Abstract
Several studies have shown that ribosomal proteins (RPs) are important mediators of p53 activation in response to nucleolar disruption; however, the pathways that control this signalling function of RPs are currently unknown. We have recently shown that RPs are targets for the ubiquitin-like molecule NEDD8, and that NEDDylation protects RPs from destabilization. Here, we identify NEDD8 as a crucial regulator of L11 RP signalling to p53. A decrease in L11 NEDDylation during nucleolar stress causes relocalization of L11 from the nucleolus to the nucleoplasm. This not only provides the signal for p53 activation, but also makes L11 susceptible to degradation. Mouse double minute 2 (MDM2) -mediated NEDDylation protects L11 from degradation and this is required for p53 stabilization during nucleolar stress. By controlling the correct localization and stability of L11, NEDD8 acts as a crucial, new regulator of nucleolar signalling to p53.
Keywords: L11 RP, MDM2, NEDD8, nucleolar stress, p53
Introduction
Stress signals such as cell contact inhibition, serum starvation or the specific inhibition of RNA polymerase I using low doses (5 nM) of actinomycin D (ActD) increase the levels of p53 and activate the p53 response. Several ribosomal proteins (RPs), including L11, L5, L23 and S7, have been identified as mediators of these signals. In response to the above stimuli, the interaction of RPs with the p53 negative regulator mouse double minute 2 (MDM2) is increased. This results in MDM2 stabilization and inhibition of MDM2-mediated p53 degradation (Lohrum et al, 2003; Bhat et al, 2004; Dai & Lu, 2004; Jin et al, 2004; Chen et al, 2007). However, the pathways that control this signalling event are not presently understood. Genetic experiments have established an important function for the ubiquitin-like molecule NEDD8 in cell growth, viability and development. The cullin family of proteins, which are scaffold components of SCF (Skip–cullin–F-box) ubiquitin ligases, are the best-characterized substrates for NEDDylation (Pan et al, 2004). However, recent studies have shown the existence of additional targets for NEDD8, suggesting that a more diverse spectrum of biological processes are controlled by NEDDylation (Chan et al, 2008; Rabut & Peter, 2008; Xirodimas, 2008). By using a proteomic approach, we recently discovered that RPs are targets for NEDDylation and that this modification protects RPs from destabilization. However, under de-NEDDylating conditions in which the levels of the L11 RP are decreased, ribosomal biogenesis is not affected, suggesting that NEDDylation is not the main mechanism of ribosome production control (Xirodimas et al, 2008). Therefore, the biological significance of the effect of NEDDylation on RP stability is presently unknown. Here, we show that NEDD8 controls the signalling of the L11 RP to p53 in response to nucleolar stress. A decrease in L11 levels owing to reduced NEDDylation compromises p53 activation on nucleolar stress. Mechanistically, NEDD8 protects L11 from degradation by ensuring its nucleolar localization. A decrease in L11 NEDDylation, induced by ActD, participates in the nucleoplasmic relocalization of L11. This not only provides an initial signal for p53 activation, but also makes L11 susceptible to degradation. MDM2 is required to maintain the NEDDylated form of L11 and protects L11 from destabilization. Interestingly, a decrease in MDM2 expression reduces L11 levels and compromises p53 stabilization on nucleolar stress. Therefore, by controlling the localization and levels of the L11 RP, NEDD8 acts as a crucial, new regulator of nucleolar signalling.
