Abstract
Dopamine oxidation has been previously demonstrated to cause dysfunction in mitochondrial respiration and membrane permeability, possibly related to covalent modification of critical proteins by the reactive dopamine quinone. However, specific mitochondrial protein targets have not been identified. In this study, we utilized proteomic techniques to identify proteins directly conjugated with 14C-dopamine from isolated rat brain mitochondria exposed to radiolabeled dopamine quinone (150 μM) and differentiated SH-SY5Y cells treated with 14C-dopamine (150 μM). We observed a subset of rat brain mitochondrial proteins that were covalently modified by 14C-dopamine, including chaperonin, ubiquinol-cytochrome c reductase core protein 1, glucose regulated protein 75/mitochondrial HSP70/mortalin, mitofilin, and mitochondrial creatine kinase. We also found the Parkinson’s disease associated proteins ubiquitin carboxy-terminal hydrolase L1 and DJ-1 to be covalently modified by dopamine in both brain mitochondrial preparations and SH-SY5Y cells. The susceptibility of the identified proteins to covalent modification by dopamine may carry implications for their role in the vulnerability of dopaminergic neurons in Parkinson’s disease pathogenesis.
Keywords: Parkinson’s disease, dopamine oxidation, protein modification, proteomics, mitochondria, SH-SY5Y, mitochondrial creatine kinase, mitofilin, UCH-L1, DJ-1
INTRODUCTION
Mitochondrial dysfunction and oxidative stress have been implicated in the pathogenesis of neurodegenerative diseases, including Parkinson’s disease (PD) (Beal, 2007; Halliwell, 2001; Halliwell, 2006; Jenner, 2003; Schapira, 2008). PD is a progressive disorder, pathologically characterized by the loss of pigmented dopaminergic neurons in substantia nigra (SN), and the formation of proteinaceous cytoplasmic inclusions called Lewy bodies (Samii et al., 2004). Though other brain regions are known to be involved in PD, the degeneration of the nigrostriatal dopaminergic neurons combined with the increased oxidative stress observed in PD suggests the neurotransmitter dopamine (DA) may be contributing to disease progression (Greenamyre and Hastings, 2004; Ogawa et al., 2005; Stokes et al., 1999).
Normal DA metabolism can lead to the production of reactive oxygen species (ROS). If not adequately stored in vesicles, DA is also prone to auto- or enzymatic oxidation in the cellular environment, leading to the formation of reactive DA quinone (DAQ) and additional ROS (Graham et al., 1978; Hastings, 1995). The electron deficient DAQ is readily susceptible to attack by cellular nucleophiles, predominantly reduced sulfhydryls abundantly found in cells on reduced glutathione, free cysteine, and cysteinyl residues of proteins (Tse et al., 1976). The interaction of protein with reactive DA metabolites will result in either covalent binding of thiols by DAQ to form 5-cysteinyl-DA, or oxidation of protein sulfhydryl groups (Graham et al., 1978; Hastings et al., 1996; Ito et al., 1988). Many vital cellular and mitochondrial proteins contain cysteine residues whose redox states are critical for function. Thus, DA-induced oxidative modifications, which may alter protein structure as well as function, could have detrimental effects on the cell (Berman and Hastings, 1999; LaVoie and Hastings, 1999; Premkumar and Simantov, 2002).
DA-induced toxicity has been demonstrated both in vitro in cell culture (Ben-Shachar et al., 2004; Jones et al., 2000; Koshimura et al., 2000; Lai and Yu, 1997; Offen et al., 1996) and in vivo (Hastings et al., 1996; Rabinovic et al., 2000), where toxicity to DA terminals was correlated to the amount of DA oxidation and modification of proteins (Hastings et al., 1996). DA and DAQ exposures also altered mitochondrial respiration in isolated intact rat brain mitochondria (Berman and Hastings, 1999; Cohen et al., 1997; Gluck et al., 2002) and triggered permeability transition (Berman and Hastings, 1999), suggesting modification of critical proteins.
Several cellular and mitochondrial proteins have been reported to have altered function following DA exposure and DA oxidation, including cytosolic and mitochondrial creatine kinase (Maker et al., 1986; Miura et al., 1999), mitochondrial aldehyde dehydrogenase (Turan et al., 1989), mitochondrial Complex I (Ben-Shachar et al., 2004; Khan et al., 2005), tyrosine hydroxylase (Kuhn et al., 1999; Xu et al., 1998), and the dopamine transporter DAT (Berman et al., 1996). Proteins associated with familial PD, parkin (LaVoie et al., 2005) and alpha-synuclein (Conway et al., 2001), have also been demonstrated to be targets of covalent modification by DA. It has also been shown that isolated brain mitochondria can accumulate exogenous radiolabeled DA (Brenner-Lavie et al., 2008). In addition, Kahn et al. demonstrated that exposure of crude mitochondrial-synaptosomal fractions to DA led to protein cross-linking and protein-bound DA (Khan et al., 2001). To date, however, the specific proteins directly modified by DAQ following exposure in mitochondria have not been identified. Thus, it was of interest to us to identify and characterize the mitochondrial targets of DA oxidation.
Recently, we have shown that several mitochondrial proteins are decreased in abundance following in vitro DAQ exposure (Van Laar et al., 2008). While demonstrating an effect of DAQ on a subset of mitochondrial proteins, the method utilized did not identify proteins directly modified by covalent binding with DAQ. In this study, we exposed isolated rat brain mitochondria to radiolabeled DA (14C-DA) quinone, and differentiated SH-SY5Y neuroblastoma cells to exogenous 14C-DA. Employing techniques that include 2-D gel electrophoresis, autoradiography, and 2-D difference in-gel electrophoresis (DIGE) fluorescent labeling technology combined with mass spectrometry (MS) analysis, we have identified proteins directly conjugated to 14C-DA. These findings further elucidate the effects of DA oxidation on cellular protein alterations, and may have implications for PD pathogenesis.
METHODS
Materials
3,4-Dihydroxyphenylethylamine, [8-14C] (14C-DA) was purchased from MP Biomedicals, Inc. (Irvine, CA). IEF DryStrips (7 cm 3-5.6pH, 4-7pH, and 6-11pH , and 18 cm 3-10pH), and CyDye™ DIGE Fluor Scarce Sample Labeling (Cysteine-reactive) and Minimal Labeling (Lysine-reactive) dye kits were purchased from GE Healthcare (Piscataway, NJ). Bradford Dye Reagent was purchased from BioRad (Hercules, CA). Promega Gold Mass Spectrometry Grade Modified Trypsin was purchased from Promega (Madison WI). Dopamine (DA), Protease inhibitor cocktail (cat#P2714), mushroom tyrosinase, retinoic acid, and most other chemicals were purchased from Sigma Chemical Co. (St. Louis, MO) unless otherwise noted. The MtCK and mitofilin polyclonal antibodies used in this study were generated for our laboratory by Genemed Synthesis, Inc. (San Antonio, TX). Dulbecco’s modified Eagle medium (DMEM; Gibco) cell culture media, fetal bovine serum (FBS; HyClone), and 10,000 U/mL penicillin/10,000 μg/mL streptomycin (pen/strep; Gibco) were purchased from Invitrogen (Carlsbad, CA). All solutions were prepared using purified water from a Milli-Q system (Millipore Corp., Bedford, MA). Solutions for in-gel and on-blot trypsin digest procedures were prepared using HPLC-grade water from Fisher Biotech (Pittsburgh, PA), and HPLC-grade acetonitrile and spectrophotometric-grade methanol from Sigma-Aldrich (St. Louis, MO).
Mitochondrial Isolation and 14C-Dopamine Exposure Reactions
All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Pittsburgh and are in accordance with guidelines put forth by the National Institutes of Health in the Guide for the Care and Use of Laboratory Animals. Mitochondria were isolated from adult male Sprague-Dawley (300-350g) rat brain tissue via differential centrifugation as previously described (Van Laar et al., 2008). Mitochondrial pellets were resuspended in isolation buffer and kept on ice. Mitochondrial protein content was determined for the total suspension by the Bradford assay (Bradford, 1976). Prior to experimental use, respiration measurements of the isolated mitochondria were made to ensure mitochondrial health, as previously described (Berman and Hastings, 1999). A state 3/state 4 ratio above 6 was considered an indication of healthy, intact mitochondria.
Mitochondrial protein (4 μg/μL) was exposed to 150 μM DA or 14C-DA (0.5-1 μCi; 150μM) and tyrosinase (300U/mL), to oxidize DA to DAQ, in reaction buffer (225 mM mannitol, 75 mM sucrose, 25 mM HEPES, and 1 mM EGTA, pH 7.4 with PIC) for 15 min at room temperature (RT). Mitochondria were then pelleted by centrifugation at 15,000 g for 15 min at 4°C. Control mitochondria underwent an identical procedure without DA present. Mitochondrial pellets were lysed by rigorous pipetting in denaturing 2-D DIGE lysis buffer (9 M urea, 2% w/v CHAPS, and 30 mM Tris-base, pH 8.5) at 100 μL buffer/mg protein. Insoluble material was pelleted by centrifugation (16,000 × g for 1-2 min at RT) and discarded. Protein concentrations in the lysed control and DAQ-exposed samples were determined by the Bradford method (Bradford, 1976).
SH-SY5Y Cell Culture and Dopamine Exposure
Proliferating SH-SY5Y cells were maintained in DMEM supplemented with 10% FBS and 1% pen/strep (SH media). For differentiation, cells were subcultured onto 6 cm or 6-well plates at 2 × 105 cells/mL. Culture media was exchanged for fresh SH media supplemented with 20 μM retinoic acid (SH differentiation media) 48 hr after plating, and every 48 hr thereafter for a total of 5 days of differentiation. On day 5, culture media was exchanged for fresh SH differentiation media supplemented with 150 μM 14C-DA (1 μCi/mL media) for 16 hr. Immediately following treatment, cells were collected by 1 min exposure to 0.25% trypsin with 2.21mM EDTA in HBSS (Mediatech; Herndon, VA) followed by force pipetting, rinsing with PBS, and centrifugation. The resulting cell pellet was rinsed with PBS, centrifuged, then lysed in DIGE lysis buffer supplemented with Chaps Cell Extract Buffer (Cell Signaling Technology; Danvers, MA) and PMSF.
2-D Gel Electrophoresis
For 2-D gel electrophoresis utilizing mini-gel SDS-PAGE, 100-250 μg total protein from 14C-DA exposed mitochondrial or SH-SY5Y cell lysate was loaded via sample cup on rehydrated 7cm pH 3-5.6, pH 4-7, or pH 6-11 DryStrips and isoelectrically focused on a Multiphor II electrophoresis unit according to manufacture’s instructions (GE Healthcare). Focused strips were equilibrated for 10 min at RT in an equilibration buffer (75 mM Tris-HCl pH 6.8, 6 M urea, 30% v/v glycerol, 1% w/v SDS) supplemented with 30 mM DTT, followed by 10 min at RT in equilibration buffer supplemented with 240 mM iodoacetamide. Equilibrated strips were then subjected to electrophoresis on 12% SDS-PAGE gels utilizing a Hoefer Mighty Small II apparatus. Precision Blue markers (BioRad) were used as molecular weight standards.
Transblotting and Autoradiography of 14C Dopamine-modified Proteins
Following 14C-DA mini 2-D electrophoresis, proteins were transferred to PVDF membrane via a BioRad Trans-Blot Semi-Dry Electrophoretic Transfer system. For autoradiography, blots were air-dried and placed in Kodak autoradiogram transcreens with BioMax MS autoradiogram film (Kodak) in a film exposure cassette for 3 days to 3 weeks at -80°C.
Radiolabeled Protein Spot Excision and Trypsin Digest
Following autoradiography, PVDF blots were aligned with the autoradiograms. Using a sterile 2 mm tissue punch, small regions corresponding with spots on the autoradiogram were excised from the PVDF, and then subjected to trypsin digest by methods adapted from Bienvenut et al. (Bienvenut et al., 1999). Briefly, spots were washed with 1:1 methanol and water, air-dried, and then submerged in 10μL 30% acetonitrile in 50 mM ammonium bicarbonate and 4μL 0.1mg/mL trypsin (Promega), and incubated 16-18 hr at RT with constant agitation. Following digestion, the supernatant was collected and saved in a separate tube. The membrane spots were then submerged in 20μL of 80% acetonitrile, and sonicated for 15 min at RT. The resulting supernatant was collected and added to the original digestion supernatant. The total supernatant was dried down by speed-vacuum without heating, and kept at 4°C under desiccation until MS analysis.
