Abstract
Wingless (Wg) signalling regulates the expression of its target genes through Pangolin, Armadillo and their interacting co-factors. In a genetic screen for Wg signalling components, we found that imitation switch (ISWI), a chromatin-remodelling ATPase, had a positive role in transducing the canonical Wg signal, promoting the expression of the Wg target senseless. ISWI is found in several chromatin-remodelling complexes, including nucleosome remodelling factor (NURF). The effect of interfering with the function of other components of the NURF complex in vivo mimics that of ISWI. The NURF complex is also required for the efficient expression of other Wg target genes. Armadillo interacts directly with the NURF complex in vitro and recruits it to Wg targets in cultured cells. Together, our results suggest that the ISWI-containing NURF complex functions as a co-activator of Armadillo to promote Wg-mediated transcription.
Keywords: ISWI, NURF, signal transduction, Wingless
Introduction
β-Catenin/Armadillo has an important role in regulating the expression of Wnt/Wingless (Wg) target genes. In the absence of the Wnt/Wg ligand, the cytosolic pool of β-catenin/Armadillo is constitutively targeted for degradation (Aberle et al, 1997; Pai et al, 1997). In the presence of Wnt/Wg, β-catenin/Armadillo is stabilized, accumulates and then enters the nucleus where it competes with co-repressors bound to T-cell factor/Pangolin (Tcf/Pan). β-Catenin/Armadillo then recruits various co-factors to form a complex that activates the expression of Wnt/Wg target genes (Stadeli et al, 2006).
The carboxy-terminal region of β-catenin/Armadillo (including the tail and the Armadillo repeats 11 and 12) recruits numerous transcriptional co-factors. Many of these, including the histone acetyltransferase CREB binding protein (CBP)/p300 (Hecht et al, 2000; Takemaru & Moon, 2000), the histone methyltransferase mixed lineage leukaemia 1 (MLL1)/SET1 (Sierra et al, 2006) and the peroxisomal assembly factor 1 (PAF1) complex component Hyrax/Parafibromin (Mosimann et al, 2006), are part of complexes involved in chromatin remodelling. In addition, the region of β-catenin comprising Armadillo repeats 7–12 recruits brahma related gene-1 (BRG-1)/Brahma, a chromatin-remodelling ATPase of the switch/sucrose nonfermentable (SWI/SNF) class (Barker et al, 2001). Here, we show that the nucleosome remodelling factor (NURF) complex (Xiao et al, 2001; Langst & Becker, 2001a), which contains another class of chromatin-remodelling ATPase, imitation switch (ISWI; Tsukiyama et al, 1995), is also recruited by the C-terminal region of Armadillo and promotes the transcription of a subset of Wg target genes.
Results And Discussion
Genetic assays show ISWI is a Wg pathway component
Legless (Lgs) is a core component of the nuclear Wg signalling complex (Kramps et al, 2002; Hoffmans & Basler, 2004; Thompson, 2004; Hoffmans et al, 2005). Expression of a dominant-negative form of Lgs (Lgs17E) in wing discs by spalt enhancer Gal4 (salE-Gal4) results in a notched-wing phenotype in the adult (Fig 1A,B; Mosimann et al, 2006), presumably owing to the compromised transcriptional output of the Armadillo activation complex. To uncover new components of the Wg signal transduction pathway, we performed an enhancer P element (EP) overexpression screen for genes that rescue this notched-wing phenotype. Hyrax, an essential co-activator of Armadillo, was identified using this set-up (Mosimann et al, 2006). We also found that the Lgs17E phenotype could be rescued by an EP insertion driving the overexpression of ISWI, a chromatin-remodelling ATPase (Fig 1C,D). A qualitatively similar rescue was obtained by using a UAS-iswi transgene (Deuring et al, 2000), confirming that ISWI was responsible for the reversion (data not shown).