Results
NEDD8 controls the signalling function of L11 to p53
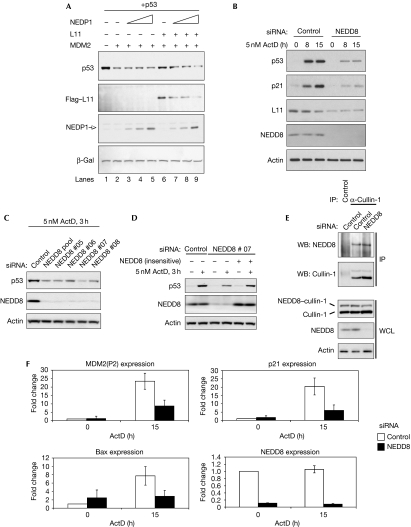
Our previous studies have shown that NEDDylation protects L11 RP from destabilization. One of the tools used to show this was the NEDD8-specific protease 1 (NEDP1 (deneddylase 1/sentrin-specific protease family, member 8)), a NEDD8-specific deconjugating enzyme that does not affect ubiquitin or small ubiquitin-like modifier (SUMO) conjugation (Xirodimas et al, 2004; Shen et al, 2005). Therefore, NEDP1 provides a specific tool for studying the function of NEDDylation in a biological process. The expression of NEDP1 results in the de-NEDDylation of L11 and decreases L11 protein levels. We used the NEDP1 protease to investigate the function of NEDD8 in the signalling of L11 to p53. In particular, we tested the effect of NEDP1 on the ability of L11 to protect p53 from MDM2-mediated degradation. As expected, MDM2 transfection resulted in reduced levels of p53, and co-expression of L11 inhibited p53 degradation by MDM2 (Fig 1A). However, increasing the amounts of NEDP1 protease reduced the levels of L11 and inhibited the protective effect of L11 on p53 (Fig 1A, compare lane 2 with lanes 6 and 9). Therefore, under conditions in which ribosome production is not affected, a decrease in NEDDylation compromises the ability of L11 to protect p53 from MDM2-mediated degradation. These data indicate that NEDDylation controls the signalling function of L11 to p53. To gain insights into the physiological function of NEDD8 in controlling p53 activation on nucleolar stress, we used a pool of four small interfering RNA (siRNA) oligomers that knock down NEDD8 (Fig 1B) but not ubiquitin (supplementary Fig S1A online), and monitored p53 stabilization after ActD treatment. As expected, low doses (5 nM) of ActD stabilized p53 in MCF7 cells, but NEDD8 knockdown compromised this stabilization effect (Fig 1B). Consistent with a protective function of NEDD8 in RP stability, the levels of L11 and its half-life were reduced on NEDD8 knockdown (Fig 1B; supplementary Fig S1B online), whereas L11 RNA levels were unaffected (supplementary Fig S1C online). Furthermore, the induction of p21 was also compromised, which is consistent with an inhibition of the p53 response. Importantly, the artificial reduction of L11 protein levels by siRNA has similar effects to NEDD8 knockdown (Bhat et al, 2004). The same effect on p53 stabilization by ActD was observed when cells were transfected using four individual siRNAs targeting different sequences in NEDD8 (Fig 1C). Importantly, we could rescue the defect in p53 stabilization by expressing a NEDD8 construct that is insensitive to one of the NEDD8 siRNAs (Fig 1D). Under these conditions, a reduction of NEDD8 levels does not cause a significant decrease in the NEDDylated form of cullin-1, a component of the SCFβTRCP (SCF β-transducin repeat-containing protein) ligase (Fig 1E; Pan et al, 2004). Furthermore, no change in the degradation of IκBα was observed, a well-described substrate for cullin-1 SCFβTRCP ligase (supplementary Fig S1D online). As cullins are efficient NEDD8 substrates, we assume that the residual NEDD8 is sufficient to conjugate cullins. The above data show that the effect we observe on p53 stabilization with the NEDD8 siRNA is specific and not due to a general effect on SCF ligases. Similar defects in p53 stabilization by ActD were observed with siRNAs against the E1 enzyme for NEDD8 (supplementary Fig S1E online). Consistent with a compromised p53 response, knockdown of NEDD8 reduced the induction of p53 target genes by ActD (Fig 1F). The above data show that the lack of NEDDylation before ActD treatment results in decreased levels of L11, and that this compromises p53 stabilization and activation on nucleolar stress.
Figure 1.
