Maintaining the fidelity of the genome is critical to the cell, and DNA lesions threaten genetic integrity.1,2 Our laboratory has explored the design and applications of metalloinsertors that target single base mismatches, a potential source of mutations in DNA.3–8 The metal complexes contain sterically expansive ligands that are too wide to fit into well-matched B-form DNA and instead bind preferentially to thermodynamically destabilized mismatched sites. These complexes, notably Rh(bpy)2(chrysi)3+ (chrysi = chrysene-5,6-quinone diimine) (Figure 1), not only selectively bind to mismatched sites but also cleave the DNA backbone upon photo-activation. Recent structural studies show that this mismatch binding occurs not by intercalation, where the metal complex binds from the major groove, increasing the base pair rise through stacking within the helix, but rather by insertion, where the bulky ligand of the complex binds from the minor groove without increasing the base rise and ejects the mismatched base pairs into the major groove.9,10 This insertion mode clearly reconciles the relationship between mismatch destabilization and site recognition: the more destabilized the mismatch, the easier the extrusion of the mismatched bases.
Figure 1.
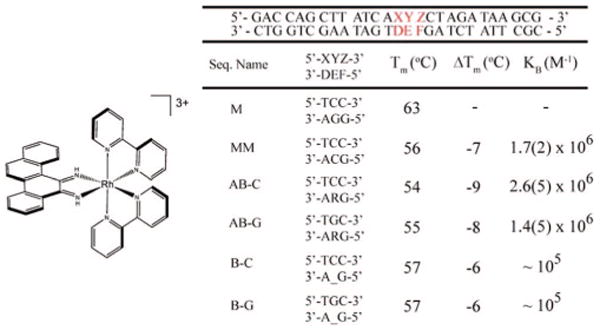
Δ-Rh(bpy)2(chrysi)3+ (left) along with sequences, melting temperatures (Tm), and binding affinities (KB) for the duplexes. Tm's were determined for 1 μM DNA in buffer (50 mM NaCl, 10 mM NaPi, pH 7.1). Photocleavage titrations of DNA (1 μM) in buffer and variable Rh(bpy)2(chrysi)3+ (0–10 μM) were employed to obtain site-specific binding constants (see Supporting Information). “R” denotes tetrahydrofuranyl abasic site; “–” indicates no nucleotide. Melting temperatures are accurate within 1 °C.
The relationship between site destabilization and Rh(bpy)2-(chrysi)3+ affinity has led us to investigate metalloinsertor recognition of two other common DNA defects: abasic sites and single base bulges. Abasic sites result from cleavage of the glycosidic bond and can arise spontaneously, as a result of exogenous carcinogens, or as an intermediate in repair.11 Spectroscopic studies have shown abasic sites to destabilize the duplex by 3–11 kcal/mol. Single base bulges, structurally related to abasic sites, are formed by replication errors. Bulged sites are more stable than abasic sites, with destabilization values of ≤4 kcal/mol.12 Left unrepaired, both structures can lead to cancer.13–15
There are, however, few reports of targeting abasic sites and bulges in DNA and none with high specificity. Higher reactivity of intercalators bearing nucleophilic amines with the abasic aldehyde intermediate has been reported, but neither specific binding nor strand cleavage at an abasic site is found.16–18 Bulky intercalators have also been seen to react at multiple base bulges or hairpin loops, but none bind to single base bulges and none with high specificity.19–22 Such specific targeting of small deformations in DNA is important both fundamentally in considering how these lesions may be detected within the cell and practically in the design of diagnostics for these lesions. Here we report that our parent metalloinsertor, Rh(bpy)2(chrysi)3+, specifically binds and, with photoactivation, cleaves abasic sites and single base bulges within duplex DNA.