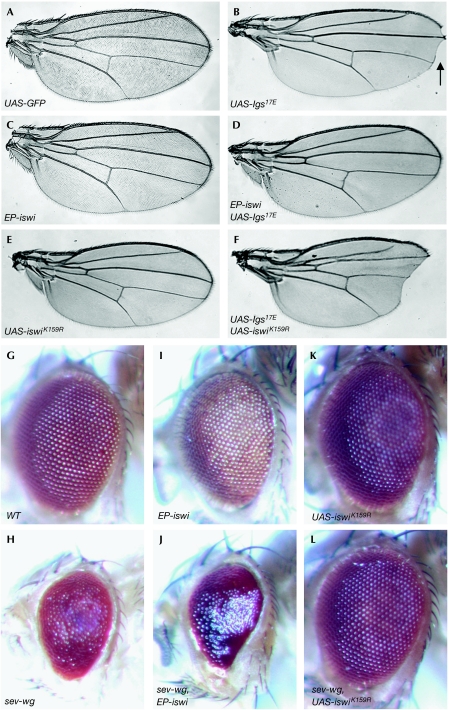
Figure 1.
The activity of ISWI is crucial when Wingless signalling is compromised in the wing and eye of Drosophila. salE-Gal4 drove the expression of (A) UAS-GFP, (B) UAS-lgs17E, (C) EP-iswi, (D) UAS-lgs17E and EP-iswi, (E) UAS-iswiK159R, and (F) UAS-lgs17E and UAS-iswiK159R. The arrow in (B) indicates the notched-wing phenotype caused by Lgs17E. (G–L) The rough eye phenotype generated by sev-Wg was affected by ISWI activity. (G) GMR-Gal4/+, (H) GMR-Gal4/+; sev-wg/+, (I) GMR-Gal4/EP-iswi, (J) GMR-Gal4/EP-iswi; sev-wg/+, (K) GMR-Gal4/+; UAS-iswiK159R/+, and (L) GMR-Gal4/+; UAS-iswiK159R/sev-wg. It has been reported that universal or strong expression of ISWIK159R can affect cell viability (Deuring et al, 2000). However, on expressing ISWIK159R alone using salE-Gal4 or GMR-Gal4, we did not observe any wing or eye phenotypes (E,K). We believe that the lack of an obvious effect on viability is due to the moderate expression levels achieved with these drivers and possibly because of the timing of the expression. EP, enhancer P element; GFP, green fluorescent protein; ISWI, imitation switch; Lgs, legless; sev, sevenless; Wg, wingless; WT, wild type.
The expression of Lgs17E generates a sensitized system in which Wg signalling is compromised. Reducing the functions of other positive regulators in this background should further impair the Wg signal. To confirm that ISWI does have a positive role, we tested whether the expression of a dominant-negative form (ISWIK159R; Deuring et al, 2000) enhances the Lgs17E phenotype. Consistent with a positive role for ISWI, the expression of ISWIK159R augmented the notched wing phenotype (Fig 1E,F).
To investigate further the function of ISWI in Wg signalling, we examined the effect of overexpressing ISWI on another sensitized system—the sevenless-Wingless (sev-Wg) assay. In this set-up, the ectopic expression of Wg (sev-Wg) resulted in a rough eye phenotype (Fig 1G,H), which could be suppressed by mutations in essential components of the Wg pathway, including Pan, Armadillo, Lgs and Pygopus (Brunner et al, 1997; Kramps et al, 2002). The use of GMR-Gal4 to drive the expression of EP-iswi or UAS-iswi enhanced the rough eye phenotype (Fig 1I,J; data not shown), whereas the expression of UAS-iswiK159R suppressed the rough eye phenotype (Fig 1K,L). Thus, through an independent series of assays, we confirmed that ISWI is a new positive regulator of Wg signalling.
