant contained twice the amount of PG compared with wild-type cells in stationary phase, while PG levels did not differ in exponential phase (Fig. 3A). Addition of physiological amounts of D-Met and D-Leu to bsrV cultures reduced the amount of PG to wild-type levels, confirming that the absence of D-aa accounted for the increased PG in the bsrV mutant. Thus, D-aa negatively regulate the amount of PG in stationary phase cells. Moreover, the structure of wild-type and bsrV PG isolated from stationary phase cells differed significantly. In comparison to wild-type, the glycan chains in stationary phase PG from the bsrV mutant were ~20% shorter, pentapeptides were reduced ~50%, and there was an increase in trimer muropeptides (Table S1). Despite being less abundant, the PG in wild-type cells appeared to be stronger than in bsrV cells. More than 20× the number of wild-type vs bsrV cells survived an osmotic challenge (Fig. 3B). Thus, D-aa production by BsrV provides a cue for V. cholerae to decrease PG synthesis and alter its cell wall in adaption to stationary phase conditions.
Figure 3.
Influence of D-aa on the amount and composition of PG and osmotic tolerance. (A) PG quantification in wild-type or bsrV cells grown in LB or LB supplemented with D-Met and D-Leu. Murein % was normalized to wild-type in each growth phase. (B) Recovery of wild-type or bsrV V. cholerae from media with the indicated concentrations of NaCl. A representative experiment is shown. (C) Portions of the HPLC profiles of muropeptides from exponentially (Exp) growing wild-type cells ± 0.5 mM D-Met or stationary phase (Stat) wild-type or bsrV cells. D-Met-containing peaks are the monomer M4-Met and the dimer D44-Met; their structures are shown schematically in (D). Muropeptide reference peaks: D3(LD)4 (Dimer crosslinked at DAP-DAP), D-43 (Dimer crosslinked at D-Ala-DAP), T444 (Trimer crosslinked at D-Ala-DAP, D-Ala-DAP).
One mechanism by which D-aa could alter PG is via incorporation into the polymer. Escherichia coli grown in 20 mM D-Met incorporated this amino acid at the fourth position of the PG-peptide bridge (4). High performance liquid chromatography (HPLC) analyses of PG isolated after the addition of physiologic D-Met (0.5 mM) to exponentially growing V. cholerae revealed two peaks that were not detected in control cultures (Fig. 3C, S4A,C). These peaks correspond to monomer and dimer muropeptides (Fig. 3D) containing D-Met, the same muropeptides observed when 20 mM (or 0.5 mM) D-Met was added to E. coli cultures (Fig. S4C,D). Therefore, physiologic production of D-aa by V. cholerae could influence the PG composition of nearby bacteria of different species. Furthermore, these D-Met-containing muropeptides were present in PG isolated from wild-type but not bsrV stationary phase cells (Fig. 3C,S4B,C). Approximately 5% of stationary phase PG contained D-Met modified muropeptides. Thus, physiological production of D-Met by BsrV results in its incorporation into PG in stationary phase. Incorporation of D-Met probably occurs in the periplasm where it is produced, perhaps via the activity of a penicillin-insensitive transpeptidase (4, 6), and may directly influence the strength or flexibility of the PG due to alterations in structure.
D-aa may also have effects on PG that are not a direct consequence of their incorporation. Indeed, 2.0 mM D-Ala stimulated the conversion of rod-shaped mrcA mutant cells to spheres, even though D-Ala is already present at the site where D-Met was incorporated (Fig. S5). Furthermore, exogenous D-Met (and not L-Met) prevented the binding of Bocillin-FL, a fluorescent penicillin derivative, to several V. cholerae cell envelope-associated PBPs, suggesting that D-aa directly bound PBPs (Fig. S6). Thus, D-aa may be regulators as well as substrates of the periplasmic enzymes that synthesize and modify PG. Moreover, the periplasmic targeting of BsrV (Fig S2) efficiently links the site of D-aa production with the cell wall targets of their action.
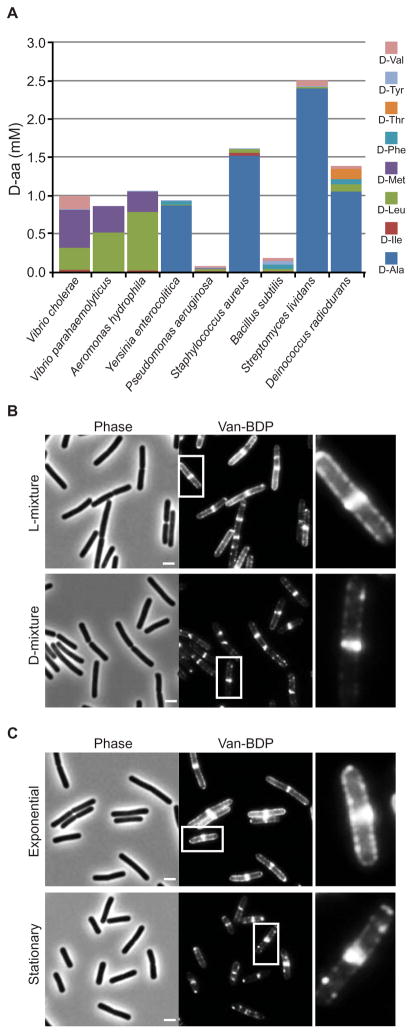
Production of D-aa was not limited to V. cholerae. Diverse bacterial species released D-aa (Fig. 4A and Table S2). All of these species appear to encode several putative racemases, suggesting that they could produce D-aa other than D-Ala and D-Glu (Table S3). The particular D-aa identified in stationary phase supernatants varied among bacterial species (Fig. 4A).