Cys- and Lys-CyDye labeling and 2-D DIGE
2-D DIGE analysis with either cysteine-reactive maleimide CyDye (Cys-CyDye) or lysine-reactive NHS-ester CyDye (Lys-CyDye) labeling of control and DAQ- or 14C-DAQ-exposed mitochondrial protein was carried out as previously described (Van Laar et al., 2008). Briefly, controls were paired with DAQ or 14C-DAQ reacted samples from the same mitochondrial isolation, and experiments were completed for sample sets from multiple mitochondrial isolations. Cy5-labeled control and Cy3-labeled DAQ- or 14C-DAQ-exposed samples were combined in equal protein amounts, generating the following DIGE gel experiments: (1) Cys CyDye Cy5 control vs. Cy3 DA, (2) Cys CyDye Cy5 control vs. Cy3 14C-DA, (3) Lys CyDye Cy5 control vs. Cy3 DA, and (4) Lys CyDye Cy5 control vs. Cy3 14C-DA. We previously determined through reciprocal labeling that there is no disparate or preferential labeling exhibited by the individual dyes (Van Laar et al., 2008). Samples were isoelectrically focused on 18cm pH 3-10 DryStrips using a Multiphor II electrophoresis system (GE Healthcare). Focused DryStrips were equilibrated as described above and proteins were separated on 12% SDS-PAGE 1.5mm thick gels using a Hoefer SE600 Ruby Electrophoresis Unit.
Fluorescence Detection, Spot Picking, and In-gel Trypsin Digest
Immediately following the second dimension run, 2-D DIGE gels were scanned for Cy3 and Cy5 dye labeling using a Typhoon 9400 scanner with ImageQuant software (GE Healthcare) to obtain a 100-200 μm resolution image of the gel. Immediately following imaging, 14C-DA 2-D DIGE gels were transblotted to PVDF or nitrocellulose membranes. Non-radioactive 2-D DIGE gels were fixed overnight in 40% MeOH, 1% acetic acid solution at 4°C. Spots of interest were excised using an automated spot picker, designed by Dr. Jonathan Minden of Carnegie Mellon University (instrumentation housed in the University of Pittsburgh Genomics and Proteomics Core Laboratories). Immediately following excision from 2-D DIGE gels, gel plugs were washed and carried through a previously described in-gel trypsin digest procedure (Van Laar et al., 2008).
Image Analysis
For 14C-DA 2-D DIGE experiments, transblots were subjected to autoradiography. Digital scan images of the autoradiogram and the corresponding DIGE gel were digitally merged and compared visually. Images were aligned based on recognizable landmarks. Fluorescence-labeled protein spots that aligned with radioactive spots were considered spots of interest. For correlating protein spots that we did not already have an identification for based on previous data (Van Laar et al., 2008), corresponding spots on non-radioactive DIGE gels were picked for identification, as described above.
MS and MS/MS Analyses for Protein Identification
For MS and MS/MS analyses, dried trypsin-digested samples were rehydrated in 2-3 μl of 0.3% trifluoroacetic acid, 1 mM ammonium citrate in 50:50 acetonitrile/water, and an equal volume of saturated α-cyano-4-hydroxycinnamic acid matrix solution, and then spotted onto a MALDI target plate at 42°C. MS and MS/MS spectra were obtained using an Applied Biosystems 4700 MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Foster City, CA) and processed by GPS Explorer™ (ver. 3) data analysis software (Applied Biosystems) coupled with Mascot™ search engine (Matrix Science) for peak list generation, database search, and statistical analyses. MS and/or combined MS and MS/MS spectra were searched against the National Center for Biotechnology Information non-redundant (NCBInr) and SwissProt databases, specifying “Rodentia” or “Rattus” species for rat brain mitochondria samples, and “Homo sapiens” species for SH-SY5Y samples. Trypsin digest was specified for searches, and MS and MS/MS peak filtering were set at a minimum S/N ratio of 10, with a peak density filter of 50 and maximum 65 peaks allowed. Allowed mass ranges were 800-4000 Da for MS peaks and 20-60 Da under precursor for MS/MS. Precursor tolerance was set at 50 to 100 ppm and MS/MS fragment tolerance was set at 0.2 to 0.4 Da, allowing 1 missed cleavage. Modifications specified included fixed or variable cysteine carbamidomethylation and methionine oxidation.
For peptide mass fingerprinting, a positive protein identification was accepted when a top ranked hit yielded: (1) a probability-based molecular weight search (MOWSE) protein score confidence interval percentage (C.I.%) > 95%, (2) peptides matched ≥ 6, (3) a predicted molecular weight that was appropriate given the migration of the protein spot on the gel or blot, and (4) MS identification for a given 14C-DA-labeled spot could be replicated across two or more separate experiments. Five statistically significant protein identities are also noted that were obtained via MS peptide mass fingerprinting analysis in only one 14C-DA exposure experiment, though corresponding 14C-DA conjugated spots were visible in blots from multiple experiments. These identities are noted in Tables 1 and 3. Identification for a particular spot was accepted for combined MS and MS/MS (MS + MS/MS) results that yielded (1) a top ranked hit with both MS probability-based MOWSE protein score and MS/MS spectra total ion score C.I.% each > 95%, (2) total peptides matched ≥ 6 and/or significant ion score for ≥ 2 unique peptides, (3) a predicted molecular weight that was appropriate given the migration of the protein spot on the blot, and (4) a corresponding 14C-DA-labeled spot was present in blots from multiple experiments. Total ion scores were calculated from weighted ion scores for individual peptides that were matched to a given spot identity. C.I.% values are derived from the probability-based MOWSE scores; values >95% suggest identities are significant, and not random matches. For each protein identity provided, the next non-homologous protein identified using Mascot did not meet our guidelines for acceptability as described above, suggesting that every identified protein was uniquely assigned to its corresponding spot (data not shown).
Table 1.
Identified DA-Conjugated Proteins from DAQ-Exposed Rat Brain Mitochondrial Fractions
| pH 3-5.6 MS Data: |
|||||||
|---|---|---|---|---|---|---|---|
| Protein Spota |
Protein Identification | Accession Number & Database |
Theoretical Protein MW |
Theoretical Protein pI |
Protein Scoreb |
Protein Score C.I.%b |
Peptide Count |
| 3a. gamma enolase | Gamma-enolase (EC 4.2.1.11) (2- phospho-D-glyceratehydro-lyase) (Neural enolase) |
P07323 SwissProt |
47110.9 | 5.03 | 227 | 100 | 25 |
| 3b. gamma enolase | Gamma-enolase (EC 4.2.1.11) (2- phospho-D-glyceratehydro-lyase) (Neural enolase) |
P07323 SwissProt |
47110.9 | 5.03 | 123 | 100 | 17 |
| 2b. HSP60 | unnamed protein product [Rattus norvegicus] |
gi|1334284 NCBI |
57889.7 | 5.35 | 130 | 100 | 18 |
| Combined MS & MS-MS Data: | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein Spota |
Protein Identification | Accession Number& Database |
Theoretical Protein MW |
Theoretical Protein pI |
Protein Scoreb |
Protein Score C.I.%b |
Total Ion Scoreb |
Total Ion C.I. %b |
Peptide Count |
| 2a. HSP60 | unnamed protein product [Rattus norvegicus] |
gi|1334284 NCBI |
57889.7 | 5.35 | 325 | 100 | 125 | 100 | 25 |
| 1. Tubulin beta | Tubulin beta chain (T beta-15) |
P04691 SwissProt |
49931 | 4.79 | 82 | 100 | 13 | 94.581 | 12 |
| pH 4-7 MS Data: |
|||||||
|---|---|---|---|---|---|---|---|
| Protein Spota |
Protein Identification | Accession Number & Database |
Theoretical Protein MW |
Theoretical Protein pI |
Protein Scoreb |
Protein Score C.I.%b |
Peptide Count |
| 8b. actin gamma | PREDICTED: similar to Actin, cytoplasmic 2Gamma-actin) [Rattus norvegicus] |
gi|109492380 NCBI |
58670 | 5.67 | 81 | 99.965 | 12 |
| 12. creatine kinase | Creatine kinase B-type (Creatine kinase B chain) |
gi|122065316 NCBI |
42725.27 | 5.39 | 146 | 100 | 16 |
| 17. DJ-1 | DJ-1 protein [Rattus norvegicus] | gi|16924002 NCBI |
20189.5 | 6.32 | 116 | 100 | 11 |
| 7. gamma-enolase | phosphopyruvate hydratase (EC 4.2.1.11) - rat |
gi|1363309 NCBI |
47495.1 | 5.07 | 85 | 99.977 | 12 |
| 4a. GRP75 | grp75 [Rattus sp.] | gi|1000439 NCBI |
73698.8 | 5.87 | 141 | 100 | 19 |
| 4b. GRP75 | grp75 [Rattus sp.] | gi|1000439 NCBI |
73698.8 | 5.87 | 95 | 100 | 14 |
|
18. guanine nucleotide- binding protein * |
Guanine nucleotide-binding protein G(I)/G(S)/G(T)subunit beta 2(Transducin beta chain 2) |
P54313 SwissProt |
37331 | 5.6 | 56 | 98.562 | 9 |
| 5a. HSP60 | unnamed protein product [Rattus norvegicus] |
gi|1334284 NCBI |
57889.7 | 5.35 | 217 | 100 | 25 |
| 5b. HSP60 | unnamed protein product [Rattus norvegicus] |
gi|1334284 NCBI |
57889.7 | 5.35 | 137 | 100 | 19 |
| 15a. isocitrate dehydrogenase | isocitrate dehydrogenase 3 (NAD+) alpha [Rattusnorvegicus] |
gi|16758446 NCBI |
40044.2 | 6.47 | 189 | 100 | 22 |
| 15b. isocitrate dehydrogenase | isocitrate dehydrogenase 3 (NAD+) alpha [Rattusnorvegicus] |
gi|16758446 NCBI |
40044.2 | 6.47 | 148 | 100 | 18 |
| 11. tubulin beta | Tubulin beta-2A chain (T beta-15) - Rattus norvegicus (Rat) |
P85108 SwissProt |
49907 | 4.78 | 226 | 100 | 25 |
| 6b. Ub-cyt C core protein 1 | Ubiquinol-cytochrome-c reductase complex core protein1, mitochondrial precursor |
Q68FY0 SwissProt |
53499.7 | 5.57 | 83 | 99.997 | 12 |
| 9. UCH-L1 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 (EC3.4.19.12) (UCH-L1) |
Q00981 SwissProt |
25164.6 | 5.14 | 80 | 99.994 | 9 |
| pH 4-7 Combined MS & MS-MS Data: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein Spota |
Protein Identification | Accession Number& Database |
Theoretical Protein MW |
Theoretical Protein pI |
Protein Scoreb |
Protein Score C.I.%b |
Total Ion Scoreb |
Total Ion C.I. %b |
Peptide Count |
| 8a. actin gamma | PREDICTED: similar to Actin, cytoplasmic 2(Gamma-actin) [Rattus norvegicus] |
gi|109492380 NCBI |
58763.5 | 5.67 | 252 | 100 | 184 | 100 | 12 |
| 13. aldehyde dehydrogenase | mitochondrial aldehyde dehydrogenase precursor [Rattus norvegicus] |
gi|45737866 NCBI |
56079.4 | 6.69 | 318 | 100 | 200 | 100 | 16 |
| 15c. isocitrate dehydrogenase | isocitrate dehydrogenase 3 (NAD+) alpha [Rattusnorvegicus] |
gi|16758446 NCBI |
40044.2 | 6.47 | 277 | 100 | 190 | 100 | 12 |
|
10. NADH- oxidoreductase 30 kDa subunit |