ISWI promotes the expression of the Wg target senseless
To confirm that the expression of Lgs17E at the centre of the wing discs by salE-Gal4 specifically weakened the transcriptional output of the Armadillo activation complex, and to test whether the co-expression of ISWI could specifically restore it, we monitored the expression of Wg and Notch targets (Giraldez & Cohen, 2003). Expression of the low-threshold Wg target Distal-less (Dll) was not affected by Lgs17E (supplementary Fig S1A online). By contrast, the high-threshold Wg target senseless (sens) was clearly repressed (supplementary Fig S1B online). This is probably because Lgs17E only reduced, but did not abolish, the transcriptional activity of the Armadillo activation complex. The residual activity was able to maintain the expression of Dll, but not that of sens, which require higher levels of Wg signal transduction. Expression of the Notch targets wg and cut at the dorsal–ventral (DV) boundary of the wing discs was not affected by Lgs17E (supplementary Fig S1B′,C online). When ISWI was co-expressed with Lgs17E at the center of the wing discs, sens expression was rescued (supplementary Fig S1E online); expression of Dll, wg and cut was not altered (supplementary Fig S1D,E′,F online). Interestingly, in a wild-type background, expression of ISWI by salE-Gal4 did not alter the expression of Dll, sens, wg or cut in wing discs (supplementary Fig S1G–I online) and caused no phenotype in adult wings (Fig 1C), suggesting that increased ISWI activity was not sufficient to enhance or ectopically induce Wg signalling during wing development. These results show that over-expression of iswi is able to ameliorate the effect of impaired transcriptional Armadillo activity, as caused by expressing Lgs17E.
Proper expression of sens requires ISWI function
Next we investigated whether ISWI function was required for expression of the Wg target sens. We created iswi null clones in the wing disc using the iswi1 allele (Deuring et al, 2000). Consistent with previous reports that the loss of ISWI affects cellular viability, iswi1 clones had a growth disadvantage. To facilitate clonal analysis, we induced clones in a Minute background. To ensure that an effect on sens expression was specific, we monitored wg expression in the same wing disc. Generally, we found that 3 days after clone induction the expression of wg was not detectably altered in the clones (Fig 2E′″). However, sens expression was reduced in the iswi clones but not in wild-type clones (Fig 2E–E″,F–F″). sens expression started at the centre of the wing disc in mid-third instar and then extended to the periphery of both antero-posterior compartments (Fang et al, 2006). Interestingly, the reduction of sens expression was only detectable in discs when sens was not fully expressed. When Sens staining extended to the periphery of the wing disc, the expression of sens was no longer detectably altered in clones (supplementary Fig S2A–A” online). These observations suggest that the ISWI function promotes, but is not essential for sens expression. Other transcriptional co-activators might compensate for the loss of ISWI at late stages. We also attenuated ISWI function in the posterior compartment of the wing discs by driving the expression of the dominant-negative ISWIK159R with en-Gal4 (Fig 2A–D). Although we observed no effect on the levels of wg (Fig 2B,C′″) or Dll expression (Fig 2A–A″) in wing discs from third-instar larvae, the expression of sens was weakened. In contrast to the null clones, expression of ISWIK159R could weaken the expression of sens even when Sens staining was fully extended (Fig 2C–C″). This difference could be due to the interference with other co-activators by the dominant-negative ISWIK159R or the residual ISWI in null clones (Deuring et al, 2000).
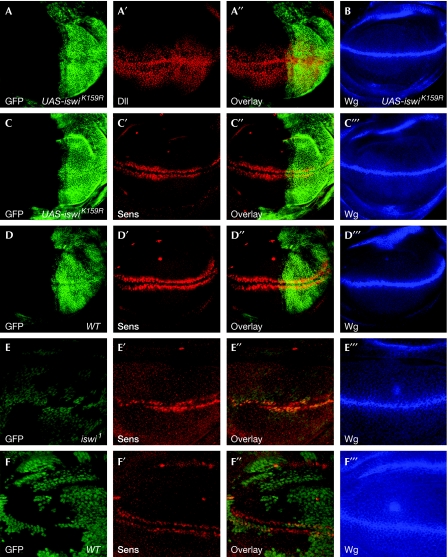
Figure 2.
