Abstract
Inhibitor of growth 2 (ING2) is a candidate tumour suppressor gene the expression of which is frequently lost in tumours. Here, we identified a new function for ING2 in the control of DNA replication and in the maintenance of genome stability. Global replication rate was markedly reduced during normal S-phase in small interfering RNA (siRNA) ING2 cells, as seen in a DNA fibre spreading experiment. Accordingly, we found that ING2 interacts with proliferating cell nuclear antigen and regulates its amount to the chromatin fraction, allowing normal replication progression and normal cell proliferation. Deregulation of DNA replication has been previously associated with genome instability. Hence, a high proportion of siRNA ING2 cells presented endoreduplication of their genome as well as an increased frequency of sister chromatid exchange. Thus, we propose for the first time that ING2 might function as a tumour suppressor gene by directly maintaining DNA integrity.
Keywords: DNA replication, genome stability, ING2, PCNA, tumour suppressor
Introduction
Inhibitor of growth 2 (ING2) belongs to the ING family of candidate tumour suppressor genes (Garkavtsev et al, 1996). Its expression is lost in human tumours (Ythier et al, 2008), but its tumour suppressor functions remain unclear. ING2 has been described as a stable component of the repressor complex mouse SIN3A–histone deacetylase 1 (HDAC1; Doyon et al, 2006). Owing to a high affinity with the histone 3 trimethylated on lysine 4 (H3K4me3), ING2 recruits SIN3A–HDAC1 to the chromatin (Shi et al, 2006) to regulate the expression of genes involved in cell-cycle control. ING2 can also promote transforming growth factor-β-induced transcription (Sarker et al, 2008) and enhance the activation of p53 in response to genotoxic stress (Gozani et al, 2003; Pedeux et al, 2005). Furthermore, ING2 has been involved in DNA repair pathways in response to ultraviolet irradiation (Wang et al, 2006). Most of the previous studies examining the role of ING2 in cellular processes have resulted from overexpression experiments. In this study, as ING2 is lost in many human tumour types, we investigated the consequences of its downregulation. Using this strategy, we characterized a new function for ING2 in the control of DNA replication and in the maintenance of genome stability. These results suggest, for the first time to our knowledge, that ING2 might act as a caretaker tumour suppressor gene by directly protecting DNA integrity, particularly during normal replication of the genome.
Results And Discussion
ING2 ensures normal DNA synthesis
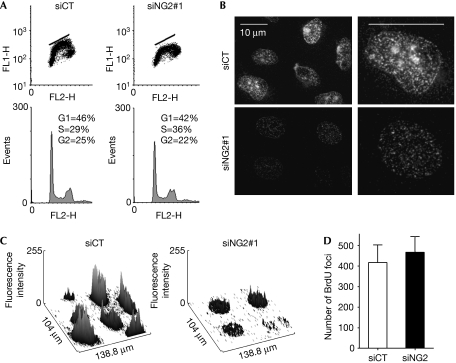
To identify the effects of ING2 downregulation on global S-phase, asynchronous populations of cells transfected with either a small interfering RNA control (siCT) or a small interfering RNA (siRNA) directed against ING2 (siING2#1) were analysed for bromodeoxyuridine (BrdU) incorporation. The efficiency of ING2 knockdown was assessed by western blotting (data not shown). In comparison with control cells, S-phase cells, downregulated for ING2, showed a deficiency in BrdU incorporation, as indicated by a flattened horseshoe when analysed by fluorescence-activated cell sorting (FACS; Fig 1A). This phenotype was confirmed with a siRNA directed against another part of ING2 (siING2#2, supplementary Fig S1A online) and was also observed in normal human telomerase reverse transcriptase (hTERT)-immortalized MRC5 cells (data not shown). Thus, a population of cells in S-phase incorporated more BrdU in the control cells than in the absence of ING2. ING2 has high homology with ING1 and it has been suggested that they might have redundant functions (He et al, 2005). However, the deficiency of BrdU incorporation was not observed in siING1 cells (supplementary Fig S1C,D online), showing that ING1 is not involved in the regulation of BrdU incorporation during S-phase. To determine whether the downregulation of ING2 affects the spatial organization and the activation of replication sites at the single-cell level, microscopic observations of cells immunostained with BrdU were carried out. This experiment again showed low BrdU incorporation in the nuclei of cells downregulated for ING2 using both siRNAs, as observed by a significant decrease of the staining intensity (Fig 1B,C; supplementary Fig S1A,B online). However, the number of replication foci was the same in siCT and siING2 cells (Fig 1D). Within each focus, many replication forks are activated. Hence, we hypothesized that the deficiency of BrdU incorporation was not due to a defect in the establishment and activation of replication foci but might be due to a slowdown of the progression of the replication fork within each active focus.