Figure 4.
D-aa are produced by diverse bacterial species and influence B. subtilis PG synthesis. (A) Concentrations of all 19 D-aa were measured and those above 0.01 mM are displayed. Vancomycin-BDP staining of (B) exponential phase B. subtilis supplemented with a mixture of physiologic D- (or the corresponding L-) amino acids or (C) unsupplemented exponential or stationary phase B. subtilis. Bar size = 2 μm. Insets are magnified on the right of each corresponding image.
We tested if physiological concentrations of D-aa also influenced cell wall synthesis in Bacillus subtilis, a gram-positive bacteria that is highly divergent from V. cholerae. To monitor synthesis, we used BodipyFL vancomycin (Van-BDP), which labels the cell wall precursor Lipid II (7). In exponential control cells grown in the presence or absence of a mixture of L-amino acids, Van-BDP predominantly stained the septum and the sidewalls in a pattern suggestive of helical and septal PG insertion (7) (Fig. 4B,C and S7). In contrast, 30 min after the addition of a mixture of D-aa found in stationary phase B. subtilis supernatants (Table S2), Van-BDP staining of the sidewalls was greatly reduced, resulting in a pattern similar to untreated stationary phase cells (Fig. 4B,C, S8). Thus, D-aa could downregulate PG synthesis in stationary phase B. subtilis as well as in V. cholerae. D-aa released in stationary phase might influence B. subtilis PG synthesis via their incorporation into PG (as shown for D-Tyr in Fig. S9) and/or by other mechanisms, such as alterations of the activity of B. subtilis PBPs. Gross changes in cell morphology occurred when B. subtilis was treated with super-physiological concentrations of D-Tyr (Fig. S7), re-enforcing the idea that D-aa regulate cell wall structure. Moreover, addition of the stationary phase D-mixture slowed the growth of exponential B. subtilis cultures (8) (Fig. S10). Thus, release of D-aa by B. subtilis during stationary phase may be a mechanism to synchronize growth inhibition and PG synthesis when population density becomes saturating.
Here, D-aa were found to be released in stationary phase by diverse bacteria as agents of a process through which cell populations synchronously controlled cell wall assembly and modification. Because the accumulation of D-aa coincides with the transition into stationary phase and appears to down regulate PG synthesis, D-aa may enable coordination of metabolic slowing in cell wall and cytoplasmic compartments when resources become scarce. Furthermore, release of D-aa may allow for interspecies regulation among bacteria or other organisms that occupy the same niche. D-aa probably affect other aspects of bacterial physiology in addition to those described here. Indeed, D-serine present in human urine regulates uropathogenic E. coli virulence gene expression (9) and D-aa induce sporulation in Myxococcus xanthus (10). Thus, bacteria have taken advantage of amino acid chirality to sense and respond to specific environmental conditions.
Supplementary Material
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References and Notes
- 1.Holtje JV. Microbiol Mol Biol Rev. 1998;62:181. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vollmer W, Blanot D, de Pedro MA. FEMS Microbiol Rev. 2008;32:149. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 3.Yoshimura T, Esak N. J Biosci Bioeng. 2003;96:103. doi: 10.1016/s1389-1723(03)90111-3. [DOI] [PubMed] [Google Scholar]
- 4.Caparros M, Pisabarro AG, de Pedro MA. J Bacteriol. 1992;174:5549. doi: 10.1128/jb.174.17.5549-5559.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lark C, Lark KG. Can J Microbiol. 1959;5:369. doi: 10.1139/m59-046. [DOI] [PubMed] [Google Scholar]
- 6.Tsuruoka T, et al. Eur J Biochem. 1985;151:209. doi: 10.1111/j.1432-1033.1985.tb09089.x. [DOI] [PubMed] [Google Scholar]
- 7.Tiyanont K, et al. Proc Natl Acad Sci U S A. 2006;103:11033. doi: 10.1073/pnas.0600829103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Champney WS, Jensen RA. J Bacteriol. 1970;104:107. doi: 10.1128/jb.104.1.107-116.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anfora AT, Haugen BJ, Roesch P, Redford P, Welch RA. Infect Immun. 2007;75:5298. doi: 10.1128/IAI.00652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Connor KA, Zusman DR. Mol Microbiol. 1997;24:839. doi: 10.1046/j.1365-2958.1997.3931757.x. [DOI] [PubMed] [Google Scholar]
- 11.We thank B. Davis, C. Jacobs-Wagner and D. Rudner for insightful comments on the manuscript. This work was supported by HHMI and NIH AI-R37-42347 (MKW), CA24487 and GM086258 (JC), MEC BFU2006-04574 and Fundacion Ramon Areces (MAP), Jane Coffin Childs Fellowship (HL), MEC Fellowship (FC), and HHMI EXROP (CNT).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.