ISWI activity is required for sens expression in the wing disc. (A–C′″) Expression of UAS-iswiK159R in the posterior compartment of the wing disc was controlled by en-Gal4, Gal80ts (second-instar larvae were preserved at 29°C temperature for 2 days). Expression of (A–A″) Dll and (B) wg was not affected by ISWIK159R. (C–C′″) The expression of sens, but not wg, was weakened by ISWIK159R in the same wing disc. (D–D′″) The control disc expressing en-Gal4, Gal80ts showed wild-type expression of sens and wg. (E–E′″) Expression of sens, but not wg, was weakened in iswi clones induced by hs-flp; FRT42 ubi-GFP Minute/FRT42: iswi1. (F–F′″) The expression of sens and wg were unaltered in wild-type clones induced by hs-flp; FRT42 ubi-GFP Minute/FRT42 +. Dll, Distal-less; FRT, flippase recognition target; GFP, green fluorescent protein; hs-flp, heat shock-flippase; ISWI, imitation switch; sens, senseless; Wg, wingless; WT, wild type.
NURF is required for expression of Wg target genes
There are at least four ISWI-containing chromatin-remodelling complexes in Drosophila: NURF, ATP-utilizing chromatin remodelling and assembly factor (ACF), chromatin remodelling and assembly factor (CHRAC) and a TATA binding protein related factor 2 (TRF2)-associated complex (Langst & Becker, 2001a; Hochheimer et al, 2002). ACF and CHRAC contribute primarily to the formation of repressive chromatin (Fyodorov et al, 2004), whereas the NURF complex (Fig 3A) is involved in both transcriptional activation and repression, and directly associates with histone H3 trimethylated at lysine 4 (H3K4me3; Wysocka et al, 2006). It has been reported that the loss of NURF301—the largest subunit of the NURF complex—results in phenotypes that are indistinguishable from those of iswi alleles (Badenhorst et al, 2002). We were therefore interested in examining whether the positive function of ISWI in Wg signalling was mediated through recruitment of the NURF complex. To test this, we examined the effect of nurf301 loss of function on sens expression using the nurf3018 null allele (Badenhorst et al, 2005). Similar to iswi clones, nurf301 clones have a growth disadvantage and were thus generated in a Minute background. At 3 days after clone induction, as for iswi, sens expression was repressed in slightly younger larvae, whereas it was only weakened in older larvae in which sens expression was fully extended (Fig 3D–D”; supplementary Fig S2C–C” online). The expression of wg was not affected in nurf301 clones (Fig 3D'”; supplementary Fig S2C'” online). We also reduced NURF301 function in the posterior compartment of wing discs by RNA-mediated interference (RNAi). Consistent with our results so far, knockdown of NURF301 weakened sens expression without reducing Wg or Dll levels (Fig 3C–C'”; supplementary Fig S2B–B″ online). These flies survived to adulthood and showed notches in the posterior wing compartment—a phenotype seen when Wg signalling is impeded (Fig 3B).
Figure 3.
The NURF complex is required for sens expression in the wing disc. (A) The NURF complex comprises four subunits. (B–C′″) Expression of UAS-nurf301dsRNA in the posterior compartment of the wing disc by en-Gal4, Gal80ts resulted in (B) a notched-wing phenotype in the posterior wing (arrow), and (C–C′″) weakening of sens expression without affecting wg expression in the posterior wing disc. (D–D′″) The expression of sens, but not wg, was weakened in nurf301 clones induced by hs-flp; ubi-GFP Minute FRT80/nurf3018 FRT80 (arrowhead). dsRNA, double-stranded RNA; GFP, green fluorescent protein; hs-flp, heat shock-flippase; NURF, nucleosome remodelling factor; sens, senseless; Wg, wingless.