NEDD8 controls L11 signalling to p53 on nucleolar stress. (A) H1299 cells were transfected with 1 μg p53, MDM2, 5 μg Flag–L11 and 2, 5 or 10 μg of NEDP1 constructs as indicated. Cells were lysed in 2 × SDS loading buffer and an equal amount of protein was analysed by western blotting. Transfection efficiency and loading was monitored by co-transfection of β-gal (3 μg). (B) MCF7 cells were transfected with control or NEDD8 siRNA and 48 h later were treated with ActD as indicated. Cells were lysed and analysed by western blotting as in (A). (C) MCF7 cells were transfected with control or individual siRNAs for NEDD8 and analysed as in (A). (D) Transfections were carried out as in (C) and cells were re-seeded the next day; the day after they were transfected with a siRNA-insensitive NEDD8 construct. After 24 h, cells were treated with ActD and used for western blotting. (E) Transfections in MCF7 cells were carried out as in (B) and cell extracts were used for IPs using anti-cullin-1. Eluates and WCL were used for western blotting as indicated. (F) MCF7 cells were transfected with control or NEDD8 siRNAs and treated with ActD. Real-time PCR for the indicated genes was carried out as described in the Methods. Data are presented as mean±s.e.m. from three independent experiments. ActD, actinomycin D; β-gal, β-galactosidase; bax, BCL2-associated X protein; IP, immunoprecipitation; LB, loading buffer; MDM2, mouse double minute 2; NEDP1, NEDD8-specific protease 1; siRNA, small interfering RNA; WCL, whole cell lysate.
NEDD8 controls the localization and mobility of L11
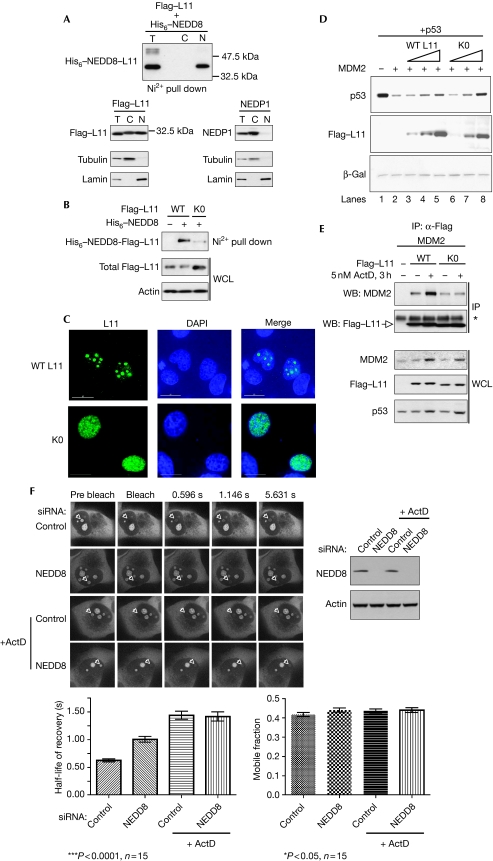
Previous studies have shown that nucleolar disruption by ActD causes relocalization of L11 in the nucleoplasm (Lohrum et al, 2003; Bhat et al, 2004); however, the mechanism responsible for this phenomenon has not been established. Subcellular fractionation showed that most of the NEDDylated form of L11 is localized in the nuclear/nucleolar fraction (Fig 2A). Interestingly, the ectopically expressed NEDP1 protease is found almost exclusively in the cytoplasm (Fig 2A). Identical localization was observed for the endogenous NEDP1 (data not shown). However, expression of NEDP1 results in complete de-NEDDylation of L11 (Xirodimas et al, 2008), suggesting that L11 can be NEDDylated in the cytoplasm, possibly before nuclear entry and nucleolar localization.
Figure 2.