Figure 1.
ING2 ensures normal DNA synthesis. U2OS cells were transfected with siCT or siING2. (A) FACS analysis showing BrdU incorporation. (B) Representative immunofluorescence images of BrdU incorporation. Pictures were acquired with the same exposure time. (C) Graphic ImageJ (NIH) representation of BrdU intensity measured for immunostained cells shown in (B). (D) Number of replication foci per nucleus counted on 10 nuclei for each condition. BrdU, bromodeoxyuridine; FACS, fluorescence-activated cell sorting; ING2, inhibitor of growth 2; siCT, small interfering RNA control.
ING2 controls replication progression rate
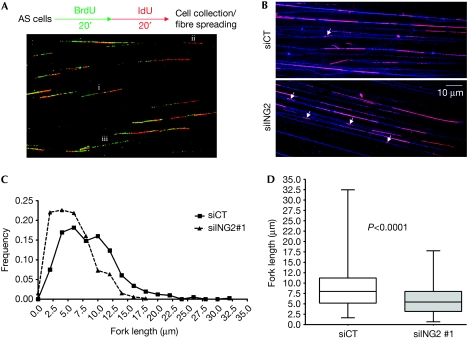
To investigate the speed of replication forks, we carried out a DNA fibre spreading experiment (Fig 2) and studied the function of ING2 on global replication fork progression rate during an ‘unperturbed' S-phase. The experiment was carried out on asynchronous cells (Fig 2A). The progression of the replication forks was visualized by sequential incorporation of BrdU and iododeoxyuridine (IdU, red) in siCT- and siING2-transfected cells (Fig 2A). DNA fibres were spread onto microscopic slides, and BrdU- and IdU-positive DNA fibres were visualized microscopically (green and red tracks, Fig 2A). The double BrdU and IdU staining allowed us to distinguish between (i) ongoing forks appearing as double-labelled tracks, (ii) newly initiated forks appearing as red-only tracks, and (iii) terminated forks appearing as green-only tracks. Thus, to calculate specifically the speed of the elongation of DNA replication, we measured the length of the IdU tracks (red tracks, Fig 2B) on double-labelled forks only. Single-strand DNA fibres were also immunostained (blue fibres, Fig 2B) to avoid the measure of aborted red tracks, which might appear because of broken fibres. Unlike control cells, siING2 cells showed a high proportion of very short IdU tracks (Fig 2B; supplementary Fig S2A online, white arrows), reflecting slowly progressing replication forks. Fork length re-partition clearly showed that most siING2 cells had shorter replication forks when compared with control cells, with many forks progressing very slowly (Fig 2C; supplementary Fig S2B online). Slow fork progression might result in a longer S-phase. Accordingly, U2OS and hTERT MRC5 siING2 cells proliferate more slowly (supplementary Fig S5A online). Global replication fork speed—determined by measuring more than 500 forks for each condition—was reduced from 20% (siING2#1, Fig 2D) to 30% (siING2#2, supplementary Fig 2SC online) when compared with the control (P<0.0001). Overall, these experiments indicate that ING2 is required to ensure normal progression of replication forks, suggesting a role in the elongation process.
Figure 2.