Next we investigated whether the NURF complex was required for the expression of Wg targets other than sens. We addressed this question by knocking down NURF complex function in Drosophila Kc cells. To target the NURF complex specifically, we selected NURF301 as it is not present in other chromatin-remodelling complexes. NURF301 RNAi had no effect on the stability of ISWI (Fig 4E), ruling out an effect on other ISWI-containing complexes. The effect of NURF301 RNAi on the transcription of Wg target genes was examined by reverse transcription–PCR, in both unstimulated and stimulated cells (Axin RNAi; Blauwkamp et al, 2008). Four Wg target genes were tested: naked cuticle (nkd; Zeng et al, 2000; Chang et al, 2008), CG5895 (Mosimann et al, 2006), frizzled 3 (fz3; Sivasankaran et al, 2000) and CG6234 (Fang et al, 2006). NURF301 RNAi had no effect on the basal expression of these genes (Fig 4A–D). In stimulated cells, the induction of CG5895, fz3 and CG6234 was reduced, but not abolished: the induced transcription of CG5895 was reduced by 30% (Fig 4B); the induction of fz3 or CG6234 was reduced by ⩾50% (Fig 4C,D). Interestingly, the induction of nkd was unaffected by NURF301 RNAi (Fig 4A). We observed similar results on using Wg-conditioned medium to stimulate the pathway (data not shown). Consistent with these results in wing discs from nurf301 mutant larvae, the expression of several Wg targets was clearly reduced, whereas that of nkd was not (supplementary Fig S3 online).
Figure 4.
The NURF complex is required for the transcriptional activation of some Wg targets in Kc cells. Cells were treated with the indicated dsRNAs for 4 days and total RNA were isolated for RT–PCR. The messenger RNA levels of actin, the ribosome protein L49 and TATA binding protein (TBP) were used as internal controls for normalization. Four Wg targets, (A) Nkd, (B) CG5895, (C) CG6234 and (D) Fz3 were measured for basal and activated transcription level. (E) The protein levels of ISWI and α-Tub were monitored in dsRNA-treated cells by Western blot. dsRNA, double-stranded RNA; Fz3, frizzled 3; GFP, green fluorescent protein; ISWI, imitation switch; Nkd, naked cuticle; NURF, nucleosome remodelling factor; RT–PCR, reverse transcription–PCR; Wg, wingless.
Together with our finding that Dll expression is unaffected by reducing ISWI or NURF301 function (Fig 2A–A″; supplementary Fig S2B–B” online), these data suggest that the NURF complex is needed to promote the expression of some, but not all, Wg targets.
Recruitment of the NURF complex by Armadillo
Is the NURF complex recruited to nuclear Armadillo? We investigated this question by testing for a direct interaction between Armadillo and components of the NURF complex. Armadillo contains an amino-terminal region, 12 Armadillo repeats and a C-terminal tail (Fig 5A). The N-terminal region is phosphorylated by the destruction complex (Peifer & Polakis, 2000), whereas the Armadillo repeats and the C-terminal tail are known to act as interaction platforms for various co-factors (Hecht et al, 2000; Takemaru & Moon, 2000; Barker et al, 2001; Mosimann et al, 2006; Sierra et al, 2006). We expressed Armadillo repeat 1–8 (ArmR1–8), ArmR1–10, ArmR9–C and ArmR11–C as glutathione S-transferase (GST) fusion proteins and used them to pull down in vitro translated components of the NURF complex (Fig 5A). Unfortunately, we could not obtain in vitro translated NURF301, presumably owing to its large size (>300 kDa). Of the smaller NURF components tested, neither NURF55 nor NURF38 bound to Arm; however, we found that ISWI interacted with the C-terminal part of Arm, being pulled down by ArmR11–C and ArmR9–C (Fig 5B; data not shown). To test further whether Arm interacts with the NURF complex, we investigated whether an intact, purified NURF complex (Xiao et al, 2001) could be pulled down by the above Arm proteins. Consistent with the above results, we found that the ISWI subunit of the NURF complex was specifically pulled down by ArmR11–C. The NURF55 subunit, which does not interact directly with Arm, was co-immunoprecipitated (Fig 5C).