NEDD8 controls the localization and mobility of L11. (A) H1299 cells were transfected with 5 μg Flag–L11, 2 μg His6–NEDD8 and 5 μg of NEDP1 constructs as indicated. Subcellular fractionation was carried out as described in the Methods. Tubulin and lamin were used as cytoplasmic and nuclear markers, respectively. (B) H1299 cells were transfected with 3 μg Flag–L11 (WT) or 12 μg Flag–K0 (K0) mutant and His6–NEDD8 as described above. NEDDylated proteins and total cell extracts were analysed by western blotting as indicated. (C) The localizations of WT L11 and the K0 mutant were analysed in MCF7 cells as described in the Methods. Fixed cells were stained with anti-Flag. (D) H1299 cells were transfected with 1 μg p53, 1 μg MDM2, 1, 3 or 5 μg of WT L11 and 4, 8 or 12 μg of K0 mutant as indicated. Cells were lysed in 2 × SDS LB and analysed by western blotting. Transfection efficiency and loading was monitored using β-gal (3 μg). (E) U2OS cells were transfected with 0.5 μg WT L11, 2 μg K0 mutant and 2 μg MDM2, and treated with ActD as indicated. IPs were carried out using anti-Flag and blotted with the indicated antibodies. WCLs were used for western blotting. The asterisk indicates a non-specific band. (F) H1299 cells, stably expressing L11–EGFP, were transfected with either control or NEDD8 siRNAs. At 48 h post-transfection, cells were treated as indicated with ActD for 1 h before the FRAP experiment. After the experiment, cells were lysed in 2 × SDS LB and analysed by western blotting (top right panel). The half-life of recovery and the mobile fraction were calculated as described in the Methods. C, cytoplasm; DAPI, 4′,6-diamidino-2-phenylindole; EGFP, enhanced green fluorescent protein; FRAP, fluorescence recovery after photobleaching; β-gal, β-galactosidase; IP, immunoprecipitation; LB, loading buffer; MDM2, mouse double minute 2; N, nucleus; NEDP1, NEDD8-specific protease 1; siRNA, small interfering RNA; T, total extract; WCL, whole cell lysate; WT, wild type.
To test whether NEDDylation controls L11 localization directly, we carried out mutagenesis to identify the required lysine residues for L11 NEDDylation. Mutation of all 16 lysines to arginines was necessary to create a NEDDylation-deficient L11 mutant, L11 K0 (Fig 2B). Consistent with a function of NEDD8 in stabilizing L11, the mutant was unstable compared with the wild-type L11 (data not shown). It was necessary to transfect four times as much DNA as the wild type to express the K0 mutant to similar levels. The residual NEDDylation of the K0 mutant could be due to amino-terminal modification. Furthermore, ribosome profiling showed that the L11 K0 mutant could be incorporated into ribosomes with an efficiency similar to that of the wild type (supplementary Fig S2 online). This is consistent with our previous studies showing that NEDD8 does not affect ribosome production and suggests that the K0 mutant retains physiological functions despite the number of mutations introduced. We carried out immunofluorescence analysis to determine the localization of the K0 mutant. An overexpressed wild-type L11 can be observed throughout the cell but, for the purpose of comparing its localization with the K0 mutant, the exposure time was set up so that L11 was found predominantly in the nucleolus. Under the same conditions, the K0 mutant showed a more diffused phenotype with clear nucleoplasmic localization (Fig 2C). This suggests that NEDDylation is implicated in the nucleolar localization of L11. Interestingly, the L11 K0 mutant phenocopies the nucleoplasmic localization of the wild-type L11 observed during nucleolar stress. Therefore, we used the K0 mutant to test the hypothesis that nucleoplasmic localization of L11 is sufficient to protect p53 from MDM2-mediated degradation in the absence of ActD. Compared with the wild-type L11, the K0 mutant is more potent in protecting p53 from MDM2-mediated degradation (Fig 2D, compare lanes 5 and 8). This shows that the relocalization of L11, which occurs during nucleolar stress, is sufficient to activate p53 and suggests that NEDDylation controls this process. As mentioned earlier, ActD treatment increases the interaction of L11 with MDM2 (Fig 2E; Lohrum et al, 2003). Interestingly, the K0 mutant is also able to interact with MDM2 but this association is not regulated by ActD (Fig 2E). This is consistent with a model in which ActD changes the localization of L11, resulting in an increased interaction with MDM2. The K0 mutant is already relocalized and consequently ActD no longer controls its interaction with MDM2. However, owing to the many mutations introduced in the K0 mutant, we also used a different approach to assess the role of NEDD8 in the localization of L11.