NADH dehydrogenase (ubiquinone) Fe-S protein 3(predicted), isoform CRA_b [Rattus norvegicus] |
gi|149022594 NCBI |
34391.7 | 8.67 | 133 | 100 | 37 | 97.67 | 13 |
| 16a. peroxiredoxin 3 | Thioredoxin-dependent peroxide reductase, mitochondrial precursor (Peroxiredoxin-3) (PRX-3) |
gi|118597399 NCBI |
28562.6 | 7.14 | 248 | 100 | 191 | 100 | 7 |
| 16b. peroxiredoxin 3 | Thioredoxin-dependent peroxide reductase, mitochondrial precursor Rattus norvegicus (Rat) |
Q9Z0V6 SwissProt |
28562.6 | 7.14 | 91 | 100 | 77 | 100 | 3 |
| 14. succinate CoA ligase | PREDICTED: similar to succinate- Coenzyme A ligase,ADP-forming, beta subunit [Rattus norvegicus] |
gi|62661722 NCBI |
50616.3 | 7.57 | 232 | 100 | 130 | 100 | 15 |
| 6a. Ub-cyt C core protein 1 | ubiquinol-cytochrome c reductase core protein I [Rattus norvegicus] |
gi|51948476 NCBI |
53499.7 | 5.57 | 364 | 100 | 229 | 100 | 16 |
| pH 6-11 MS Data: |
|||||||
|---|---|---|---|---|---|---|---|
| Protein Spota |
Protein Identification | Accession Number & Database |
Theoretical Protein MW |
Theoretical Protein pI |
Protein Scoreb |
Protein Score C.I.%b |
Peptide Count |
| 26. ATP synthase b * | ATP synthase B chain, mitochondrial |
Q9CQQ7 SwissProt |
28930.4 | 9.11 | 50 | 99.06 | 10 |
|
20. glutamate oxaloacetate transaminase 2 |
glutamate oxaloacetate transaminase 2 [Rattusnorvegicus] |
gi|6980972 NCBI |
47683.2 | 9.13 | 181 | 100 | 19 |
| 27. triosephosphate isomerase | Triosephosphate isomerase (EC 5.3.1.1) |
P48500 SwissProt |
26772.7 | 6.51 | 44 | 98.941 | 7 |
| 24. SOD2 * | Superoxide dismutase [Mn], mitochondrial precursor (EC 1.15.1.1) - Rattus norvegicus |
P07895 SwissProt |
24658.6 | 8.96 | 59 | 99.327 | 7 |
| 23. VDAC1 * | Unknown (protein for IMAGE:7462139) [Rattusnorvegicus] |
gi|75773332 NCBI |
31920.2 | 8.35 | 87 | 99.993 | 10 |
| Combined MS & MS-MS Data: | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein Spota |
Protein Identification | Accession Number& Database |
Theoretical Protein MW |
Theoretical Protein pI |
Protein Scoreb |
Protein Score C.I.%b |
Total Ion Scoreb |
Total Ion C.I. %b |
Peptide Count |
| 22. ATP synthase gamma | ATP synthase gamma chain, mitochondrial - Rattusnorvegicus (Rat) |
P35435 SwissProt |
30228.7 | 8.87 | 54 | 97.427 | 25 | 98.625 | 6 |
|
20. fumarylacetoacetate hydrolase |
PREDICTED: similar to fumarylacetoacetate hydrolasedomain containing 2A [Rattus norvegicus] |
gi|34858672 NCBI |
40884.2 | 8.49 | 105 | 100 | 55 | 99.989 | 8 |
|
19. mitochondrial creatine kinase |
creatine kinase, mitochondrial 1, ubiquitous [Rattusnorvegicus] |
gi|60678254 NCBI |
46932.2 | 8.58 | 175 | 100 | 70 | 99.998 | 15 |
| 25. Rieske Fe-S | Rieske Fe-S protein precursor | gi|206681 NCBI |
27671.3 | 8.9 | 119 | 100 | 74 | 100 | 7 |
= Protein Spots correspond with Figure 1
= Probability-based MOWSE score (Protein Score), Protein Score Confidence Interval (C.I.), Total Ion Score, and Total Ion C.I.% represent the top score and C.I. pairing obtained across all blots from which the protein was confidently identified
= Identified by MS-based peptide mass fingerprinting in only one blot, but corresponding spots were observed in blots from 2 or more separate experiments For all other identities provided, n = 2-4 for MS analyses, n = 1-3 for MS/MS analyses
Table 3.
Identified DA-Conjugated Proteins from DA-Exposed SH-SY5Y Cells
| MS Data: | |||||||
|---|---|---|---|---|---|---|---|
| Protein Spota |
Protein Identification | Accession Number |
Theoretical Protein MW |
Theoretical Protein pI |
Protein Scoreb |
Protein Score C.I.%b |
Peptide Count |
| 2a. Actin * | ACTB protein [Homo sapiens] | gi|15277503 | 40194 | 5.55 | 108 | 100 | 15 |
| 2b. Actin * | ACTB protein [Homo sapiens] | gi|15277503 | 40194 | 5.55 | 146 | 100 | 16 |
| 2c. Actin * | actin, gamma 1 propeptide [Homo sapiens] |
gi|4501887 | 41662 | 5.31 | 138 | 100 | 17 |
| 4. Cloride intracellular channel* | chloride intracellular channel 1 [Homo sapiens] |
gi|14251209 | 26792 | 5.09 | 119 | 100 | 13 |
| 7a. ER-60 | protein disulfide isomerase family A, member 3, isoformCRA_a [Homo sapiens] |
gi|119597640 | 54454 | 6.78 | 249 | 100 | 28 |
|
1. protein disulfide isomerase related protein 5 * |
protein disulfide isomerase-related protein 5 [Homosapiens] |
gi|1710248 | 46170.2 | 4.95 | 131 | 100 | 8 |
| 3. Tropomyosin | hypothetical protein [Homo sapiens] |
gi|57997573 | 27387 | 4.71 | 199 | 100 | 22 |
| 5. UCH-L1 | ubiquitin carboxyl-terminal esterase L1 |
gi|21361091 | 25151 | 5.33 | 148 | 100 | 15 |
| Combined MS & MS-MS Data: | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein Spota |
Protein Identification | Accession Number |
Theoretical Protein MW |
Theoretical Protein pI |
Protein Scoreb |
Protein Score C.I.%b |
Total Ion Scoreb |
Total Ion C.I. %b |
Peptide Count |
| 6. GRP75 | heat shock 70kDa protein 9B precursor |
gi|24234688 | 73920 | 5.87 | 145 | 100 | 94 | 100 | 10 |
| 7b. ER-60 | protein disulfide isomerase [Homo sapiens] |
gi|860986 | 57043 | 6.1 | 393 | 100 | 210 | 100 | 21 |
| 9. DJ-1 | (Q99497) Protein DJ-1 (Oncogene DJ1) |
Q99497 ** | 19879 | 6.33 | 172 | 100 | 78 | 100 | 10 |
| 8. Peroxiredoxin 2 | TSA [Homo sapiens] | gi|1617118 | 18486 | 5.19 | 325 | 100 | 236 | 100 | 9 |
| 10. Nucleoside diphosphate kinase A | NME1-NME2 protein [Homo sapiens] |
gi|66392203 | 30346 | 9.06 | 228 | 100 | 147 | 100 | 8 |
| 11. SOD1 | Chain A, A4v Mutant Of Human Sod1 |
gi|47169370 | 16051 | 5.7 | 95 | 99.993 | 52 | 99.86 | 4 |
= Protein Spots correspond with Figure 5
= Probability-based MOWSE score (Protein Score) Protein Score Confidence Interval (C.I.) and Total Ion Score and Total Ion C.I.% represent the top score and C.I. pairing obtained across all blots from which the protein was confidently identified
= Identified by MS-based peptide mass fingerprinting in only one blot, but corresponding spots were observed in blots from 2 or more separate experiments For all other identities provided, n = 2-3 for MS analyses, n = 1-3 for MS/MS analyses
= Accession Number from SwissProt database. All others are from NCBI database.
Western Blot Detection of Mitofilin & MtCK
Following autoradiography, the transblots generated from 14C-DA 2-D DIGE gels were carried through the Western blot detection procedure. The membrane was washed and placed in a 1:1000 dilution of rabbit anti-MtCK or 1:5000 dilution of rabbit anti-mitofilin primary antibody 16-18 hr at 4°C. Membranes were developed using the BioRad Immune-Star® goat-anti-rabbit Chemiluminescence Detection kit, exposed to Biomax MR film (Kodak), and developed for imaging. Using recognizable landmarks for alignment, scanned images of the Western blot and the autoradiogram were digitally merged with the ImageQuant scan of the fluorescent DIGE gel for visual comparison.
RESULTS
14C-DA-conjugated mitochondrial proteins were identified directly from transblots of 2-D gels
Following exposure to 14C-DAQ, mitochondria were lysed and proteins separated by 2-D gel electrophoresis using three separate pI ranges for the first dimension, pH 3-5.6 (n = 5 blots, from 4 independent mitochondrial experiments), pH 4-7 (n = 9 blots, from 6 independent mitochondrial experiments), and pH 6-11 (n = 7 blots, from 6 independent mitochondrial experiments). 2-D gels were transblotted to PVDF, and the membranes used to generate autoradiograms (Figure 1). Distinct spots of radioactivity indicated protein targets covalently modified by 14C-DAQ. The autoradiograms were then aligned with their PVDF blots, corresponding 14C-DA-associated spots were excised from the blot, and proteins were identified by mass spectrometry (MS) analysis (Figure 1, Table 1).
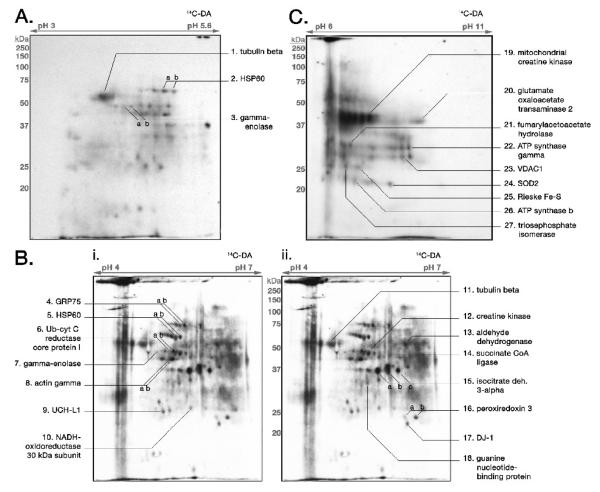
Figure 1. 14C-DAQ Exposure of Isolated Brain Mitochondria Results in DA Conjugation to Select Mitochondrial Proteins.
Isolated rat brain mitochondria were exposed to 14C-DA/tyrosinase (150μM, 0.5-1μCi; 300 U/mL) and subjected to 2-D gel electrophoresis, followed by transblotting to PVDF membrane and autoradiography. Samples were analyzed at pH ranges of (A) pH 3-5.6, (B) pH 4-7, and (C) pH 6-11. Select radioactive spots displaying protein conjugated with 14C-DA were picked from the PVDF membrane and subjected to MS analysis for protein identification. Protein identities are provided for each pI range. The autoradiographic image of a representative pH 4-7 2-D gel protein blot was duplicated in (B-i.) and (B-ii.) for ease of accurate labeling of identified proteins.
Proteins involved in various mitochondrial functions were identified as being covalently modified by 14C-DAQ, including aldehyde dehydrogenase, the tricarboxylic acid (TCA) cycle protein subunit isocitrate dehydrogenase 3-alpha, TCA cycle associated protein glutamate oxaloacetate transaminase 2, the NADH-ubiquinone oxidoreductase 30 kDa subunit of Complex I, the ubiquinol-cytochrome c reductase core protein 1 and Rieske Fe-S protein subunits of Complex III, ubiquitous mitochondrial creatine kinase (MtCK), and the chaperone proteins heat shock protein 60 (HSP60, chaperonin), and mortalin, also known as glucose regulated protein 75 or mitochondrial heat shock protein 70 (mortalin/GRP75/mtHSP70). Additionally, three proteins previously demonstrated to interact with mitochondrial membranes, the glycolytic enzyme enolase (Brandina et al., 2006; Entelis et al., 2006; Giege et al., 2003) and the cytoskeletal proteins tubulin (Carre et al., 2002) and actin (Boldogh and Pon, 2006; Boldogh and Pon, 2007), were also identified to be covalently modified following exposure to 14C-DAQ. Mitochondrial voltage-dependent anion channel 1 (VDAC1), mitochondrial manganese superoxide dismutase (SOD2), and guanine nucleotide-binding protein subunit beta 2 were confirmed by MS peptide mass fingerprinting analysis from only one blot each, though corresponding radiolabeled spots were present on blots from multiple experiments. Several other proteins not specifically mitochondrial were also found to be covalently modified by 14C-DA, including the glycolytic enzyme triosephosphate isomerase, cytosolic creatine kinase, and the PD-associated proteins DJ-1 and ubiquitin carboxy-terminal hydrolase L1 (UCH-L1). The significance of the presence and modification of these proteins is discussed below.