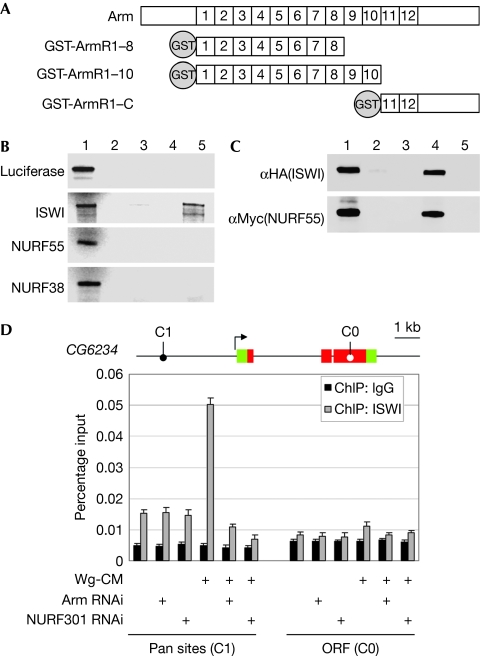
Figure 5.
Recruitment of ISWI and the NURF complex by Armadillo. (A) Domain structure of full-length Armadillo and truncated Armadillo fusion constructs. (B) In vitro translated samples were pulled down by Armadillo constructs and detected by autoradiography. Lane 1, 10% input. Lane 2, pulled down by GST. Lane 3, pulled down by GST-ArmR1–8. Lane 4, pulled down by GST-ArmR1–10. Lane 5, pulled down by GST-ArmR11–C. (C) The NURF complex was pulled down by Armadillo constructs. HA-tagged ISWI and Myc-tagged NURF55 were detected by western blot. Lane 1, 20% input. Lane 2, pulled down by GST-ArmR1–8. Lane 3, pulled down by GST-ArmR1–10. Lane 4, pulled down by GST-ArmR11–C. Lane 5, pulled down by GST. (D) The binding of ISWI at the CG6234 locus. Kc cells were treated with control medium or Wg-conditioned medium (Wg-CM), combined with control RNAi, Arm RNAi or NURF301 RNAi. A locus containing Pan binding sites (C1) and a locus in the open reading frame (C0) were monitored for ISWI binding. ArmR, Armadillo repeat; ChIP, chromatin immunoprecipitation; GST, glutathione S-transferase; HA, haemagglutinin; ISWI, imitation switch; NURF, nucleosome remodelling factor; ORF, open reading frame; Pan, Pangolin; RNAi, RNA-mediated interference; Wg, wingless.
To test whether Arm can recruit the NURF complex to the Wg target genes, we performed chromatin immunoprecipitation (ChIP) experiments and monitored the binding of ISWI to the loci of CG6234 and nkd, which contain validated clusters of Pan binding sites (Fang et al, 2006). At Pan sites in the CG6234 locus, we observed a moderate ISWI signal in unstimulated cells that was not affected by NURF301 RNAi, suggesting the binding of another ISWI-containing complex. Wg stimulation increased the binding of ISWI at the Pan sites. This binding was greatly reduced by both Arm and NURF301 RNAi, confirming that on Wg signalling ISWI, in the context of the NURF complex, was recruited through Arm (Fig 5D). At the nkd locus, we did not observe the ISWI signal in unstimulated cells, whereas a moderate signal, which also depended on Arm and NURF301, was detected at the Pan sites on stimulation (supplementary Fig S4 online).
A recent report has found that, in the absence of Wg signalling, ISWI and the ACF/CHRAC component, ACF1, are required for the Pan-mediated basal repression of Wg target genes (Liu et al, 2008). Our data suggest that, in the presence of Wg signalling, the activation of Wg target genes also requires ISWI activity but in the presence of another complex, the NURF complex, which is recruited by Arm. Interestingly, it has been reported that SNF2H, a mammalian homologue of ISWI, also interacts with β-catenin in an equivalent domain (Sierra et al, 2006); in this case the functional relevance has not been noted.