Quantitative proteomic analysis and live-cell imaging studies using fluorescence recovery after photobleaching (FRAP) have shown that on their synthesis, most RPs are rapidly imported into the nucleolus (Chen & Huang, 2001; Andersen et al, 2005; Lam et al, 2007). We used FRAP to gain further insights into the function of NEDD8 in the regulation of L11 mobility. Nucleoli of H1299 cells, stably expressing L11–EGFP (enhanced green fluorescent protein), were photobleached and the recovery of nucleolar fluorescence was measured as described in the Methods. As a control, we treated cells with ActD as it is predicted to affect L11 mobility. NEDD8 knockdown caused a reduction in the mobility of L11, which is consistent with the nucleoplasmic localization of the K0 mutant. Importantly, ActD had the same effect on L11 mobility as NEDD8 knockdown (Fig 2F; supplementary Fig S3 online). Interestingly, the combination of ActD and NEDD8 knockdown was not additive, suggesting that the NEDD8 pathway is part of the ActD response. The mobile fraction of L11 was unaffected by any treatment, suggesting that L11 complex formation is not altered. The combination of L11 K0 mutant localization and the effect of NEDD8 knockdown on L11 mobility strongly suggests that NEDD8 controls L11 nucleolar localization.
NEDDylation of L11 is specifically reduced by ActD
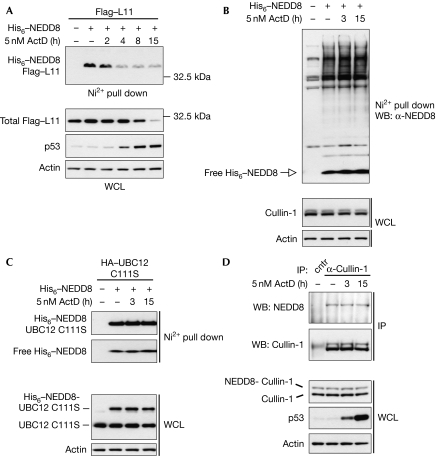
The above data indicate that NEDDylation regulates the localization and stability of L11. Next, we investigated whether nucleolar stress regulates the NEDDylation of L11. We found that L11 NEDDylation is decreased within the first 2 h of ActD treatment, before p53 stabilization (Fig 3A). This suggests that by decreasing L11 NEDDylation, ActD causes relocalization of L11 and that this participates in p53 activation. Prolonged application of ActD (8 and 15 h) also decreased the levels of L11 protein (Fig 3A). Similar results were obtained in other cell lines such as H1299 and U2OS (data not shown). Interestingly, ActD had the same effect on L11 modification and protein stability as NEDP1 expression. This suggests that ActD reduces the levels of L11 through a decrease in NEDDylation. Under similar conditions, ActD did not affect the overall NEDDylation in cells (Fig 3B). Consistent with this, ActD had no effect on the initial transfer step of NEDD8 from the E1 activating enzyme to the E2 conjugating enzyme ubiquitin conjugating enzyme 12 (UBC12) (Fig 3C). For analysing this, we used the C111S UBC12 mutant, which forms a stable covalent bond with NEDD8 and easily allows the monitoring of UBC12 charging with NEDD8 (Wada et al, 2000). Furthermore, NEDDylation of cullin-1 was also unaffected by ActD (Fig 3D). This shows that L11 NEDDylation is specifically reduced during nucleolar stress, and suggests that this participates in nucleoplasmic localization of L11 and p53 activation.
Figure 3.
NEDDylation of L11 is specifically reduced by ActD. (A) MCF7 cells were transfected with 3 μg Flag–L11 and 1 μg of His6–NEDD8 constructs, and ActD was applied as indicated. Western blot analysis on the NEDDylated eluates and WCL was carried out using the indicated antibodies. (B) H1299 cells were transfected with 2 μg His6–NEDD8 and treated with ActD as indicated. NEDDylated proteins or WCL were analysed by western blotting. (C) Experiment carried out as in (B), including 5 μg UBC12C111S mutant. (D) MCF7 cells were treated with ActD as indicated and IPs were carried out using anti-cullin-1. Eluates and WCL were analysed by western blotting. ActD, actinomycin D; HA, haemagglutinin; IP, immunoprecipitation; UBC12, ubiquitin conjugating enzyme 12; WB, western blot; WCL, whole cell lysate.