ING2 controls the progression of the replication fork. (A) U2OS cells transfected with siCT or siING2 were pulsed sequentially with BrdU and IdU, as shown in the experimental protocol. DNA fibres were spread onto microscopic slides (Jackson & Pombo, 1998) and immunostained with antibodies recognizing BrdU or IdU to distinguish between (i) ongoing forks, (ii) newly initiated forks, and (iii) terminated forks. (B) Representative immunofluorescence images showing IdU-positive tracks (pink) on DNA fibres (blue). White arrows indicate short IdU-positive tracks. (C) Fork length re-partition measured in micrometres (μm) using ImageJ software (NIH) for siCT (n=520) and siING2#1 (n=601). (D) Boxplots represent interquartile ranges and extreme values of fork lengths for siCT and siING2. Horizontal bar denotes the median. The P-value was measured using analysis of a variance test followed by a Mann–Whitney test. AS, asynchronous; BrdU, bromodeoxyuridine; IdU, iododeoxyuridine; ING2, inhibitor of growth 2; siCT, small interfering RNA control.
ING2 interacts with PCNA
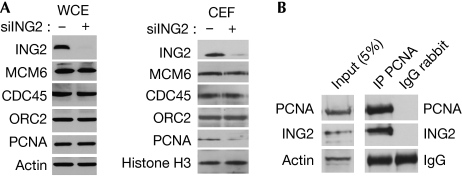
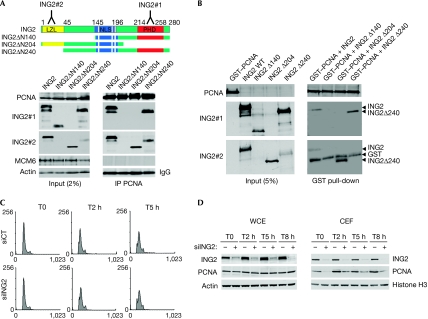
A decrease in the speed of the replication fork prompted us to study whether ING2 regulates proteins of the replication machinery. Indeed, DNA replication is a multi-step process, requiring efficient coordination between DNA unwinding and DNA synthesis. The synchronization of these events is crucial to maintain normal replication rate and subsequent genome stability. The firing of new replication forks requires the regulated loading of many proteins. In early G1-phase, the mini chromosome maintenance 2–7 (MCM2–7) helicase complex binds to the origin recognition complex proteins at the replication forks (Bell & Dutta, 2002). The MCM proteins are assembled in the pre-replicative complex and are required for DNA unwinding. CDC45 protein is then recruited to the pre-replicative complex during the G1–S transition to initiate DNA replication (Pacek et al, 2006). The processing of DNA synthesis at these forks then requires the loading of proliferating cell nuclear antigen (PCNA), which acts during elongation until the end of S-phase by increasing the processivity of DNA polymerases (Hao et al, 2008). The level of these proteins was assessed by western blotting in siCT- or siING2-transfected cells in the whole-cell extract and in the chromatin-enriched fraction (CEF; Fig 3A; supplementary Fig S3A online). In the whole-cell extract, the amount of these proteins was not altered after ING2 downregulation (Fig 3A). In the CEF, the same amount of MCM6, CDC45 and origin recognition complex 2 proteins was present in control or in siING2-transfected cells (CEF, Fig 3A). By contrast, we saw a marked decrease in the chromatin-bound fraction of PCNA in cells downregulated for ING2, also apparent by immunofluorescence (supplementary Fig S3C online). Similar results were obtained in normal human fibroblasts (supplementary Fig S3B online). This is in accordance with the known ability of ING2 in recruiting or regulating proteins such as SIN3A–HDAC1 to the chromatin (Shi et al, 2006). To investigate whether the regulation of PCNA by ING2 could occur through an interaction between the two proteins, we carried out an immunoprecipitation experiment. Indeed, we found that PCNA co-immunoprecipitated endogenous ING2 (Fig 3B). The use of different truncated forms of ING2, transfected in the cells, showed that the region located between the leuzine zipper-like motif and the nuclear localization sequence domains of ING2 is required for PCNA binding (Fig 4A). The interaction between PCNA and ING2 was confirmed in vitro by a glutathione S-transferase pull-down experiment using recombinant proteins (Fig 4B). It is interesting to point out that ING1 has been previously described as interacting with PCNA. However, this interaction was only seen in response to the stress induced by ultraviolet irradiation and was involved in DNA repair pathways (Scott et al, 2001). Furthermore, ING1 interacts with PCNA through a PCNA-interacting protein motif, whereas ING2 seems to interact with PCNA through another motif—as is the case for the protein GADD45, for example (Vairapandi et al, 2000)—which has very low homology with ING1 (He et al, 2005). This suggests that the ING1–PCNA and ING2–PCNA interactions have different roles in the cells. The ING2–PCNA interaction would occur during normal DNA synthesis to regulate the amount of PCNA chromatin, enhancing elongation complex processivity, whereas ING1 might interact with PCNA in response to DNA damage. This is in accordance with our observation that siING1 cells show a normal rate of BrdU incorporation, compared with that in siING2 cells (supplementary Fig S1 online). However, this does not rule out the possibility that the ING2–PCNA interaction might also have a function in DNA repair.