Concluding remarks
The finding that two types of chromatin-remodelling ATPases—Brahma, of the SWI/SNF type (Barker et al, 2001), and ISWI (this report)—can be recruited by Armadillo to enhance target gene expression suggests the existence of more complex regulatory mechanisms in Wg-mediated transcription than previously anticipated. Both SWI/SNF and ISWI are able to rearrange chromatin structure, but they have distinct chromatin-remodelling activities (Racki & Narlikar, 2008). For example, it has been reported that SWI/SNF promotes the release of nucleosomes from DNA, whereas ISWI promotes their sliding (Langst & Becker, 2001b; Fan et al, 2003). In addition, it has been suggested that Brahma functions at a step before Pol II recruitment (Armstrong et al, 2002), while the preferential binding of NURF301 to H3K4me3 suggested the recruitment of the NURF complex at or near transcriptional initiation (Wysocka et al, 2006). We propose that Brahma and the ISWI-containing NURF complex are recruited sequentially by Armadillo. The later recruitment of the NURF complex is stabilized by binding to histone tails and functions to fine-tune Wg-mediated transcriptional activation, which is a crucial event for the expression of a subset of Wg target genes. An important step in the future will be to study the timing of the individual recruitment events and to identify the reasons that underlie the heterogeneity in the requirement of various co-factors.
Methods
Protein interaction studies. [S35]methionine-labelled proteins or the purified NURF complex was diluted in binding buffer (20 mM phosphate, pH 7.4, 200 mM NaCl, 2 mM EDTA, 5% glycerol, 0.2% Nonidet P-40, 0.5 mM dithiothreitol and protease inhibitor cocktail) and incubated with glutathione beads bearing GST or GST fusion proteins on a nutator for 3 h at 4°C. After an extensive wash, the bound proteins were eluted and resolved by SDS–polyacrylamide gel electrophoresis and analysed by autoradiography or Western blotting.
ChIP. Kc cells were pre-treated with 5 mM dimethyl-3, 3′-dithiobispropionimidate dihydrochloride (DTBP; Sigma, St Louis, MO, USA) on ice for 30 min and washed with 100 mM Tris (pH 8.0) and 150 mM NaCl. The cells were crosslinked with 1% formaldehyde at 25°C, for 20 min, and ChIP assays were performed with the EZ-Magna ChIP kit (Upstate; Billerica, MA, USA). In all, 8 × 106 cells were used for each immunoprecipitation. Quantitative PCR primers for CG6234 and nkd loci are described in Fang et al (2006) and sequence information is available upon request.
More Methods are available in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary information
Acknowledgments
We thank G. Hausmann for providing experimental advice and critically reading the paper, J. Tamkun and P. Badenhorst for providing fly strains, C. Wu and H. Xiao for providing the purified NURF complex, and I. Duncan, J. Kadonaga and H. Bellen for providing the antibodies. This work was supported by the Swiss National Science Foundation and the Kanton of Zürich.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R (1997) Beta-catenin is a target for the ubiquitin–proteasome pathway. EMBO J 16: 3797–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong JA, Papoulas O, Daubresse G, Sperling AS, Lis JT, Scott MP, Tamkun JW (2002) The Drosophila BRM complex facilitates global transcription by RNA polymerase II. EMBO J 21: 5245–5254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst P, Voas M, Rebay I, Wu C (2002) Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev 16: 3186–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst P, Xiao H, Cherbas L, Kwon SY, Voas M, Rebay I, Cherbas P, Wu C (2005) The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev 19: 2540–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H (2001) The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J 20: 4935–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blauwkamp TA, Chang MV, Cadigan KM (2008) Novel TCF-binding sites specify transcriptional repression by Wnt signalling. EMBO J 27: 1436–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner E, Peter O, Schweizer L, Basler K (1997) Pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature 385: 829–833 [DOI] [PubMed] [Google Scholar]