MDM2 promotes L11 NEDDylation
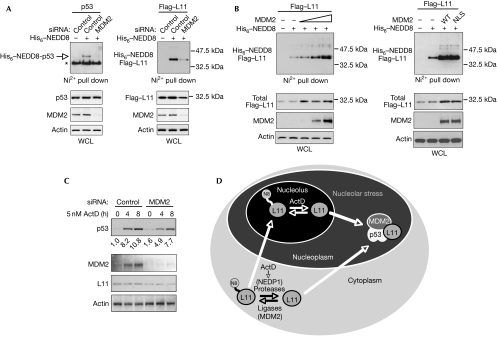
As already discussed, MDM2 has an important function in the nucleolar signalling of RPs to p53, as RPs protect p53 from MDM2-mediated degradation. As an E3 ligase, MDM2 promotes NEDDylation of p53 (Fig 4A; Xirodimas et al, 2004). The reduction of MDM2 levels caused a decrease in L11 NEDDylation and in the levels of L11, suggesting that MDM2 controls L11 stability through NEDDylation (Fig 4A). Furthermore, overexpressed MDM2 increased both the unmodified and the NEDDylated fraction of L11 (Fig 4B). This suggests that MDM2 protects L11 from destabilization by promoting L11 NEDDylation. Furthermore, an MDM2 nuclear localization signal mutant, which is localized exclusively in the cytoplasm (Xirodimas et al, 2001a), is able to promote L11 NEDDylation to levels comparable with wild-type MDM2 (Fig 4B). This further supports the idea that L11 NEDDylation can occur in the cytoplasm.
Figure 4.
MDM2 controls the stability of L11 through NEDDylation and is required for p53 stabilization on nucleolar stress. (A) H1299 cells were transfected with control or MDM2 siRNA. The next day, cells were re-seeded and the day after were transfected with 1 μg His6–NEDD8 and either 0.5 μg p53 (left panel) or 3 μg Flag–L11 (right panel) with Fugene as indicated. NEDDylated proteins and WCL were analysed by Western blotting. The asterisk indicates the presence of unmodified p53. (B) Left panel: wild-type MDM2 was transfected (2, 5, 10 μg) as indicated with 5 μg Flag–L11 and 2 μg His6–NEDD8 in H1299 cells. Western blot analysis on the NEDDylated proteins and WCL was carried out as before. Right panel: the experiment was carried out as in the left panel, including 5 μg WT MDM2 or the NLS mutant. (C) MCF7 cells were transfected with the indicated siRNAs and treated with ActD as indicated. Cells were lysed and analysed by Western blotting as before. Signals were quantified with Image Gauge. (D) Model for the role of NEDD8 in nucleolar signalling. NEDDylation of L11 is required for the correct localization of L11 in the nucleolus. Stress signals, such as ActD treatment, result in rapid de-NEDDylation of L11 causing either its release from the nucleolus and/or impairment in nucleolar import. This results in L11 nucleoplasmic localization, not only providing the signal for p53 activation, but also making L11 susceptible to degradation. The MDM2–p53 complex acts as a sensor of mislocalized L11, leading to p53 activation. ActD, actinomycin D; MDM2, mouse double minute 2; NEDP1, NEDD8-specific protease 1; NLS, nuclear localization signal; siRNA, small interfering RNA; WCL, whole cell lysate; WT, wild type.
The above data show that MDM2 regulates the levels of the crucial mediator of nucleolar signalling, L11. We tested the function of MDM2 in p53 stabilization by ActD. As expected, knockdown of MDM2 caused an increase in p53 levels in unstressed conditions. Surprisingly, a decrease in MDM2 levels compromised p53 stabilization induced by ActD. Consistent with a stabilizing function of MDM2 for L11, knockdown of MDM2 reduced the levels of L11 (Fig 4C). These data show that MDM2 participates in p53 stabilization on nucleolar stress, possibly through the regulation of L11 levels.