The proteins tubulin, HSP60, gamma-enolase, and UCH-L1 were found in both pH 3-5.6 (Figure 1A) and pH 4-7 (Figure 1B) blots due to overlap of the pI ranges examined, dually confirming the identity of these proteins as targets of covalent modification by 14C-DAQ. The identity of some 14C-DA-associated spots, however, remained unidentified, presumably due to limitations with protein recovery and MS analysis.
DA conjugated proteins can be identified by comparing 2-D DIGE Fluorescent Dye Labeling and 14C-DA autoradiography
Previously, utilizing 2-D DIGE and MS techniques, we demonstrated that in vitro exposure of rat brain mitochondria to DAQ resulted in decreased abundance of several mitochondrial proteins (Van Laar et al., 2008). We reasoned here that by combining 2-D DIGE fluorescent labeling technology with autoradiographic techniques, we could take advantage of the protein identification map we already established for 2-D DIGE analysis of rat brain mitochondria. This methodology would provide both identities of covalently modified proteins and information as to whether covalent modification by DA correlated with the previously observed loss of abundance in select mitochondrial proteins following DAQ exposure.
Cysteine CyDye DIGE gels of rat brain mitochondria (Cys-CyDye Cy5 control vs. Cys-CyDye Cy3 14C-DAQ-exposed protein; Figure 2A) demonstrated a spot patterning and differential fluorescence identical to that previously described (Van Laar et al., 2008), as did the Lysine CyDye labeled DIGE gels of 14C-DAQ-exposed protein (data not shown). DIGE gels of 14C-DA-exposed mitochondrial proteins (Cys-CyDye DIGE, n = 5 gels from 5 separate experiments; Lys-CyDye DIGE, n = 4 gels from 3 separate experiments) were imaged and then transblotted to a membrane for autoradiography. The resulting autoradiograms were compared back to images of their parent gels. Figure 2B shows the autoradiogram generated from a blot of the representative 14C-DA Cys-CyDye DIGE gel in Figure 2A. Figure 2C shows the digital merge of the corresponding fluorescent and autoradiographic images.
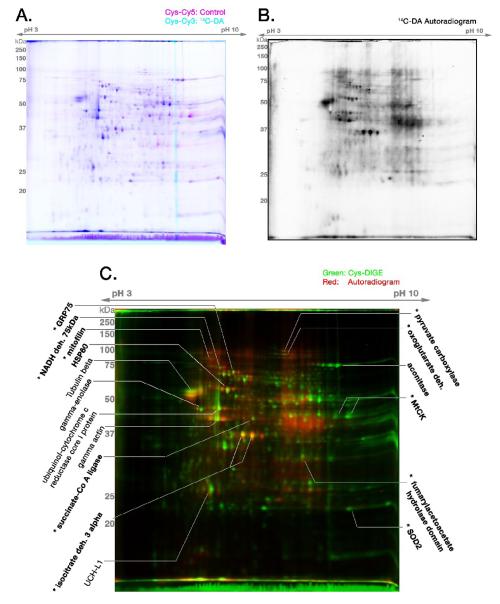
Figure 2. 14C-DAQ Exposure of Isolated Brain Mitochondria Combined with 2-D DIGE.
Lysed Control (C) and 14C-DAQ exposed (14C-DA) mitochondrial proteins were reacted separately with a minimal concentration of cysteine-reactive CyDyes, and then analyzed by 2-D DIGE and autoradiography. (A) Cysteine-labeled (maleimide dye) DIGE gel of Control mitochondrial protein (Cy5, magenta) and 14C-DAQ exposed mitochondrial protein (Cy3, cyan). (B) Autoradiogram generated by the PVDF transblot of the 14C-DIGE gel shown in (A). (C) Merged pseudocolor image of the fluorescent cysteine-labeled 14C-DA DIGE (Cys-CyDye DIGE) image in (A) (green) and its corresponding autoradiogram of the PVDF transblot (red). Yellow spots demonstrate proteins visualized by DIGE that are conjugated to 14C-DA. Protein identifications were obtained from parallel non-radioactive DIGE gels treated identically to 14C-DA DIGE experiments. BOLD protein identifications were previously described (Van Laar et al., 2008). * indicates proteins found to be decreased in abundance following DAQ exposure based on previous DeCyder analysis of DIGE gels (Van Laar et al., 2008). Figure is representative of n = 5 separate gels.
The identities of several proteins associated with 14C-DA labeling were determined based on identities established for analogous spots from our previous DIGE experiments (Van Laar et al., 2008) (Figure 2C; bold text). Several proteins that we previously identified as decreased in abundance following exposure of brain mitochondria to DAQ were identified as being covalently modified by 14C-DAQ (Figure 2C; marked with *). These proteins include mitofilin, 75kDa subunit of Complex I, mortalin/GRP75/mtHSP70, fumarylacetoacetate hydrolase domain protein, superoxide dismutase 2 (SOD2), MtCK, and the TCA cycle proteins isocitrate dehydrogenase 3-alpha subunit and succinate-CoA ligase. 14C-DA modification was also correlated with several previously identified protein spots whose abundances did not appear to be altered following DAQ exposure (Van Laar et al., 2008), including the chaperone HSP60 and TCA cycle protein aconitase (Figure 2C; bold text).
In this study, we also observed several 14C-DA conjugated protein spots that were not previously identified in our other DIGE experiments (Figure 2C; regular text). Additionally, these did not appear decreased in abundance. To identify these proteins, Cys- and Lys-CyDye 2-D DIGE gels with control and non-radiolabeled DA exposed mitochondrial protein were run in parallel with the 14C-DAQ DIGE gels, as described in the methods. Protein spots identified in these experiments include actin, ubiquinol cytochrome-C reductase core protein 1, tubulin, and UCH-L1 (Figure 2C, Table 2). DeCyder analysis confirmed that the overall normalized abundance of these proteins does not decrease following DAQ exposure (data not shown). It should also be pointed out that several abundant proteins, based on DIGE labeling, were not associated with DA labeling, including the previously-identified protein glutamate dehydrogenase 1 (Van Laar, et al., 2008), a cysteine-containing protein which was also previously found to be unaltered in abundance following DAQ exposure. This suggests that reactivity of proteins to DAQ is not based on sheer abundance alone.
Table 2.
Identified Proteins from Control vs. 14C-DA DIGE Corresponding with Radiolabeling
| Protein Spota |
Protein Identification | Accession Number & Database |
Theoretical Protein MW |
Theoretical Protein pI |
Protein Scoreb |
Protein Score C.I.%b |
Peptide Count |
|---|---|---|---|---|---|---|---|
| tubulin beta | Tubulin, beta, 2 [Rattus norvegicus] |
gi|38014578 NCBI |
49769 | 4.79 | 225 | 100 | 24 |
| gamma enolase | enol_cds [Rattus norvegicus] | gi|1619609 NCBI |
47110.9 | 5.03 | 139 | 100 | 19 |
| Ub-cyt C red core 1 protein | Ubiquinol-cytochrome c reductase core protein I [Rattusnorvegicus] |
gi|51259340 NCBI |
52815.4 | 5.57 | 91 | 99.997 | 14 |
| actin gamma | similar to gamma actin-like protein [Rattus norvegicus] |
gi|34875636 NCBI |
43082.3 | 5.11 | 143 | 100 | 17 |
| UCH-L1 | Ubiquitin carboxy-terminal hydrolase L1 [Mus musculus] (Protein Group: ubiquitin carboxyl- terminal hydrolase PGP9.5 Rattus |
gi|25058057 NCBI |
25164.6 | 5.14 | 87 | 99.996 | 9 |
= Protein Spots correspond with Figure 2
= Probability-based MOWSE score (Protein Score) and Protein Score Confidence Interval (C.I.) represent the top score and C.I. pairing obtained across all blots from which the protein was confidently identified (n = 3-5)
Comparing 14C-DA autoradiography, 2-D DIGE, and Western blot detection demonstrates DA conjugation with MtCK and mitofilin
Following autoradiography, blots of 14C-DA 2-D DIGE gels were used for immunochemical detection of MtCK and mitofilin. Both proteins were previously identified from Cys- and Lys-CyDye DIGE gels comparing DAQ-exposed and control rat brain mitochondria (Van Laar et al., 2008). Here, Western blot analysis confirmed the positions of mitofilin (Figure 3) and MtCK (Figure 4) within the 14C-DA 2-D DIGE gel blots. Alignment of digital images of the parent fluorescent gels and the resulting autoradiograms demonstrated the association of covalently bound 14C-DA with these two proteins (Figures 3D and 4D).
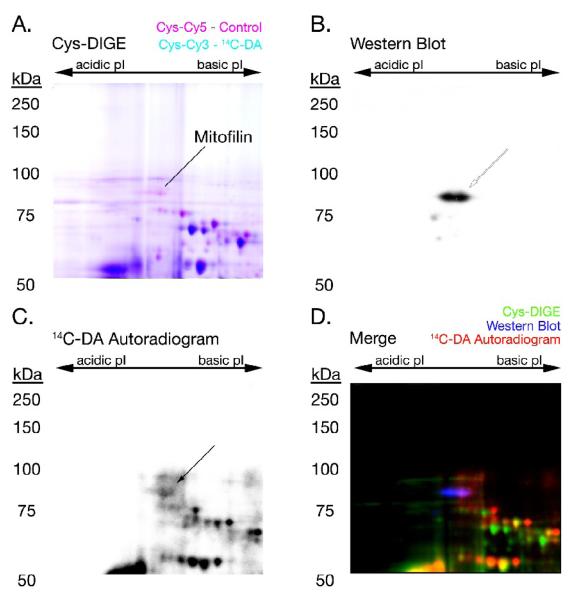
Figure 3. 14C-DA Conjugation with Mitofilin.
A 14C-DA Cys-CyDye DIGE gel was transferred to PVDF membrane, followed by autoradiography and Western blot analysis to detect mitofilin. The fluorescent scan of the DIGE gel (A), Western blot (B), and autoradiogram (C) of 14C-DA-labeled mitochondrial mitofilin were merged (D) to demonstrate the association between 14C-DA-modification (red), immunodetection (blue), and the DIGE-MS spot identification (green) of mitofilin. Arrows point to the location of mitofilin in (B) and (C).
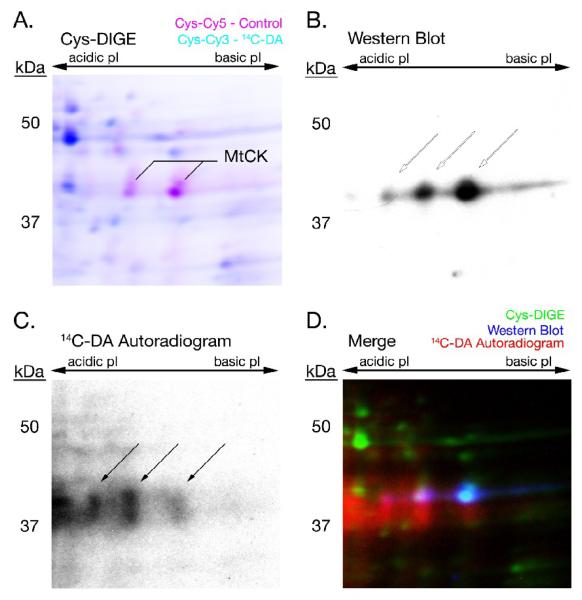
Figure 4. 14C-DA Conjugation with MtCK.
A 14C-DA Cys-CyDye DIGE gel was transferred to PVDF membrane, followed by autoradiography and Western blot analysis to detect MtCK. The fluorescent scan of the DIGE gel (A), Western blot (B), and autoradiogram (C) of 14C-DA-labeled mitochondrial creatine kinase (MtCK) were merged (D) to demonstrate the association between 14C-DA-modification (red), immunodetection (blue), and the DIGE-MS identification (green) of MtCK. Arrows point to the location of MtCK in (B) and (C).