- Chang JL, Chang MV, Barolo S, Cadigan KM (2008) Regulation of the feedback antagonist naked cuticle by Wingless signaling. Dev Biol 321: 446–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuring R et al. (2000) The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell 5: 355–365 [DOI] [PubMed] [Google Scholar]
- Fan HY, He X, Kingston RE, Narlikar GJ (2003) Distinct strategies to make nucleosomal DNA accessible. Mol Cell 11: 1311–1322 [DOI] [PubMed] [Google Scholar]
- Fang M, Li J, Blauwkamp T, Bhambhani C, Campbell N, Cadigan KM (2006) C-terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. EMBO J 25: 2735–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT (2004) Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev 18: 170–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cohen SM (2003) Wingless and Notch signaling provide cell survival cues and control cell proliferation during wing development. Development 130: 6533–6543 [DOI] [PubMed] [Google Scholar]
- Hecht A, Vleminckx K, Stemmler MP, van Roy F, Kemler R (2000) The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J 19: 1839–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R (2002) TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature 420: 439–445 [DOI] [PubMed] [Google Scholar]
- Hoffmans R, Basler K (2004) Identification and in vivo role of the Armadillo–Legless interaction. Development 131: 4393–4400 [DOI] [PubMed] [Google Scholar]
- Hoffmans R, Stadeli R, Basler K (2005) Pygopus and legless provide essential transcriptional coactivator functions to armadillo/beta-catenin. Curr Biol 15: 1207–1211 [DOI] [PubMed] [Google Scholar]
- Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, Murone M, Zullig S, Basler K (2002) Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin–TCF complex. Cell 109: 47–60 [DOI] [PubMed] [Google Scholar]
- Langst G, Becker PB (2001a) Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J Cell Sci 114: 2561–2568 [DOI] [PubMed] [Google Scholar]
- Langst G, Becker PB (2001b) ISWI induces nucleosome sliding on nicked DNA. Mol Cell 8: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Liu YI, Chang MV, Li HE, Barolo S, Chang JL, Blauwkamp TA, Cadigan KM (2008) The chromatin remodelers ISWI and ACF1 directly repress Wingless transcriptional targets. Dev Biol 323: 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K (2006) Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell 125: 327–341 [DOI] [PubMed] [Google Scholar]
- Pai LM, Orsulic S, Bejsovec A, Peifer M (1997) Negative regulation of Armadillo, a Wingless effector in Drosophila. Development 124: 2255–2266 [DOI] [PubMed] [Google Scholar]
- Peifer M, Polakis P (2000) Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science 287: 1606–1609 [DOI] [PubMed] [Google Scholar]
- Racki LR, Narlikar GJ (2008) ATP-dependent chromatin remodeling enzymes: two heads are not better, just different. Curr Opin Genet Dev 18: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra J, Yoshida T, Joazeiro CA, Jones KA (2006) The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev 20: 586–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasankaran R, Calleja M, Morata G, Basler K (2000) The Wingless target gene Dfz3 encodes a new member of the Drosophila Frizzled family. Mech Dev 91: 427–431 [DOI] [PubMed] [Google Scholar]
- Stadeli R, Hoffmans R, Basler K (2006) Transcription under the control of nuclear Arm/beta-catenin. Curr Biol 16: R378–R385 [DOI] [PubMed] [Google Scholar]
- Takemaru KI, Moon RT (2000) The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J Cell Biol 149: 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ (2004) A complex of Armadillo, Legless, and Pygopus coactivates dTCF to activate wingless target genes. Curr Biol 14: 458–466 [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Daniel C, Tamkun J, Wu C (1995) ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell 83: 1021–1026 [DOI] [PubMed] [Google Scholar]
- Wysocka J et al. (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442: 86–90 [DOI] [PubMed] [Google Scholar]
- Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, Fu D, Wu C (2001) Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell 8: 531–543 [DOI] [PubMed] [Google Scholar]
- Zeng W, Wharton KA Jr, Mack JA, Wang K, Gadbaw M, Suyama K, Klein PS, Scott MP (2000) Naked cuticle encodes an inducible antagonist of Wnt signalling. Nature 403: 789–795 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information