A series of oligonucleotides were synthesized to probe recognition of abasic sites and single base bulges by Rh(bpy)2(chrysi)3+ (Figure 1).8 Six duplexes, identical except for a central site, were employed: well-matched (M), mismatched (MM), abasic site with unpaired cytosine (AB-C), abasic site with unpaired guanine (AB-G), cytosine single base bulge (B-C), and guanine single base bulge (B-G). Given the instability of the natural hemiacetal abasic site, the tetrahydrofuranyl analogue was employed for the abasic strands.23 Examination of the helix melting points, Tm, of the DNA assemblies reveals that the abasic site and bulged base significantly destabilize the duplex, in fact to a degree comparable to the CC mismatch (Figure 1).11,12
Site-specific targeting by Rh(bpy)2(chrysi)3+ was examined in DNA photocleavage experiments. As illustrated in Figure 2, Rh(bpy)2(chrysi)3+ specifically binds and, with irradiation, cleaves not only mismatches but also abasic sites and single base bulges. In all cases, cleavage neighbors the unpaired base and is evident only on one strand. The intensities of the photocleavage bands for the abasic sites are comparable to that at the mismatch site; photocleavage at the bulge sites is significantly diminished. This reduced intensity results both from a lower binding affinity and from lower photocleavage efficiency; in fact, the poor cleavage efficiency permits the determination of only an approximate affinity. For the abasic sites, photocleavage titrations yield site-specific binding affinities of ∼2 × 106 M−1, values comparable to that for the CC mismatch (see Figure 1). As expected for binding through metalloinsertion, affinity correlates with the local destabilization.
Figure 2.
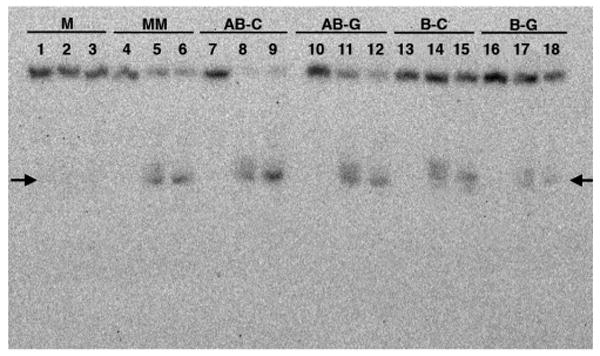
Autoradiogram of a denaturing 20% polyacrylamide gel revealing photocleavage for Rh(bpy)2(chrysi)3+ with the different DNA duplexes. 5′-32P-labeling is on the strand containing 5′-GAC. Conditions are duplex (1 μM), Rh (3 μM) in 50 mM NaCl, 10 mM NaPi, pH 7.1 for 30 min at ambient temperature followed by irradiation (0 or 30 min) with a solar simulator (325–450 nm) and incubation at 60 °C (0 or 30 min). For matched DNA, lanes 1–3 contain DNA with Rh without irradiation, DNA with Rh and 30 min irradiation, and DNA with Rh, 30 min irradiation, and 30 min at 60 °C, respectively. This set of three conditions is repeated for each duplex: MM (lanes 4–6), AB-C (7–9), AB-G (10–12), B-C (13–15), and B-G (16–18). The arrows mark the mispaired site.
In addition, the DNA cleavage products appear to mirror those seen with duplexes containing the mismatch (Figure 2 and Supporting Information), consistent with an analogous binding mode. Assayed by gel mobility, for each unpaired DNA assembly, we find a mixture of cleavage products consistent with reaction to the 3′-side of the unpaired base to form 15-mers containing a 3′-terminal phosphate along with 15-mers containing partially degraded 3′-phosphoesters; heating or overnight incubation yields a tighter band that results from decomposition of the mixture to the 3′-phosphate. Mass spectrometry has earlier been used to identify products of Rh photocleavage of mismatched DNA.24 Here, mass spectrometry results for an abasic assembly yield a product distribution pattern analogous to that of mismatched DNA. These products are consistent with H1′ abstraction by the photoactivated metal complex from the minor groove side, as expected with metalloinsertion.25 For the bulged DNA assemblies, gel analysis reveals bands of mobility similar to that created by mismatch cleavage, indicative of reaction to the 3′-side of the bulged base.26 Mass spectral data for the bulged assemblies were inconclusive, however, owing to the low product yield.