Figure 3.
ING2 interacts with PCNA. (A) Western blotting analysis of the WCE or the CEF proteins in U2OS cells. Actin and histone H3 were used as loading controls. (B) Immunoprecipitation (IP) of endogenous PCNA. Actin and IgG were used as the loading control. CEF, chromatin-enriched fraction; ING2, inhibitor of growth 2; PCNA, proliferating cell nuclear antigen; WCE, whole-cell extract.
Figure 4.
ING2 regulates the amount of PCNA in the chromatin fraction. (A) PCNA was immunoprecipitated in U2OS cells transfected with the truncated forms of ING2 (top panel). The proteins were detected using the indicated antibodies. MCM6 was used as a negative control of the IP. (B) GST pull-down experiment of recombinant GST–PCNA with ING2 in vitro translated constructs. Actin and IgG were used as loading controls for the Input and the IP, respectively. (C) Cells were collected from early (T0) to the end (T8 h) of S-phase. (D) The total expression of ING2 and PCNA was assessed in WCE. In parallel, the chromatin-bound proteins were detected in CEF. CEF, chromatin-enriched fraction; GST, glutathione S-transferase; ING2, inhibitor of growth 2; IP, immunoprecipitation; MCM6, mini chromosome maintenance 6; PCNA, proliferating cell nuclear antigen, WCE, whole-cell extract.
ING2 regulates the amount of PCNA to the chromatin
ING2 might be important for the presence of PCNA in the chromatin fraction when the elongation complex is assembling in early S-phase or, alternatively, ING2 might intervene later during S-phase progression to stabilize PCNA at the chromatin until replication is complete. To distinguish between these two possibilities, cells transfected with siCT or siING2 were synchronized in G1–S-phase and collected for 8 h during S-phase progression (T0–T8 h), as previously established (supplementary Fig S4A online). FACS analysis confirmed that siCT and siING2 cells entered the cell cycle with the same kinetics (Fig 4C), and presented the same amount of replication protein A (a single-strand DNA binding protein) to the CEF (data not shown), suggesting that initiation might occur properly in siING2 cells. However, siCT cells reached late S-phase slightly faster than siING2 cells, showing that completion of DNA synthesis is delayed by ING2 downregulation (supplementary Fig S4B online). The expression of ING2 and PCNA was assessed by western blot in the whole-cell extract and in the CEF (Fig 4D). In the whole-cell extract, we observed that ING2 downregulation had no consequence on the overall amount of PCNA, regardless of S-phase progression. In the CEF of siCT cells, a small amount of PCNA was present in the chromatin at T0. At 2 h after G1/S release, when cells entered S-phase, the amount of PCNA to the chromatin had markedly increased and remained high and stable throughout S-phase progression (from T2 h to T8 h). By contrast, in siING2 cells the amount of PCNA in the CEF was barely detectable at T0. At T2 h, the amount of PCNA increased but remained markedly lower throughout S-phase until T8 h. These results show that ING2 regulates the amount of PCNA to the chromatin fraction from the moment the elongation complex forms at the chromatin until replication is complete. Altogether, these data show that the downregulation of ING2 leading to the decrease of chromatin-bound PCNA during S-phase progression is associated with a defect in the elongation process. To exclude any function of ING2 in the initiation process, further investigations are necessary to determine whether siING2 cells would present the same density of active forks as do control cells.