Discussion
Here, we have identified a biological function for L11 NEDDylation. Our data show that a prolonged lack of NEDDylation results in decreased levels of L11. Although this does not affect ribosome production, L11 signalling to p53 on nucleolar stress is compromised. The rapid decrease in L11 NEDDylation, induced by ActD, participates in the relocalization of L11 to the nucleoplasm. This is not only sufficient to activate p53, but also makes L11 susceptible to degradation. This provides a mechanism for the nucleoplasmic localization of L11 observed with ActD and explains the decrease in L11 levels observed on prolonged ActD treatment. It also provides a mechanism for previous studies suggesting that nucleoplasmic RPs are targeted for degradation (Lam et al, 2007). Although a decrease in L11 NEDDylation during nucleolar disruption is part of p53 activation (through the relocalization of L11), prolonged de-NEDDylation and a decrease of L11 levels before stress compromises p53 activation on ActD treatment.
The mechanism by which NEDD8 protects L11 from destabilization is to ensure its correct nucleolar localization. The data show that a lack of NEDD8 or ActD treatment decreases the mobility of L11. Therefore, the observed nucleoplasmic localization of L11 during nucleolar stress could be due to its relocalization from the nucleolus and/or due to deficiency in correct nucleolar import. As our data suggest that NEDDylation of L11 can occur in the cytoplasm, this could control the rate of L11 import and nucleolar localization in unstressed conditions. On nucleolar stress, attenuation of MDM2 activity and/or increase in the deconjugation reaction might contribute to the decrease in L11 NEDDylation. Indeed, especially in overexpression experiments, we observed a consistent increase in the levels of NEDP1 (in the cytoplasm; data not shown) on ActD treatment (supplementary Fig S4 online). Therefore, regulation of L11 NEDDylation in the cytoplasm might control correct and/or fast nucleolar entry of L11. The nucleoplasmic MDM2–p53 complex might act as a sensor of mislocalized L11 during stress, leading to p53 activation (Fig 4D); however, as MDM2 is a shuttling protein, we cannot exclude its nuclear function as a NEDD8 E3 ligase for L11.
A possible mechanism by which NEDD8 protects L11 from degradation is to compete with ubiquitin for lysine modifications. This, however, does not seem to be the case as knock down of NEDD8 decreases L11 ubiquitination, which can be rescued by proteasome inhibition (supplementary Fig S5 online). Furthermore, L11 NEDDylation and ubiquitination show differential lysine requirements, as L11 with three carboxy-terminal lysines mutated (3CKR) is still NEDDylated but not ubiquitinated. Importantly, the 3CKR mutant is nucleolar (supplementary Fig S6 online), suggesting that the nucleoplasmic localization of the K0 mutant is due to the lack of NEDDylation. Although these data suggest a regulatory function of NEDD8 in the degradation of ubiquitinated L11, ubiquitin-independent mechanisms for L11 degradation cannot be excluded. NEDDylated L11 could be prevented from being recognized by the proteasome or by alternative degradation pathways such as the recently discovered selective process of autophagy, named ribophagy (Kraft et al, 2008).
MDM2 will have an important function in this process as it is required to maintain the NEDDylated form of L11 and to protect L11 from degradation. This also explains the surprising result for the requirement of MDM2 in p53 stabilization. Recent studies have shown that MDM2 can promote ubiquitination and degradation of the L26 RP in the cytoplasm (Ofir-Rosenfeld et al, 2008). The dual and potentially differential function of MDM2 as a NEDD8 and ubiquitin E3 ligase might determine the fate and/or rate of degradation of RPs.
In summary, our studies identify NEDD8 as a key, new component of nucleolar signalling. By controlling the localization and levels of key mediators such as L11 RP, NEDD8 controls transcriptional activity during nucleolar stress.
Methods
Transfections. Calcium phosphate was used to transfect H1299 cells in 10 cm dishes. Fugene 6 (Roche, Basel, Schweiz, Switzerland) was used for MCF7 and U2OS transfections. The total amount of cytomegalovirus-transfected plasmids was normalized using empty vector. For siRNA transfections, 30 nM of oligomers were transfected with oligofectamine (Invitrogen, Carlsbad, CA, USA).
Purification of His6–NEDDylated proteins. His6–NEDDylated proteins were purified as described by Xirodimas et al (2001b). A total of 20% of the cells were used for lysis in 2 × SDS loading buffer.
Gene expression. RNA isolated from MCF7 cells, transfected with siRNAs and treated with ActD, was used for complimentary DNA synthesis and TaqMan real-time PCR.