The high site specificity in targeting abasic sites and bulged bases by Rh(bpy)2(chrysi)3+ coupled with the correspondence in affinities and cleavage products to mismatch targeting all point to an analogous binding interaction of the metal complex with these structures versus mismatched DNA. Figure 3 illustrates our model for Rh binding to an abasic site based upon the crystal structure of Rh(bpy)2(chrysi)3+ bound to a CA mismatch.9 In this model, the metal complex inserts from the minor groove, displacing the unpaired base. The binding affinities for mismatched DNA and an abasic site are equivalent, as are the Tm's, reflecting the thermodynamic destabilization at the unpaired sites, the basis for ejecting the unpaired base. The shared asymmetric cleavage and resultant products furthermore support a similar orientation for the complex within the binding sites, with the Rh bound from the minor groove, positioning the activated ligand for H1′ abstraction. For the bulged DNA, the metal complex is likely similarly positioned, but the binding pocket must differ, accounting for the poor photocleavage efficiency. Nonetheless, for the bulged assemblies, based upon the cleavage strand asymmetry and estimated binding affinities, a generally comparable binding mode and orientation are likely.
Figure 3.
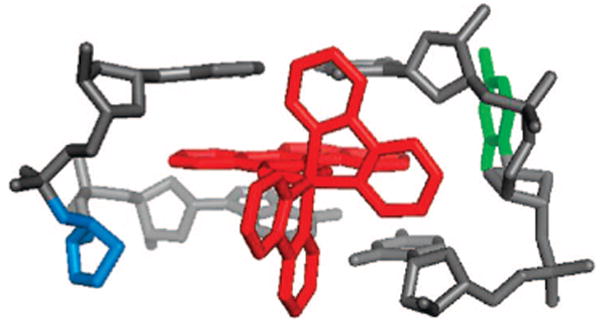
Model of Rh(bpy)2(chrysi)3+ inserted at an abasic site. The metalloinsertor is in red, the extruded base in green, and the abasic deoxyribose in blue.
These results establish that Rh(bpy)2(chrysi)3+, a sterically bulky metalloinsertor, binds and photocleaves abasic sites and single base bulges within duplex DNA with high affinity and selectivity. The work thus expands the family of unpaired DNA structures targeted by Rh(bpy)2(chrysi)3+. Moreover, these data highlight the generality of site-specific binding to destabilized structures within a DNA duplex through metalloinsertion.
Supplementary Material
Acknowledgments
We are grateful to the NIH (GM33309) for their financial support, the SURF program for a summer internship for J.A.B, and the NSF for a graduate fellowship to B.M.Z.
Footnotes
Supporting Information Available: Photocleavage titrations, gel analysis, and mass spectral data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Marnett LJ, Pastaras JP. Trends Genet. 2001;17:214–221. doi: 10.1016/s0168-9525(01)02239-9. [DOI] [PubMed] [Google Scholar]
- 2.a Iyer RR, Pluciennik A, Burdett V, Modrich PK. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]; b David SS, O'Shea VL, Kundu S. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeglis BM, Pierre VP, Barton JK. Chem Commun. 2007;44:4565. doi: 10.1039/b710949k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Jackson BA, Barton JK. J Am Chem Soc. 1997;119:12987. [Google Scholar]; b Jackson BA, Barton JK. Biochemistry. 2000;39:157–161. doi: 10.1021/bi9927033. [DOI] [PubMed] [Google Scholar]; c Jackson BA, Alekseyev VY, Barton JK. Biochemistry. 1999;38:4655–4662. doi: 10.1021/bi990255t. [DOI] [PubMed] [Google Scholar]
- 5.a Junicke H, Hart JR, Kisko J, Glebov O, Kirsch IR, Barton JK. Proc Natl Acad Sci USA. 2003;100:3737–3742. doi: 10.1073/pnas.0537194100. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Ruba E, Hart JR, Barton JK. Inorg Chem. 2004;43:4570–4578. doi: 10.1021/ic0499291. [DOI] [PubMed] [Google Scholar]