ING2 maintains genome stability
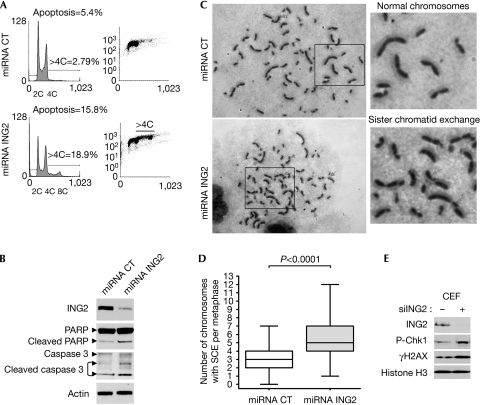
PCNA has been shown previously to be involved in normal S-phase progression and in the maintenance of genome stability (Nedelcheva-Veleva et al, 2006; Urtishak et al, 2009). To determine the consequences of the deregulated replication observed in siING2 cells after several rounds of DNA replication, we established stable clones downregulated for ING2. The cell-cycle profiles of three different stable U2OS clones maintained in culture for two weeks were assessed and the representative cell-cycle profile of one clone is presented in Fig 5A (micro RNA (miRNA) ING2). FACS analysis of propidium iodide (left panel) and BrdU incorporation (right panel) clearly showed that the downregulation of Ing2 (see western blot, Fig 5B) markedly increased the percentage of cells with more than 4C in DNA content to almost 20% compared with only 3% in miRNA CT cells. More specifically, we detected a distinct population of 8C cells, which are indicators of genome endoreduplication. Endoreduplication of the genome can initiate chromosomal aberrations. This was assessed by a sister chromatid exchange (SCE) experiment (Fig 5C), showing an increase in the number of SCE counted per metaphase in miRNA ING2 cells with a mean of 5.4 SCE per cell compared with a mean of 2.9 SCE per cell in miRNA CT cells (Fig 5C,D). Furthermore, this genome instability can generate DNA double-strand breaks. Indeed, in siING2 cells, we detected an increase of H2AX phosphorylation (Fig 5E), a marker of DNA double-strand breaks (Bonner et al, 2008), as well as the activation of the DNA damage checkpoint Chk1, seen by its phosphorylation. Activation of DNA damage checkpoint pathways can drive cells into apoptosis or senescence, which are two mechanisms that protect cells against neoplastic transformation; FACS analysis showed that nearly 16% of cells downregulated for ING2 underwent apoptosis (Fig 5A), and western blotting revealed the cleavage of poly(ADP-ribose) polymerase and caspase 3 (Fig 5B). This 10% increase of cells undergoing apoptosis might account, in part, for the 30% decrease in proliferation (supplementary Fig S5A online). Senescence-associated β-galactosidase assay showed that the occurrence of senescence was almost double in these cells compared with control cells (supplementary Fig S5B online), as previously described for another cell line downregulated for ING2 (Kumamoto et al, 2008). Thus, ING2 downregulation enhances genome instability.
Figure 5.