Immunoprecipitations. Cells pellets were lysed in GE buffer (50 mM HEPES, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 25 mM NaF, 10 μM ZnCl2, 10% glycerol and 1% Triton X-100) containing Roche cocktail protease inhibitors, and extracts were incubated with the appropriate antibodies. Protein G beads were added and immunoprecipitates were then used for western blot analysis.
Immunofluorescence and FRAP analysis. At 48 h post-transfection, cells were fixed with paraformaldehyde and stained with Flag antibodies and Jackson (West Grove, PA, USA) fluorescein isothiocyanate anti-mouse antibodies. For FRAP analysis, H1299 cells stably expressing L11 EGFP were transfected with siRNAs and treated with ActD for 1 h before the experiment. Nucleoli were photobleached and nucleolar recovery of fluorescence was measured in a 15 s period.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Figs S1–S7
Acknowledgments
We thank K. Vousden and H. Horn for providing useful reagents and discussing the data. We also thank M. Saville and N. Allende-Vega for real time PCR primers. We thank S. Swift for his help in the fluorescence recovery after photobleaching (FRAP) experiment. D.P.X. is a research fellow of the Association for International Cancer Research (AICR).
Footnotes
The authors declare that they have no conflict of interest.
References
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M (2005) Nucleolar proteome dynamics. Nature 433: 77–83 [DOI] [PubMed] [Google Scholar]
- Bhat KP, Itahana K, Jin A, Zhang Y (2004) Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J 23: 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y, Yoon J, Wu JT, Kim HJ, Pan KT, Yim J, Chien CT (2008) DEN1 deneddylates non-cullin proteins in vivo. J Cell Sci 121: 3218–3223 [DOI] [PubMed] [Google Scholar]
- Chen D, Huang S (2001) Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J Cell Biol 153: 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, Wang H, Zhang R (2007) Ribosomal protein S7 as a novel modulator of p53–MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene 26: 5029–5037 [DOI] [PubMed] [Google Scholar]
- Dai MS, Lu H (2004) Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem 279: 44475–44482 [DOI] [PubMed] [Google Scholar]
- Jin A, Itahana K, O'Keefe K, Zhang Y (2004) Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol 24: 7669–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M (2008) Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 10: 602–610 [DOI] [PubMed] [Google Scholar]
- Lam YW, Lamond AI, Mann M, Andersen JS (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol 17: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH (2003) Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3: 577–587 [DOI] [PubMed] [Google Scholar]
- Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M (2008) MDM2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell 32: 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K (2004) Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23: 1985–1997 [DOI] [PubMed] [Google Scholar]
- Rabut G, Peter M (2008) Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects' review series. EMBO Rep 9: 969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen LN, Liu H, Dong C, Xirodimas D, Naismith JH, Hay RT (2005) Structural basis of NEDD8 ubiquitin discrimination by the deNEDDylating enzyme NEDP1. EMBO J 24: 1341–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Yeh ET, Kamitani T (2000) A dominant-negative UBC12 mutant sequesters NEDD8 and inhibits NEDD8 conjugation in vivo. J Biol Chem 275: 17008–17015 [DOI] [PubMed] [Google Scholar]
- Xirodimas DP (2008) Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem Soc Trans 36: 802–806 [DOI] [PubMed] [Google Scholar]
- Xirodimas DP, Stephen CW, Lane DP (2001a) Cocompartmentalisation of p53 and MDM2 is a major determinant for MDM2-mediated degradation of p53. Exp Cell Res 270: 66–77 [DOI] [PubMed] [Google Scholar]
- Xirodimas D, Saville MK, Edling C, Lane DP, Lain S (2001b) Different effects of p14ARF on the levels of ubiquitinated p53 and MDM2 in vivo. Oncogene 20: 4972–4983 [DOI] [PubMed] [Google Scholar]
- Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP (2004) MDM2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 118: 83–97 [DOI] [PubMed] [Google Scholar]
- Xirodimas DP, Sundqvist A, Nakamura A, Shen L, Botting C, Hay RT (2008) Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep 9: 280–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs S1–S7