- 6.Hart JR, Glebov O, Ernst RJ, Kirsch IR, Barton JK. Proc Natl Acad Sci USA. 2006;103:15359–15363. doi: 10.1073/pnas.0607576103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a Zeglis BM, Barton JK. J Am Chem Soc. 2006;128:5654–5655. doi: 10.1021/ja061409c. [DOI] [PubMed] [Google Scholar]; b Schatzschneider U, Barton JK. J Am Chem Soc. 2004;126:8630–8631. doi: 10.1021/ja048543m. [DOI] [PubMed] [Google Scholar]; c Petitjean A, Barton JK. J Am Chem Soc. 2004;126:14728–14729. doi: 10.1021/ja047235l. [DOI] [PubMed] [Google Scholar]; d Lim MH, Lau I, Barton JK. Inorg Chem. 2007;46:9528. doi: 10.1021/ic701598k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeglis BM, Barton JK. Nat Prot. 2007;2:357–371. doi: 10.1038/nprot.2007.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pierre VP, Kaiser JT, Barton JK. Proc Natl Acad Sci USA. 2007;104:429–434. doi: 10.1073/pnas.0610170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordier C, Pierre VP, Barton JK. J Am Chem Soc. 2007;129:12287–12296. doi: 10.1021/ja0739436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a Vesnaver G, Chang CN, Eisenberg M, Grollman AP, Breslauer KJ. Proc Natl Acad Sci USA. 1989;86:3614–3618. doi: 10.1073/pnas.86.10.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Gelfand CA, Plum GE, Grollman AP, Johnson F, Breslauer KJ. Biochemistry. 1998;37:7321–7327. doi: 10.1021/bi9803372. [DOI] [PubMed] [Google Scholar]
- 12.a Leblanc DA, Morden KM. Biochemistry. 1991;30:4042–4047. doi: 10.1021/bi00230a031. [DOI] [PubMed] [Google Scholar]; b Tanaka F, Kameda A, Yamamoto M, Ohuchi A. Biochemistry. 2004;43:7143–7150. doi: 10.1021/bi036188r. [DOI] [PubMed] [Google Scholar]
- 13.Boiteux S, Guillet M. DNA Repair. 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Lovett ST. Mol Microbiol. 2004;52:1243–1253. doi: 10.1111/j.1365-2958.2004.04076.x. [DOI] [PubMed] [Google Scholar]
- 15.Jacob S, Praz F. Biochimie. 2002;84:27–47. doi: 10.1016/s0300-9084(01)01362-1. [DOI] [PubMed] [Google Scholar]
- 16.Boturyn D, Boudali A, Constant JF, Lhomme J. Tetrahedron. 1997;53:5485–5492. [Google Scholar]
- 17.Bertrand JR, Vasseur JJ, Gouyette A, Rayner B, Imbach KD, Paoletti C, Malvy C. J Biol Chem. 1989;264:14172–14178. [PubMed] [Google Scholar]
- 18.Berthet N, Boudali A, Constant JF, Decout JL, Demeunynck M, Fkyerat A, Garcia J, Laayoun A, Michon P, Lhomme JJ. J Mol Recognit. 1994;7:99–107. doi: 10.1002/jmr.300070207. [DOI] [PubMed] [Google Scholar]
- 19.Kappen LS, Goldberg IH. Biochemistry. 1993;32:13138–13145. doi: 10.1021/bi00211a024. [DOI] [PubMed] [Google Scholar]
- 20.Woodson SA, Crothers DM. Biochemistry. 1988;27:8904–8914. doi: 10.1021/bi00425a006. [DOI] [PubMed] [Google Scholar]
- 21.Cheng CC, Kuo YN, Chuang KS, Luo CF, Wang WJ. Angew Chem. 1999;38:1255–1257. doi: 10.1002/(SICI)1521-3773(19990503)38:9<1255::AID-ANIE1255>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 22.a Morgan JL, Buck DP, Turley AG, Collins JG, Keene FR. J Biol Inorg Chem. 2006;11:824–834. doi: 10.1007/s00775-006-0130-9. [DOI] [PubMed] [Google Scholar]; b Morgan JL, Buck DP, Turley AG, Collins JG, Keene FR. Inorg Chim Acta. 2006;359:888. doi: 10.1007/s00775-006-0130-9. [DOI] [PubMed] [Google Scholar]
- 23.Takeshita M, Chang CN, Johnson F, Will S, Grollman AP. J Biol Chem. 1987;262:10171–10179. [PubMed] [Google Scholar]
- 24.Brunner J, Barton JK. J Am Chem Soc. 2006;128:6772–6773. doi: 10.1021/ja0612753. [DOI] [PubMed] [Google Scholar]
- 25.For major groove intercalation, photocleavage products are instead consistent with H3′ radical formation. See reference 24 and Keilkopf CL, Erkkila KE, Hudson BP, Barton JK, Rees DC. Nat Struct Biol. 2000;7:117–121. doi: 10.1038/72385.
- 26.For the B-C duplex, bulge migration is possible, but this would result in a product of slower mobility.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