ING2 protects DNA integrity. Stable U2OS clones were established using miRNA. (A) Propidium iodide (left panels) and BrdU incorporation (right panels) were analysed by FACS to detect genome instability (>4C cells), endoreduplication (8C cells) and apoptosis (sub-G1 population). (B) Western blot analysis of caspase 3 and PARP in miRNA cells shown in (A). (C) Representative images of SCEs assessed in BrdU-labelled, giemsa-stained chromosome spreads from miRNA CT and miRNA ING2 cells. (D) At least 45 metaphases were counted for each condition and the number of SCE was normalized to the number of chromosomes per metaphase. The boxplots represent interquartile ranges and extreme values of SCEs per cell. The horizontal bars denote the median. The P-value was measured using an analysis of variance test followed by a Mann–Whitney U-test. (E) Western blot analysis of γH2AX and phospho-Chk1 (P-Chk1) in the CEF in siCT and siING2 cells. Histone H3 was used as a loading control. BrdU, bromodeoxyuridine; FACS, fluorescence-activated cell sorting; PARP, poly(ADP-ribose) polymerase; miRNA, microRNA; SCE, sister chromatid exchange; siCT, small interfering RNA control.
Our study shows, for the first time to our knowledge, that loss of ING2, which occurs in human tumours, results in an impaired DNA replication process and increased genome instability. Thus, ING2 could function as a caretaker tumour suppressor protein to prevent tumorigenesis. A previous study of ING5 has shown its interaction with the MCM helicase complex (Doyon et al, 2006), suggesting a potential model in which ING5 would be involved in DNA unwinding, whereas ING2 would simultaneously enhance the processivity of the elongation complex in association with PCNA. As each ING protein associates with a specific histone acetyltransferase or HDAC complex, we can hypothesize that different ING proteins in complex with various histone acetyltransferases and HDACs might be required at different steps during the initiation and progression of DNA replication.
Methods
Cell lines. U2OS osteosarcoma cells and hTERT MRC5 normal lung fibroblasts were used.
RNA-mediated interference. Stealth RNA-mediated interference (RNAi) (Invitrogen, Carlsbad, CA, USA) was used for ING2 downregulation (siING2). The universal RNAi negative control (#12935-110, Invitrogen) was used (siCT). Transfections were carried out using lipofectAMINE RNAimax (#13778-075, Invitrogen). For stable clones downregulated for ING2, we used the BLOCK-iT Pol II miR RNAi Expression Vector Kit (# K4936-00, Invitrogen). The miRNA duplex was inserted into the pcDNA 6.2-GW miR. Stable transfected clones were selected by the addition of 5 mg/ml of blasticidin to the culture medium.
Western blotting of chromatin-enriched fraction. Nuclear fraction was isolated using the NE-PER kit (#78833, Pierce, Rockford, IL, USA). The CEF was then isolated by re-suspending cell nuclei in an ice-cold hypotonic buffer (3 mM EDTA, 0.2 mM. EGTA and 1 mM DTT (pH 7.85) with protease inhibitor cocktail). Soluble fraction was removed and the pellet (CEF) was washed twice in the hypotonic buffer. The CEF was re-suspended and sonicated in a sonication buffer (10 mM Tris, 10% sucrose, 10 mM MgCl2 (pH 7.4) with protease inhibitor cocktail).
DNA fibre spreading. Asynchronous cells were pulsed sequentially with 100 μM BrdU (Zymed Laboratories, San Francisco, CA, USA) and 100 μM IdU (Sigma Aldrich, St Louis, MO, USA) for 20 min each. Cells were collected and DNA fibres were spread onto microscope slides as described by Jackson & Pombo (1998).
Immunofluorescence staining. Immunofluorescences were carried out as described previously by Sengupta et al (2003). An AxioImager Apotome (Zeiss, Germany) microscope was used.
Sister chromatid exchange experiment. SCE was carried out on stable U2OS clones as described previously by Perry & Wolff (1974). At least 45 metaphases were counted for each condition.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Material
Acknowledgments
We thank O. Gozani for providing the ING2 antibody and T. Gostan (Institut de Génétique Moléculaire de Montpellier) for the fibre spreading analysis using the IDeFIx software. This study was supported by the International Association for the Study of Lung Cancer, an ‘Agir à dom' grant and a Marie Curie International Reintegration Grant (MIRG-CT-2006-042148) (RP); DL, DY and RB are funded by the French Ministry of Education and Research, Institut National du Cancer and Fondation pour la Recherche Médicale, respectively. SS was supported by National Institute of Immunology core funds and the National Institutes of Health (BT/PR5936/BRB/10/408/2005).