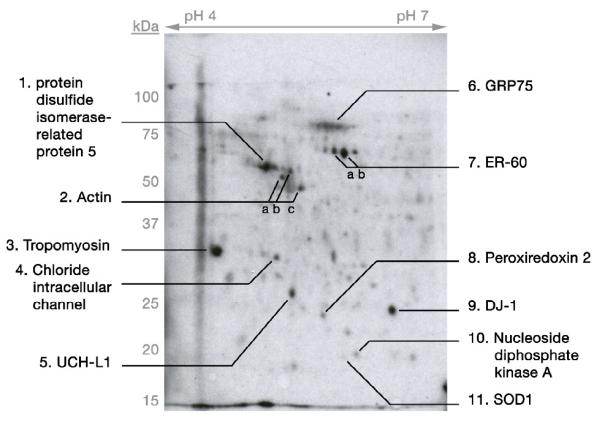
14C-DA-conjugated proteins were identified from differentiated SH-SY5Y cells
Differentiated SH-SY5Y cells were exposed to 150 μM 14C-DA (1 μCi/mL media) for 16 hrs, a concentration and time point established in our laboratory to induce slight (∼10%) yet significant cell death (unpublished data). Whole cell lysates from three separate platings of cells with 14C-DA treatment were independently separated by 2-D gel electrophoresis, and the resulting blots of the gels were used to generate autoradiograms to map DA-conjugated proteins. A representative autoradiogram showing covalently modified proteins following SH-SY5Y cell exposure to exogenous radiolabeled DA is presented in Figure 5. Using MS and MS + MS/MS analyses, we identified several 14C-DA conjugated proteins (Figure 5; Table 3). These proteins include peroxiredoxin 2 isoform A, nucleoside diphosphate kinase A, superoxide dismutase 1 (SOD1), ER-60, mortalin/GRP75/mtHSP70, tropomyosin, UCH-L1, and DJ-1. Actin, protein disulfide isomerase-related protein 5, and chloride intracellular channel protein were confirmed by MS peptide mass fingerprinting analysis in only one blot, though corresponding radiolabeled spots were present in blots from all three experiments. Interestingly, some of the modified proteins identified in SH-SY5Y cells, actin, DJ-1, UCH-L1, and mortalin/GRP75/mtHSP70, were also detected as covalently modified by 14C-DA in the rat brain mitochondrial model of DAQ exposure.
Figure 5. Exposure of Differentiated SH-SY5Y Cells to Exogenous 14C-DA Results in DA Conjugation to Cellular Proteins.
Differentiated SH-SY5Y cells were treated with 14C-DA (150μM; 10μCi/mL media) and subjected to 2-D gel electrophoresis, followed by transblotting to PVDF membrane and autoradiography. Samples were analyzed at a pH range of 4-7 pI. Select radioactive spots displaying protein conjugated with 14C-DA were picked from the PVDF membrane and subjected to MS analysis for protein identification. Protein identities are indicated on autoradiogram (details in Table 3). Representative gel of n = 3.
DISCUSSION
Summary
Utilizing proteomic analyses with the DA oxidation model of PD, we identified protein targets of covalent modification by DAQ. Analysis of isolated rat brain mitochondria exposed to 14C-DAQ in vitro revealed a subset of mitochondrial proteins conjugated to DA, including mortalin/GRP75/mtHSP70, HSP60, MtCK, mitofilin, glutamate oxaloacetate transaminase 2, isocitrate dehydrogenase 3 subunit alpha, the ubiquinol-cytochrome c reductase core protein 1 and Rieske Fe-S protein subunits of Complex III, and the 75 kDa and 30 kDa subunits of Complex I. We also identified several DA-conjugated proteins from whole-cell lysate of 14C-DA-exposed differentiated SH-SY5Y cells. Some of the proteins identified from SH-SY5Y cells, including actin, UCH-L1, DJ-1, and mortalin/GRP75/mtHSP70, correlated with the mitochondrial 14C-DAQ-exposure model, suggesting that DA-modified proteins identified from DAQ-exposed mitochondria are also targets of modification in living cells. Covalent modification of these proteins by DA may lead to altered structure or inactivation of function, and may play a role in the vulnerability of dopaminergic neurons in PD pathogenesis.
We utilized two methods for identification of 14C-DA-modified proteins in the mitochondrial model. First, using MS analyses we carried out direct identification of protein spots conjugated to radiolabeled DA from the PVDF membrane. Second, we compared known spot patterns from fluorescent-labeled 2-D DIGE gels of radiolabeled protein to the autoradiographic images generated from blots of those same gels. In the second method, identities were determined by MS analyses of protein spots from parallel, non-radioactive DIGE gels. Most of the covalently-modified proteins identified were similar between the two methods, validating both the protein identities and the two methods for identifying DA-modified proteins. One inherent limitation to the methods utilized in this study is that they favor the detection of higher-abundance proteins (reviewed in Ahmed and Rice, 2005; Shi et al., 2008). Therefore, it is possible that other, less-abundant proteins were also modified by DAQ, but were below our level of detection.
Noting that isolated brain mitochondria were intact when exposed to 14C-DAQ, it is interesting that we identified covalently-modified proteins associated with the mitochondrial matrix as well as the intermembrane space. This suggests that the electrophilic DAQ gains access to both compartments in the intact mitochondrion at a physiological pH (pH 7.4). This finding also has implications for the reactivity of specific mitochondrial proteins to DA quinone. Thus, the 14C-DA labeled spots observed in this study represent proteins that are the most reactive and/or accessible to DAQ as compared to the remainder of the mitochondrial proteome.
Some proteins identified in DAQ-exposed rat brain mitochondria, such as triosephosphate isomerase and UCH-L1, are not typically associated with the mitochondria. Mitochondrial-enriched fractions isolated from brain by our described procedure typically contain approximately 10% synaptosomes (Berman et al., 2000), and thus we attribute the identification of a small number of non-mitochondrial proteins to possible association with these components. The identification of DA-conjugated non-mitochondrial proteins in samples does not diminish the significance of their modification. We demonstrate here that the cytosolic proteins actin, tubulin, UCH-L1, DJ-1, and triosephosphate isomerase in their native state are targets of DA modification when exposed to DAQ in vitro. Further, actin, UCH-L1, and DJ-1 were also observed to be covalently modified in DA-exposed SH-SY5Y cells, which may have implications for the role of these proteins in neurodegeneration.
Several mitochondrial proteins covalently modified by DA displayed changes in abundance
Several proteins we identified here as being covalently modified by DAQ were also identified in our previous study as being decreased in abundance following mitochondrial exposure to DAQ (Van Laar et al., 2008). The proteins MtCK and mitofilin, in particular, were previously confirmed to be decreased in DAQ-exposed rat brain mitochondria and DA-exposed PC12 cells via Western blot analysis. By comparison of Western blot and autoradiography, MtCK and mitofilin are demonstrated here to also be covalently modified by 14C-DA. We previously hypothesized that the loss of protein may involve either rapid aggregation or proteolytic degradation of oxidatively modified proteins. However, other proteins identified as DA-conjugated in this study were not associated with any measurable change in abundance, such as HSP60 and aconitase. Thus, covalent modification by DAQ does not necessarily correspond with decreases in protein abundance. Additional factors such as the site of modification or location of the protein within the mitochondria may play a role in defining the susceptibility of specific mitochondrial proteins for proteolytic degradation or aggregation following oxidative modification. Further study will be necessary to evaluate the differences in individual protein responses to DA oxidation both in mitochondria and in cells.
Protein Targets of DA Conjugation Encompass Multiple Critical Functions, and are Known Targets for Oxidative Modification
The protein targets of covalent modification by DAQ that we have identified are involved in a range of critical mitochondrial and cellular functions. Because they are susceptible to modification by the electrophilic DAQ, many of the proteins identified in this study are likely vulnerable to other oxidative agents that induce modification. Indeed, several proteins identified here have been reported in other studies to be oxidatively modified in association with disease, disease models, and oxidative stress. One such protein is mitofilin, which we previously identified as exhibiting decreased abundance following DAQ-exposure (Van Laar et al., 2008). Various studies have shown that mitofilin is susceptible to oxidative stress, demonstrating oxidatively-modified cysteine residues following alcohol exposure in hepatoma cells (Suh et al., 2004), as well as a ROS-induced reduction of protein levels (Magi et al., 2004). Given the proposed role of mitofilin in maintaining mitochondrial cristae morphology (John et al., 2005) and interaction with key mitochondrial import proteins (Xie et al., 2007), oxidative modification could have a detrimental impact on proper function or protein-protein interactions, and thus on mitochondrial stability.
Protein import and processing could also be impacted by the identified modification of two mitochondrial protein chaperones, HSP60 and mortalin/GRP75/mtHSP70. HSP60 and mortalin/GRP75/mtHSP70 are key protein processing chaperones in the mitochondria, with roles in matrix protein folding and mitochondrial protein import, respectively (Wadhwa et al., 2005; Yaguchi et al., 2007). Mortalin/GRP75/mtHSP70, in particular, is a known target of oxidative stress, and has previously been linked to neurodegenerative diseases PD and AD based on altered expression (Jin et al., 2006; Osorio et al., 2007). A recent study found that both HSP60 and Mortalin/GRP75/mtHSP70 interacted with frataxin, a protein involved in iron-sulfur (Fe-S) cluster biogenesis for Fe-S cluster-dependent enzymes (Shan et al., 2007). The authors also noted that Mortalin/GRP75/mtHSP70 shares homology with the HSP70-family protein Ssq1, a mitochondrial matrix protein required for Fe-S cluster assembly (Lutz et al., 2001; Shan et al., 2007). Interestingly, we identified several Fe-S cluster-containing proteins covalently modified by 14C-DA, including the 30 kDa subunit of Complex I, Rieske Fe-S protein subunit of Complex III, and aconitase. Thus, DA oxidation may potentially contribute to an impaired Fe-S protein system in dopaminergic neurons, both in Fe-S protein biogenesis and function.
Multiple subunits of Complex I (75 kDa and 30 kDa subunits) and Complex III (ubiquinol-cytochrome c reductase core protein 1 and Rieske Fe-S protein) of the mitochondrial electron transport chain (ETC) were identified as targets of covalent DA modification. Several studies have demonstrated that incubation of isolated brain mitochondria with DA or DAQ inhibits mitochondrial respiration (Berman and Hastings, 1999; Cohen et al., 1997; Gluck et al., 2002; Gluck and Zeevalk, 2004). Study in disrupted mitochondria have also suggested DA can directly interact with and inhibit Complex I (Ben-Shachar et al., 2004; Brenner-Lavie et al., 2008). Decreased NADH dehydrogenase activity of Complex I has been observed in both the SN (Janetzky et al., 1994; Orth and Schapira, 2002; Schapira et al., 1990) and the periphery (Blandini et al., 1998; Shoffner et al., 1991) of PD patients. PD models utilizing Complex I inhibitors rotenone and 1-methyl 4-phenyl 1,2,3,6-tetrahydropyridine (MPTP) replicate characteristics of the disease, including nigrostriatal dopaminergic cell death (Przedborski et al., 2000; Sherer et al., 2003). Modification of critical ETC proteins by DA oxidation may potentially inhibit these proteins, possibly leading to an increase in mitochondrial dysfunction and play a role in the increased susceptibility of dopaminergic neurons in PD.
Both the cytosolic and mitochondrial isoforms of CK were identified as targets of DA modification in isolated brain mitochondria. Previous studies have shown that exposure to dopamine and dopamine oxidation can inhibit the activity of CK proteins (Maker et al., 1986; Miura et al., 1999). Cytosolic creatine kinase (CK) was shown to be susceptible to increased oxidation in the hippocampus of aged mice, as well as in young and old ApoE-KO mice, as compared to brain tissue of young mice (Choi et al., 2004a). CK was also identified as having increased carbonyl modification in AD brain (Castegna et al., 2002). Given the critical role of CK and MtCK in ATP level maintenance (Eder et al., 2000), and the association of MtCK with the proteins involved in the permeability transition pore (Vyssokikh and Brdiczka, 2003), oxidative modification of CK proteins may carry implications for energy maintenance and mitochondrial function in dopaminergic neurons. In a similar vein, we found 14C-DA covalent modification of the protein nucleoside diphosphate kinase A in SH-SY5Y cells. Considered a multifunctional protein, the assigned function of this enzyme is to maintain a balance between ADP, GDP, ATP, and GTP levels in the cell, and its activity is modulated by disulfide cross-linking of two oxidation-sensitive cysteine residues (Cumming et al., 2004; Song et al., 2000). Thus, DA modification of this protein may alter the energy balance in dopaminergic neurons.
Though not necessarily mitochondrial, the glycolysis proteins enolase 2 and triosephosphate isomerase were also shown in this study to be modified by DA in DAQ-exposed mitochondria preparations. These proteins have previously been shown to be targets of oxidative modification in association with neurodegenerative disease. Triosephosphate isomerase, alpha- and gamma-enolase, and beta-actin were found to be targets of protein nitration in AD brain (Castegna et al., 2003). Oxidative modification of these proteins by DAQ could have implications for dysfunction in cellular metabolism and energy generation upstream of the ETC, which may have detrimental effects given the high-energy demands of neurons.
Antioxidant and Thiol Oxidoreductase Enzymes are covalently modified by DAQ
We observed covalent DA modification of mitochondrial SOD2 and peroxiredoxin 3 in 14C-DAQ-exposed brain mitochondria, and cytosolic SOD1 and peroxiredoxin 2 in 14C-DA-exposed differentiated SH-SY5Y cells. Both enzyme types are integral in managing ROS levels and protecting against oxidative stress. SOD enzymes catalyze conversion of the free radical superoxide to oxygen and H2O2, and peroxiredoxins catalyze reduction of H2O2 to water. Alterations in expression and activity of both enzymes have been linked to PD and PD models. Post mortem PD brain tissue displayed increased SOD activity (Saggu et al., 1989), and elevated levels of SOD2 have been observed in PD patient CSF (Yoshida et al., 1994), suggestive of a response to increased levels of superoxide. Peroxiredoxin 2, which is highly abundant in neurons, showed increased abundance in PD brain SN (Basso et al., 2004), again suggestive of an oxidative stress response. Peroxiredoxin 2 was also found to be S-nitrosylated in rotenone and MPP+ treated SH-SY5Y cells and in PD brain (Fang et al., 2007). In addition, silencing expression of mitochondrial peroxiredoxin 3 and 5 increased the vulnerability of SH-SY5Y cells to MPP+ toxicity (De Simoni et al., 2008). Thus, DA modification of these enzymes could compromise the antioxidant defense mechanisms of dopaminergic neurons.
DA modification of the endoplasmic reticulum chaperones ER-60 (also known as GRP58 or ERp57) and protein disulfide isomerase-related protein 5 (also known as ERp5 or PDIA6) were also observed in SH-SY5Y cells exposed to DA. Both proteins are members of the protein disulfide isomerase family, which employ thiol-disulfide oxidoreductase activity to form disulfide bonds in substrates, and thus mediate proper protein folding (Ellgaard and Ruddock, 2005). These proteins are primarily localized to the endoplasmic reticulum, and each contains two cysteine-glycine-histidine-cysteine, or C-X-X-C, thioredoxin-like activity motifs integral in enzymatic disulfide bond formation (Ellgaard and Ruddock, 2005). Proteins containing the C-X-X-C motif are very susceptible to modification by electrophilic compounds such as DAQ (Lame et al., 2003; Lame et al., 2005). ER-60 also has demonstrated cysteine protease activity (Okudo et al., 2000; Urade and Kito, 1992; Urade et al., 1992) and a role in degradation of misfolded proteins (Otsu et al., 1995). We have previously shown the upregulation of ER-60 expression along with other ER chaperones following DA exposure in differentiated PC12 cells, suggesting DA toxicity induces ER stress and the unfolded protein response (Dukes et al., 2008). As aberrant protein folding and aggregation are associated with PD, DA may be contributing by modification and inactivation of key ER chaperone proteins.
DJ-1 and UCH-L1 Are Targets for Covalent Modification by DAQ
An intriguing finding was the covalent DA modification of two proteins directly linked to genetic forms of Parkinson’s disease, UCH-L1 and DJ-1. DJ-1, linked to familial PARK7 (Bonifati et al., 2003), is found widely distributed in the brain (Bandopadhyay et al., 2004). While the specific function of DJ-1 remains unknown, data suggest a role for the protein in oxidative stress regulation, possibly through the protein’s own susceptibility to oxidation of a critical cysteine residue (Canet-Aviles et al., 2004). UCH-L1 is linked to PARK5 through a mutation that leads to a decrease in the deubiquinating activity of the protein (Leroy et al., 1998; Nishikawa et al., 2003). The exact mechanism by which this mutation leads to decreased activity is unknown. However, it has been shown that the active site of UCH-L1 contains cysteine and histidine residues that are sensitive to modification by 4-hydroxynonenal (4-HNE), a reactive byproduct of lipid peroxidation (Nishikawa et al., 2003). Additionally, oxidative damage of both DJ-1 and UCH-L1, in the form of carbonylation and direct oxidation at cysteines and methionines, has been identified in both PD and AD brain (Castegna et al., 2002; Choi et al., 2004b; Choi et al., 2006). Thus covalent modification by DAQ is likely to be detrimental to DJ-1 and UCH-L1 function, and may lead to disruption of antioxidant and ubiquitin-proteasome system functions, respectively. It is important to note that DA-induced modifications have been associated with alterations in two other PD-linked proteins, alpha-synuclein (Conway et al., 2001) and parkin (LaVoie et al., 2005). The susceptibility of multiple PD-linked proteins to DA modification further supports a contributing role for DA oxidation in dopaminergic neuron degeneration and PD pathogenesis.
DAQ Binds to Cytoskeletal Proteins
We found that cytoskeletal proteins actin and tubulin were targets of covalent modification by DAQ in isolated mitochondria preparations, and actin and interacting protein tropomyosin were covalently modified by DA in DA-exposed differentiated SH-SY5Y cells. Tubulin protein is highly abundant, contains multiple free thiol groups critical for microtubule assembly, and is a known target of oxidative modification (Landino et al., 2002; Luduena and Roach, 1991). Tubulin was modified in vitro at specific cysteine residues by 4-HNE (Stewart et al., 2007), which has been shown to disrupt neuronal microtubules and neurite outgrowth in cultured Neuro 2A cells (Neely et al., 2005; Neely et al., 1999). Likewise, beta-actin was observed to be modified by 4-HNE in brain tissue of patients with mild cognitive impairment patient (Reed et al., 2008). Increased carbonylation and nitration of actin was observed in post mortem AD brain (Aksenov et al., 2001; Castegna et al., 2003). Relevant to our findings, other cytoskeletal proteins have been demonstrated as targets of DA oxidation in vitro. Exposure to oxidized DA promoted polymerization of tau protein (Santa-Maria et al., 2005) and covalent cross-linking of neurofilament proteins (Montine et al., 1995). It is well known that the transport and localization of mitochondria within the cell relies upon their close interaction with intact cytoskeletal structures (Boldogh and Pon, 2006; Boldogh and Pon, 2007). Thus, oxidation-induced damage to cytoskeletal proteins could have drastic effects on cellular structure, and on critical transport, localization, and ultimately function of organelles such as mitochondria.
Conclusions
We identified proteins in this study that are targets of DA quinone modification following intracellular DA oxidation, which may lead to altered protein function, disrupted protein-protein interactions, targeting of proteins for degradation, or promoting aggregation of damaged proteins. The susceptibility of these proteins to modification may play a role in the enhanced vulnerability of DAergic neurons, and should be considered for further study in connection with the pathophysiology of PD.
In this regard, it is important to note that many of the proteins we identify as targets of DA modification have previously been identified as targets of oxidative modification in conjunction with various neurodegenerative diseases and disease models, as discussed above. The identification of these same proteins across multiple independent studies not only validates their characteristic susceptibility to oxidative modification but also demonstrates their potential significance in neurodegenerative disease pathogenesis. Such proteins deserve critical attention to elucidate their roles in disease progression and their potential as targets for novel therapeutic strategies.
ACKNOWLEDGMENTS
We thank Dr. Ashraf Elamin, Mirunalni Thangavelu, proteomics manager Dr. Manimalha Balasubramani, and proteomics director Dr. Billy Day of the University of Pittsburgh Genomics and Proteomics Core Laboratories for their excellent technical expertise, advice, and assistance.
* This work was supported by grants from the National Institutes of Health, NS44076, DA09601, and AG20899.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
BIBLIOGRAPHY
- Ahmed N, Rice GE. Strategies for revealing lower abundance proteins in twodimensional protein maps. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;815:39–50. doi: 10.1016/j.jchromb.2004.10.070. [DOI] [PubMed] [Google Scholar]
- Aksenov MY, Aksenova MV, Butterfield DA, Geddes JW, Markesbery WR. Protein oxidation in the brain in Alzheimer’s disease. Neuroscience. 2001;103:373–83. doi: 10.1016/s0306-4522(00)00580-7. [DOI] [PubMed] [Google Scholar]
- Bandopadhyay R, Kingsbury AE, Cookson MR, Reid AR, Evans IM, Hope AD, Pittman AM, Lashley T, Canet-Aviles R, Miller DW, McLendon C, Strand C, Leonard AJ, Abou-Sleiman PM, Healy DG, Ariga H, Wood NW, de Silva R, Revesz T, Hardy JA, Lees AJ. The expression of DJ-1 (PARK7) in normal human CNS and idiopathic Parkinson’s disease. Brain. 2004;127:420–30. doi: 10.1093/brain/awh054. [DOI] [PubMed] [Google Scholar]
- Basso M, Giraudo S, Corpillo D, Bergamasco B, Lopiano L, Fasano M. Proteome analysis of human substantia nigra in Parkinson’s disease. Proteomics. 2004;4:3943–52. doi: 10.1002/pmic.200400848. [DOI] [PubMed] [Google Scholar]
- Beal MF. Mitochondria and neurodegeneration. Novartis Found Symp. 2007;287:183–92. doi: 10.1002/9780470725207.ch13. discussion 192-6. [DOI] [PubMed] [Google Scholar]
- Ben-Shachar D, Zuk R, Gazawi H, Ljubuncic P. Dopamine toxicity involves mitochondrial complex I inhibition: implications to dopamine-related neuropsychiatric disorders. Biochem Pharmacol. 2004;67:1965–74. doi: 10.1016/j.bcp.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Berman SB, Hastings TG. Dopamine oxidation alters mitochondrial respiration and induces permeability transition in brain mitochondria: implications for Parkinson’s disease. J Neurochem. 1999;73:1127–37. doi: 10.1046/j.1471-4159.1999.0731127.x. [DOI] [PubMed] [Google Scholar]
- Berman SB, Watkins SC, Hastings TG. Quantitative biochemical and ultrastructural comparison of mitochondrial permeability transition in isolated brain and liver mitochondria: evidence for reduced sensitivity of brain mitochondria. Exp Neurol. 2000;164:415–25. doi: 10.1006/exnr.2000.7438. [DOI] [PubMed] [Google Scholar]
- Berman SB, Zigmond MJ, Hastings TG. Modification of dopamine transporter function: effect of reactive oxygen species and dopamine. J Neurochem. 1996;67:593–600. doi: 10.1046/j.1471-4159.1996.67020593.x. [DOI] [PubMed] [Google Scholar]
- Bienvenut WV, Sanchez JC, Karmime A, Rouge V, Rose K, Binz PA, Hochstrasser DF. Toward a clinical molecular scanner for proteome research: parallel protein chemical processing before and during western blot. Anal Chem. 1999;71:4800–7. doi: 10.1021/ac990448m. [DOI] [PubMed] [Google Scholar]