Footnotes
The authors declare that they have no conflict of interest.
References
- Bell SP, Dutta A (2002) DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y (2008) GammaH2AX and cancer. Nat Rev Cancer 8: 957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J (2006) ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell 21: 51–64 [DOI] [PubMed] [Google Scholar]
- Garkavtsev I, Kazarov A, Gudkov A, Riabowol K (1996) Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat Genet 14: 415–420 [DOI] [PubMed] [Google Scholar]
- Gozani O et al. (2003) The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 114: 99–111 [DOI] [PubMed] [Google Scholar]
- Hao H, Xin T, Nancai Y, Yanxia W, Qian L, Wei M, Yandong Y, Hanju H (2008) Short-interfering RNA-mediated silencing of proliferating cell nuclear antigen inhibit proliferation and induce apoptosis in HeLa cells. Int J Gynecol Cancer 18: 36–42 [DOI] [PubMed] [Google Scholar]
- He GH, Helbing CC, Wagner MJ, Sensen CW, Riabowol K (2005) Phylogenetic analysis of the ING family of PHD finger proteins. Mol Biol Evol 22: 104–116 [DOI] [PubMed] [Google Scholar]
- Jackson DA, Pombo A (1998) Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 140: 1285–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto K, Spillare EA, Fujita K, Horikawa I, Yamashita T, Appella E, Nagashima M, Takenoshita S, Yokota J, Harris CC (2008) Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res 68: 3193–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelcheva-Veleva MN, Krastev DB, Stoynov SS (2006) Coordination of DNA synthesis and replicative unwinding by the S-phase checkpoint pathways. Nucleic Acids Res 34: 4138–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC (2006) Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21: 581–587 [DOI] [PubMed] [Google Scholar]
- Pedeux R et al. (2005) ING2 regulates the onset of replicative senescence by induction of p300-dependent p53 acetylation. Mol Cell Biol 25: 6639–6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry P, Wolff S (1974) New Giemsa method for the differential staining of sister chromatids. Nature 251: 156–158 [DOI] [PubMed] [Google Scholar]
- Sarker KP, Kataoka H, Chan A, Netherton SJ, Pot I, Huynh MA, Feng X, Bonni A, Riabowol K, Bonni S (2008) ING2 as a novel mediator of transforming growth factor-beta-dependent responses in epithelial cells. J Biol Chem 283: 13269–13279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M, Bonnefin P, Vieyra D, Boisvert FM, Young D, Bazett-Jones DP, Riabowol K (2001) UV-induced binding of ING1 to PCNA regulates the induction of apoptosis. J Cell Sci 114: 3455–3462 [DOI] [PubMed] [Google Scholar]
- Sengupta S et al. (2003) BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J 22: 1210–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X et al. (2006) ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442: 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urtishak KA, Smith KD, Chanoux RA, Greenberg RA, Johnson FB, Brown EJ (2009) Timeless maintains genomic stability and suppresses sister chromatid exchange during unperturbed DNA replication. J Biol Chem 284: 8777–8785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vairapandi M, Azam N, Balliet AG, Hoffman B, Liebermann DA (2000) Characterization of MyD118, Gadd45, and proliferating cell nuclear antigen (PCNA) interacting domains. PCNA impedes MyD118 AND Gadd45-mediated negative growth control. J Biol Chem 275: 16810–16819 [DOI] [PubMed] [Google Scholar]
- Wang J, Chin MY, Li G (2006) The novel tumor suppressor p33ING2 enhances nucleotide excision repair via inducement of histone H4 acetylation and chromatin relaxation. Cancer Res 66: 1906–1911 [DOI] [PubMed] [Google Scholar]
- Ythier D, Larrieu D, Brambilla C, Brambilla E, Pedeux R (2008) The new tumor suppressor genes ING: genomic structure and status in cancer. Int J Cancer 123: 1483–1490 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material