- Blandini F, Nappi G, Greenamyre JT. Quantitative study of mitochondrial complex I in platelets of parkinsonian patients. Mov Disord. 1998;13:11–5. doi: 10.1002/mds.870130106. [DOI] [PubMed] [Google Scholar]
- Boldogh IR, Pon LA. Interactions of mitochondria with the actin cytoskeleton. Biochim Biophys Acta. 2006;1763:450–62. doi: 10.1016/j.bbamcr.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Boldogh IR, Pon LA. Mitochondria on the move. Trends Cell Biol. 2007;17:502–10. doi: 10.1016/j.tcb.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–9. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brandina I, Graham J, Lemaitre-Guillier C, Entelis N, Krasheninnikov I, Sweetlove L, Tarassov I, Martin RP. Enolase takes part in a macromolecular complex associated to mitochondria in yeast. Biochim Biophys Acta. 2006;1757:1217–28. doi: 10.1016/j.bbabio.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Brenner-Lavie H, Klein E, Zuk R, Gazawi H, Ljubuncic P, Ben-Shachar D. Dopamine modulates mitochondrial function in viable SH-SY5Y cells possibly via its interaction with complex I: relevance to dopamine pathology in schizophrenia. Biochim Biophys Acta. 2008;1777:173–85. doi: 10.1016/j.bbabio.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Canet-Aviles RM, Wilson MA, Miller DW, Ahmad R, McLendon C, Bandyopadhyay S, Baptista MJ, Ringe D, Petsko GA, Cookson MR. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci U S A. 2004;101:9103–8. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carre M, Andre N, Carles G, Borghi H, Brichese L, Briand C, Braguer D. Tubulin is an inherent component of mitochondrial membranes that interacts with the voltage-dependent anion channel. J Biol Chem. 2002;277:33664–9. doi: 10.1074/jbc.M203834200. [DOI] [PubMed] [Google Scholar]
- Castegna A, Aksenov M, Aksenova M, Thongboonkerd V, Klein JB, Pierce WM, Booze R, Markesbery WR, Butterfield DA. Proteomic identification of oxidatively modified proteins in Alzheimer’s disease brain. Part I: creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radic Biol Med. 2002;33:562–71. doi: 10.1016/s0891-5849(02)00914-0. [DOI] [PubMed] [Google Scholar]
- Castegna A, Thongboonkerd V, Klein JB, Lynn B, Markesbery WR, Butterfield DA. Proteomic identification of nitrated proteins in Alzheimer’s disease brain. J Neurochem. 2003;85:1394–401. doi: 10.1046/j.1471-4159.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- Choi J, Forster MJ, McDonald SR, Weintraub ST, Carroll CA, Gracy RW. Proteomic identification of specific oxidized proteins in ApoE-knockout mice: relevance to Alzheimer’s disease. Free Radic Biol Med. 2004a;36:1155–62. doi: 10.1016/j.freeradbiomed.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, Li L. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J Biol Chem. 2004b;279:13256–64. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- Choi J, Sullards MC, Olzmann JA, Rees HD, Weintraub ST, Bostwick DE, Gearing M, Levey AI, Chin LS, Li L. Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases. J Biol Chem. 2006;281:10816–24. doi: 10.1074/jbc.M509079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen G, Farooqui R, Kesler N. Parkinson disease: a new link between monoamine oxidase and mitochondrial electron flow. Proc Natl Acad Sci U S A. 1997;94:4890–4. doi: 10.1073/pnas.94.10.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway KA, Rochet JC, Bieganski RM, Lansbury PT., Jr. Kinetic stabilization of the alpha-synuclein protofibril by a dopamine-alpha-synuclein adduct. Science. 2001;294:1346–9. doi: 10.1126/science.1063522. [DOI] [PubMed] [Google Scholar]
- Cumming RC, Andon NL, Haynes PA, Park M, Fischer WH, Schubert D. Protein disulfide bond formation in the cytoplasm during oxidative stress. J Biol Chem. 2004;279:21749–58. doi: 10.1074/jbc.M312267200. [DOI] [PubMed] [Google Scholar]
- De Simoni S, Goemaere J, Knoops B. Silencing of peroxiredoxin 3 and peroxiredoxin 5 reveals the role of mitochondrial peroxiredoxins in the protection of human neuroblastoma SH-SY5Y cells toward MPP+ Neurosci Lett. 2008;433:219–24. doi: 10.1016/j.neulet.2007.12.068. [DOI] [PubMed] [Google Scholar]
- Dukes AA, Van Laar VS, Cascio M, Hastings TG. Changes in endoplasmic reticulum stress proteins and aldolase A in cells exposed to dopamine. J Neurochem. 2008;106:333–46. doi: 10.1111/j.1471-4159.2008.05392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder M, Stolz M, Wallimann T, Schlattner U. A conserved negatively charged cluster in the active site of creatine kinase is critical for enzymatic activity. J Biol Chem. 2000;275:27094–9. doi: 10.1074/jbc.M004071200. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entelis N, Brandina I, Kamenski P, Krasheninnikov IA, Martin RP, Tarassov I. A glycolytic enzyme, enolase, is recruited as a cofactor of tRNA targeting toward mitochondria in Saccharomyces cerevisiae. Genes Dev. 2006;20:1609–20. doi: 10.1101/gad.385706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Nakamura T, Cho DH, Gu Z, Lipton SA. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson’s disease. Proc Natl Acad Sci U S A. 2007;104:18742–7. doi: 10.1073/pnas.0705904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giege P, Heazlewood JL, Roessner-Tunali U, Millar AH, Fernie AR, Leaver CJ, Sweetlove LJ. Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell. 2003;15:2140–51. doi: 10.1105/tpc.012500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluck M, Ehrhart J, Jayatilleke E, Zeevalk GD. Inhibition of brain mitochondrial respiration by dopamine: involvement of H(2)O(2) and hydroxyl radicals but not glutathione-protein-mixed disulfides. J Neurochem. 2002;82:66–74. doi: 10.1046/j.1471-4159.2002.00938.x. [DOI] [PubMed] [Google Scholar]
- Gluck MR, Zeevalk GD. Inhibition of brain mitochondrial respiration by dopamine and its metabolites: implications for Parkinson’s disease and catecholamine-associated diseases. J Neurochem. 2004;91:788–95. doi: 10.1111/j.1471-4159.2004.02747.x. [DOI] [PubMed] [Google Scholar]
- Graham DG, Tiffany SM, Bell WR, Jr., Gutknecht WF. Autoxidation versus covalent binding of quinones as the mechanism of toxicity of dopamine, 6-hydroxydopamine, and related compounds toward C1300 neuroblastoma cells in vitro. Mol Pharmacol. 1978;14:644–53. [PubMed] [Google Scholar]
- Greenamyre JT, Hastings TG. Biomedicine. Parkinson’s--divergent causes, convergent mechanisms. Science. 2004;304:1120–2. doi: 10.1126/science.1098966. [DOI] [PubMed] [Google Scholar]
- Halliwell B. Role of free radicals in the neurodegenerative diseases: therapeutic implications for antioxidant treatment. Drugs Aging. 2001;18:685–716. doi: 10.2165/00002512-200118090-00004. [DOI] [PubMed] [Google Scholar]
- Halliwell B. Oxidative stress and neurodegeneration: where are we now? J Neurochem. 2006;97:1634–58. doi: 10.1111/j.1471-4159.2006.03907.x. [DOI] [PubMed] [Google Scholar]
- Hastings TG. Enzymatic oxidation of dopamine: the role of prostaglandin H synthase. J Neurochem. 1995;64:919–24. doi: 10.1046/j.1471-4159.1995.64020919.x. [DOI] [PubMed] [Google Scholar]
- Hastings TG, Lewis DA, Zigmond MJ. Role of oxidation in the neurotoxic effects of intrastriatal dopamine injections. Proc Natl Acad Sci U S A. 1996;93:1956–61. doi: 10.1073/pnas.93.5.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Kato T, Fujita K. Covalent binding of catechols to proteins through the sulphydryl group. Biochem Pharmacol. 1988;37:1707–10. doi: 10.1016/0006-2952(88)90432-7. [DOI] [PubMed] [Google Scholar]
- Janetzky B, Hauck S, Youdim MB, Riederer P, Jellinger K, Pantucek F, Zochling R, Boissl KW, Reichmann H. Unaltered aconitase activity, but decreased complex I activity in substantia nigra pars compacta of patients with Parkinson’s disease. Neurosci Lett. 1994;169:126–8. doi: 10.1016/0304-3940(94)90372-7. [DOI] [PubMed] [Google Scholar]
- Jenner P. Oxidative stress in Parkinson’s disease. Ann Neurol. 2003;53(Suppl 3):S26–36. doi: 10.1002/ana.10483. discussion S36-8. [DOI] [PubMed] [Google Scholar]
- Jin J, Hulette C, Wang Y, Zhang T, Pan C, Wadhwa R, Zhang J. Proteomic identification of a stress protein, mortalin/mthsp70/GRP75: relevance to Parkinson disease. Mol Cell Proteomics. 2006;5:1193–204. doi: 10.1074/mcp.M500382-MCP200. [DOI] [PubMed] [Google Scholar]
- John GB, Shang Y, Li L, Renken C, Mannella CA, Selker JM, Rangell L, Bennett MJ, Zha J. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol Biol Cell. 2005;16:1543–54. doi: 10.1091/mbc.E04-08-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DC, Gunasekar PG, Borowitz JL, Isom GE. Dopamine-induced apoptosis is mediated by oxidative stress and Is enhanced by cyanide in differentiated PC12 cells. J Neurochem. 2000;74:2296–304. doi: 10.1046/j.1471-4159.2000.0742296.x. [DOI] [PubMed] [Google Scholar]
- Khan FH, Saha M, Chakrabarti S. Dopamine induced protein damage in mitochondrial-synaptosomal fraction of rat brain. Brain Res. 2001;895:245–9. doi: 10.1016/s0006-8993(00)03284-4. [DOI] [PubMed] [Google Scholar]
- Khan FH, Sen T, Maiti AK, Jana S, Chatterjee U, Chakrabarti S. Inhibition of rat brain mitochondrial electron transport chain activity by dopamine oxidation products during extended in vitro incubation: implications for Parkinson’s disease. Biochim Biophys Acta. 2005;1741:65–74. doi: 10.1016/j.bbadis.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Koshimura K, Tanaka J, Murakami Y, Kato Y. Effects of dopamine and L-DOPA on survival of PC12 cells. J Neurosci Res. 2000;62:112–9. doi: 10.1002/1097-4547(20001001)62:1<112::AID-JNR12>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Kuhn DM, Arthur RE, Jr., Thomas DM, Elferink LA. Tyrosine hydroxylase is inactivated by catechol-quinones and converted to a redox-cycling quinoprotein: possible relevance to Parkinson’s disease. J Neurochem. 1999;73:1309–17. doi: 10.1046/j.1471-4159.1999.0731309.x. [DOI] [PubMed] [Google Scholar]
- Lai CT, Yu PH. Dopamine- and L-beta-3,4-dihydroxyphenylalanine hydrochloride (L-Dopa)-induced cytotoxicity towards catecholaminergic neuroblastoma SH-SY5Y cells. Effects of oxidative stress and antioxidative factors. Biochem Pharmacol. 1997;53:363–72. doi: 10.1016/s0006-2952(96)00731-9. [DOI] [PubMed] [Google Scholar]
- Lame MW, Jones AD, Wilson DW, Segall HJ. Protein targets of 1,4-benzoquinone and 1,4-naphthoquinone in human bronchial epithelial cells. Proteomics. 2003;3:479–95. doi: 10.1002/pmic.200390062. [DOI] [PubMed] [Google Scholar]
- Lame MW, Jones AD, Wilson DW, Segall HJ. Monocrotaline pyrrole targets proteins with and without cysteine residues in the cytosol and membranes of human pulmonary artery endothelial cells. Proteomics. 2005;5:4398–413. doi: 10.1002/pmic.200402022. [DOI] [PubMed] [Google Scholar]
- Landino LM, Hasan R, McGaw A, Cooley S, Smith AW, Masselam K, Kim G. Peroxynitrite oxidation of tubulin sulfhydryls inhibits microtubule polymerization. Arch Biochem Biophys. 2002;398:213–20. doi: 10.1006/abbi.2001.2729. [DOI] [PubMed] [Google Scholar]
- LaVoie MJ, Hastings TG. Dopamine quinone formation and protein modification associated with the striatal neurotoxicity of methamphetamine: evidence against a role for extracellular dopamine. J Neurosci. 1999;19:1484–91. doi: 10.1523/JNEUROSCI.19-04-01484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVoie MJ, Ostaszewski BL, Weihofen A, Schlossmacher MG, Selkoe DJ. Dopamine covalently modifies and functionally inactivates parkin. Nat Med. 2005;11:1214–21. doi: 10.1038/nm1314. [DOI] [PubMed] [Google Scholar]
- Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein MJ, Jonnalagada S, Chernova T, Dehejia A, Lavedan C, Gasser T, Steinbach PJ, Wilkinson KD, Polymeropoulos MH. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395:451–2. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- Luduena RF, Roach MC. Tubulin sulfhydryl groups as probes and targets for antimitotic and antimicrotubule agents. Pharmacol Ther. 1991;49:133–52. doi: 10.1016/0163-7258(91)90027-j. [DOI] [PubMed] [Google Scholar]
- Lutz T, Westermann B, Neupert W, Herrmann JM. The mitochondrial proteins Ssq1 and Jac1 are required for the assembly of iron sulfur clusters in mitochondria. J Mol Biol. 2001;307:815–25. doi: 10.1006/jmbi.2001.4527. [DOI] [PubMed] [Google Scholar]
- Magi B, Ettorre A, Liberatori S, Bini L, Andreassi M, Frosali S, Neri P, Pallini V, Di Stefano A. Selectivity of protein carbonylation in the apoptotic response to oxidative stress associated with photodynamic therapy: a cell biochemical and proteomic investigation. Cell Death Differ. 2004;11:842–52. doi: 10.1038/sj.cdd.4401427. [DOI] [PubMed] [Google Scholar]
- Maker HS, Weiss C, Brannan TS. Amine-mediated toxicity. The effects of dopamine, norepinephrine, 5-hydroxytryptamine, 6-hydroxydopamine, ascorbate, glutathione and peroxide on the in vitro activities of creatine and adenylate kinases in the brain of the rat. Neuropharmacology. 1986;25:25–32. doi: 10.1016/0028-3908(86)90054-7. [DOI] [PubMed] [Google Scholar]
- Miura T, Muraoka S, Fujimoto Y. Inactivation of creatine kinase induced by dopa and dopamine in the presence of ferrylmyoglobin. Chem Biol Interact. 1999;123:51–61. doi: 10.1016/s0009-2797(99)00124-6. [DOI] [PubMed] [Google Scholar]
- Montine TJ, Farris DB, Graham DG. Covalent crosslinking of neurofilament proteins by oxidized catechols as a potential mechanism of Lewy body formation. J Neuropathol Exp Neurol. 1995;54:311–9. doi: 10.1097/00005072-199505000-00004. [DOI] [PubMed] [Google Scholar]
- Neely MD, Boutte A, Milatovic D, Montine TJ. Mechanisms of 4-hydroxynonenal-induced neuronal microtubule dysfunction. Brain Res. 2005;1037:90–8. doi: 10.1016/j.brainres.2004.12.027. [DOI] [PubMed] [Google Scholar]
- Neely MD, Sidell KR, Graham DG, Montine TJ. The lipid peroxidation product 4-hydroxynonenal inhibits neurite outgrowth, disrupts neuronal microtubules, and modifies cellular tubulin. J Neurochem. 1999;72:2323–33. doi: 10.1046/j.1471-4159.1999.0722323.x. [DOI] [PubMed] [Google Scholar]
- Nishikawa K, Li H, Kawamura R, Osaka H, Wang YL, Hara Y, Hirokawa T, Manago Y, Amano T, Noda M, Aoki S, Wada K. Alterations of structure and hydrolase activity of parkinsonism-associated human ubiquitin carboxyl-terminal hydrolase L1 variants. Biochem Biophys Res Commun. 2003;304:176–83. doi: 10.1016/s0006-291x(03)00555-2. [DOI] [PubMed] [Google Scholar]
- Offen D, Ziv I, Sternin H, Melamed E, Hochman A. Prevention of dopamine-induced cell death by thiol antioxidants: possible implications for treatment of Parkinson’s disease. Exp Neurol. 1996;141:32–9. doi: 10.1006/exnr.1996.0136. [DOI] [PubMed] [Google Scholar]
- Ogawa N, Asanuma M, Miyazaki I, Diaz-Corrales FJ, Miyoshi K. L-DOPA treatment from the viewpoint of neuroprotection. Possible mechanism of specific and progressive dopaminergic neuronal death in Parkinson’s disease. J Neurol. 2005;252(Suppl 4):IV23–IV31. doi: 10.1007/s00415-005-4006-7. [DOI] [PubMed] [Google Scholar]
- Okudo H, Urade R, Moriyama T, Kito M. Catalytic cysteine residues of ER-60 protease. FEBS Lett. 2000;465:145–7. doi: 10.1016/s0014-5793(99)01721-4. [DOI] [PubMed] [Google Scholar]
- Orth M, Schapira AH. Mitochondrial involvement in Parkinson’s disease. Neurochem Int. 2002;40:533–41. doi: 10.1016/s0197-0186(01)00124-3. [DOI] [PubMed] [Google Scholar]
- Osorio C, Sullivan PM, He DN, Mace BE, Ervin JF, Strittmatter WJ, Alzate O. Mortalin is regulated by APOE in hippocampus of AD patients and by human APOE in TR mice. Neurobiol Aging. 2007;28:1853–62. doi: 10.1016/j.neurobiolaging.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Otsu M, Urade R, Kito M, Omura F, Kikuchi M. A possible role of ER-60 protease in the degradation of misfolded proteins in the endoplasmic reticulum. J Biol Chem. 1995;270:14958–61. doi: 10.1074/jbc.270.25.14958. [DOI] [PubMed] [Google Scholar]
- Premkumar A, Simantov R. Mitochondrial voltage-dependent anion channel is involved in dopamine-induced apoptosis. J Neurochem. 2002;82:345–52. doi: 10.1046/j.1471-4159.2002.00966.x. [DOI] [PubMed] [Google Scholar]
- Przedborski S, Jackson-Lewis V, Djaldetti R, Liberatore G, Vila M, Vukosavic S, Almer G. The parkinsonian toxin MPTP: action and mechanism. Restor Neurol Neurosci. 2000;16:135–142. [PubMed] [Google Scholar]
- Rabinovic AD, Lewis DA, Hastings TG. Role of oxidative changes in the degeneration of dopamine terminals after injection of neurotoxic levels of dopamine. Neuroscience. 2000;101:67–76. doi: 10.1016/s0306-4522(00)00293-1. [DOI] [PubMed] [Google Scholar]
- Reed T, Perluigi M, Sultana R, Pierce WM, Klein JB, Turner DM, Coccia R, Markesbery WR, Butterfield DA. Redox proteomic identification of 4-hydroxy-2-nonenal-modified brain proteins in amnestic mild cognitive impairment: insight into the role of lipid peroxidation in the progression and pathogenesis of Alzheimer’s disease. Neurobiol Dis. 2008;30:107–20. doi: 10.1016/j.nbd.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Saggu H, Cooksey J, Dexter D, Wells FR, Lees A, Jenner P, Marsden CD. A selective increase in particulate superoxide dismutase activity in parkinsonian substantia nigra. J Neurochem. 1989;53:692–7. doi: 10.1111/j.1471-4159.1989.tb11759.x. [DOI] [PubMed] [Google Scholar]
- Samii A, Nutt JG, Ransom BR. Parkinson’s disease. Lancet. 2004;363:1783–93. doi: 10.1016/S0140-6736(04)16305-8. [DOI] [PubMed] [Google Scholar]
- Santa-Maria I, Hernandez F, Smith MA, Perry G, Avila J, Moreno FJ. Neurotoxic dopamine quinone facilitates the assembly of tau into fibrillar polymers. Mol Cell Biochem. 2005;278:203–12. doi: 10.1007/s11010-005-7499-6. [DOI] [PubMed] [Google Scholar]
- Schapira AH. Mitochondria in the aetiology and pathogenesis of Parkinson’s disease. Lancet Neurol. 2008;7:97–109. doi: 10.1016/S1474-4422(07)70327-7. [DOI] [PubMed] [Google Scholar]
- Schapira AH, Cooper JM, Dexter D, Clark JB, Jenner P, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. J Neurochem. 1990;54:823–7. doi: 10.1111/j.1471-4159.1990.tb02325.x. [DOI] [PubMed] [Google Scholar]
- Shan Y, Napoli E, Cortopassi G. Mitochondrial frataxin interacts with ISD11 of the NFS1/ISCU complex and multiple mitochondrial chaperones. Hum Mol Genet. 2007;16:929–41. doi: 10.1093/hmg/ddm038. [DOI] [PubMed] [Google Scholar]
- Sherer TB, Betarbet R, Testa CM, Seo BB, Richardson JR, Kim JH, Miller GW, Yagi T, Matsuno-Yagi A, Greenamyre JT. Mechanism of toxicity in rotenone models of Parkinson’s disease. J Neurosci. 2003;23:10756–64. doi: 10.1523/JNEUROSCI.23-34-10756.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Caudle WM, Zhang J. Biomarker discovery in neurodegenerative diseases: A proteomic approach. Neurobiol Dis. 2008 doi: 10.1016/j.nbd.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoffner JM, Watts RL, Juncos JL, Torroni A, Wallace DC. Mitochondrial oxidative phosphorylation defects in Parkinson’s disease. Ann Neurol. 1991;30:332–9. doi: 10.1002/ana.410300304. [DOI] [PubMed] [Google Scholar]
- Song EJ, Kim YS, Chung JY, Kim E, Chae SK, Lee KJ. Oxidative modification of nucleoside diphosphate kinase and its identification by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Biochemistry. 2000;39:10090–7. doi: 10.1021/bi000267a. [DOI] [PubMed] [Google Scholar]
- Stewart BJ, Doorn JA, Petersen DR. Residue-specific adduction of tubulin by 4-hydroxynonenal and 4-oxononenal causes cross-linking and inhibits polymerization. Chem Res Toxicol. 2007;20:1111–9. doi: 10.1021/tx700106v. [DOI] [PubMed] [Google Scholar]
- Stokes AH, Hastings TG, Vrana KE. Cytotoxic and genotoxic potential of dopamine. J Neurosci Res. 1999;55:659–65. doi: 10.1002/(SICI)1097-4547(19990315)55:6<659::AID-JNR1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Suh SK, Hood BL, Kim BJ, Conrads TP, Veenstra TD, Song BJ. Identification of oxidized mitochondrial proteins in alcohol-exposed human hepatoma cells and mouse liver. Proteomics. 2004;4:3401–12. doi: 10.1002/pmic.200400971. [DOI] [PubMed] [Google Scholar]
- Tse DC, McCreery RL, Adams RN. Potential oxidative pathways of brain catecholamines. J Med Chem. 1976;19:37–40. doi: 10.1021/jm00223a008. [DOI] [PubMed] [Google Scholar]
- Turan SC, Shah P, Pietruszko R. Inactivation of aldehyde dehydrogenase in intact rat liver mitochondria by dopamine. Alcohol. 1989;6:455–60. doi: 10.1016/0741-8329(89)90051-7. [DOI] [PubMed] [Google Scholar]
- Urade R, Kito M. Inhibition by acidic phospholipids of protein degradation by ER-60 protease, a novel cysteine protease, of endoplasmic reticulum. FEBS Lett. 1992;312:83–6. doi: 10.1016/0014-5793(92)81415-i. [DOI] [PubMed] [Google Scholar]
- Urade R, Nasu M, Moriyama T, Wada K, Kito M. Protein degradation by the phosphoinositide-specific phospholipase C-alpha family from rat liver endoplasmic reticulum. J Biol Chem. 1992;267:15152–9. [PubMed] [Google Scholar]
- Van Laar VS, Dukes AA, Cascio M, Hastings TG. Proteomic analysis of rat brain mitochondria following exposure to dopamine quinone: implications for Parkinson disease. Neurobiol Dis. 2008;29:477–89. doi: 10.1016/j.nbd.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyssokikh MY, Brdiczka D. The function of complexes between the outer mitochondrial membrane pore (VDAC) and the adenine nucleotide translocase in regulation of energy metabolism and apoptosis. Acta Biochim Pol. 2003;50:389–404. [PubMed] [Google Scholar]
- Wadhwa R, Takano S, Kaur K, Aida S, Yaguchi T, Kaul Z, Hirano T, Taira K, Kaul SC. Identification and characterization of molecular interactions between mortalin/mtHsp70 and HSP60. Biochem J. 2005;391:185–90. doi: 10.1042/BJ20050861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Marusich MF, Souda P, Whitelegge J, Capaldi RA. The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 2007;581:3545–9. doi: 10.1016/j.febslet.2007.06.052. [DOI] [PubMed] [Google Scholar]
- Xu Y, Stokes AH, Roskoski R, Jr., Vrana KE. Dopamine, in the presence of tyrosinase, covalently modifies and inactivates tyrosine hydroxylase. J Neurosci Res. 1998;54:691–7. doi: 10.1002/(SICI)1097-4547(19981201)54:5<691::AID-JNR14>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Yaguchi T, Aida S, Kaul SC, Wadhwa R. Involvement of mortalin in cellular senescence from the perspective of its mitochondrial import, chaperone, and oxidative stress management functions. Ann N Y Acad Sci. 2007;1100:306–11. doi: 10.1196/annals.1395.032. [DOI] [PubMed] [Google Scholar]
- Yoshida E, Mokuno K, Aoki S, Takahashi A, Riku S, Murayama T, Yanagi T, Kato K. Cerebrospinal fluid levels of superoxide dismutases in neurological diseases detected by sensitive enzyme immunoassays. J Neurol Sci. 1994;124:25–31. doi: 10.1016/0022-510x(94)90006-x. [DOI] [PubMed] [Google Scholar]