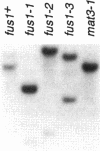
Abstract
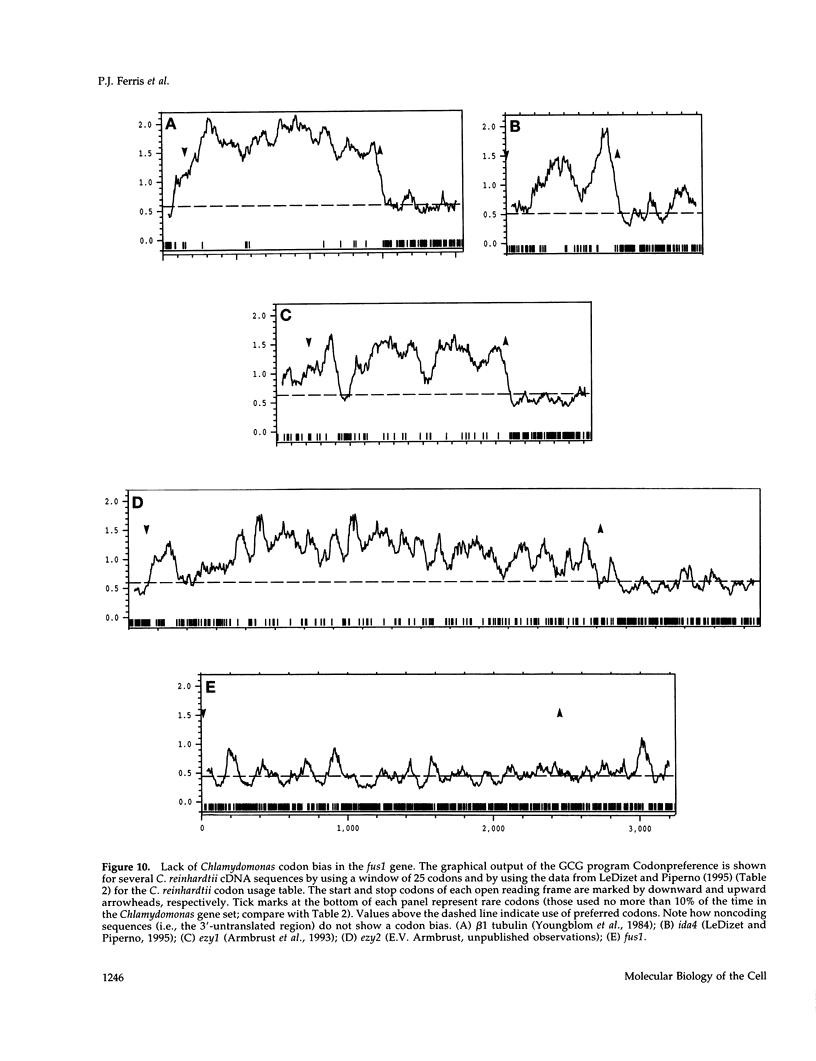
Sexual fusion between plus and minus gametes of the unicellular green alga Chlamydomonas reinhardtii entails adhesion between plus-specific and minus-specific "fringe" proteins displayed on the plasma membrane of gametic mating structures. We report the identification of the gene (fus1) encoding the plus fringe glycoprotein, which resides in a unique domain of the mating-type plus (mt+) locus, and which was identified by transposon insertions in three fusion-defective mutant strains. Transformation with fus1+ restores fringe and fusion competence to these mutants and to the pseudo-plus mutant imp11 mt-, defective in minus differentiation. The fus1 gene is remarkable in lacking the codon bias found in all other nuclear genes of C. reinhardtii.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adair W. S., Hwang C., Goodenough U. W. Identification and visualization of the sexual agglutinin from the mating-type plus flagellar membrane of Chlamydomonas. Cell. 1983 May;33(1):183–193. doi: 10.1016/0092-8674(83)90347-1. [DOI] [PubMed] [Google Scholar]
- Akashi H. Inferring weak selection from patterns of polymorphism and divergence at "silent" sites in Drosophila DNA. Genetics. 1995 Feb;139(2):1067–1076. doi: 10.1093/genetics/139.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida E. A., Huovila A. P., Sutherland A. E., Stephens L. E., Calarco P. G., Shaw L. M., Mercurio A. M., Sonnenberg A., Primakoff P., Myles D. G. Mouse egg integrin alpha 6 beta 1 functions as a sperm receptor. Cell. 1995 Jun 30;81(7):1095–1104. doi: 10.1016/s0092-8674(05)80014-5. [DOI] [PubMed] [Google Scholar]
- Armbrust E. V., Ferris P. J., Goodenough U. W. A mating type-linked gene cluster expressed in Chlamydomonas zygotes participates in the uniparental inheritance of the chloroplast genome. Cell. 1993 Sep 10;74(5):801–811. doi: 10.1016/0092-8674(93)90460-8. [DOI] [PubMed] [Google Scholar]
- Armbrust E. V., Ibrahim A., Goodenough U. W. A mating type-linked mutation that disrupts the uniparental inheritance of chloroplast DNA also disrupts cell-size control in Chlamydomonas. Mol Biol Cell. 1995 Dec;6(12):1807–1818. doi: 10.1091/mbc.6.12.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölker M., Urban M., Kahmann R. The a mating type locus of U. maydis specifies cell signaling components. Cell. 1992 Feb 7;68(3):441–450. doi: 10.1016/0092-8674(92)90182-c. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin-Osdoby P., Adair W. S. Characterization of the purified Chlamydomonas minus agglutinin. J Cell Biol. 1985 Sep;101(3):1144–1152. doi: 10.1083/jcb.101.3.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. B., Adair W. S., Mecham R. P., Heuser J. E. Chlamydomonas agglutinin is a hydroxyproline-rich glycoprotein. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5898–5901. doi: 10.1073/pnas.80.19.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersold W. T. Chlamydomonas reinhardi: heterozygous diploid strains. Science. 1967 Jul 28;157(3787):447–449. doi: 10.1126/science.157.3787.447. [DOI] [PubMed] [Google Scholar]
- Fernández E., Schnell R., Ranum L. P., Hussey S. C., Silflow C. D., Lefebvre P. A. Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6449–6453. doi: 10.1073/pnas.86.17.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P. J., Goodenough U. W. The mating-type locus of Chlamydomonas reinhardtii contains highly rearranged DNA sequences. Cell. 1994 Mar 25;76(6):1135–1145. doi: 10.1016/0092-8674(94)90389-1. [DOI] [PubMed] [Google Scholar]
- Ferris P. J., Goodenough U. W. Transcription of novel genes, including a gene linked to the mating-type locus, induced by Chlamydomonas fertilization. Mol Cell Biol. 1987 Jul;7(7):2360–2366. doi: 10.1128/mcb.7.7.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P. J. Localization of the nic-7, ac-29 and thi-10 genes within the mating-type locus of Chlamydomonas reinhardtii. Genetics. 1995 Oct;141(2):543–549. doi: 10.1093/genetics/141.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forest C. L. Genetic control of plasma membrane adhesion and fusion in Chlamydomonas gametes. J Cell Sci. 1987 Dec;88(Pt 5):613–621. doi: 10.1242/jcs.88.5.613. [DOI] [PubMed] [Google Scholar]
- Galloway R. E., Goodenough U. W. Genetic analysis of mating locus linked mutations in Chlamydomonas reinhardii. Genetics. 1985 Nov;111(3):447–461. doi: 10.1093/genetics/111.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloeckner G., Beck C. F. Genes involved in light control of sexual differentiation in Chlamydomonas reinhardtii. Genetics. 1995 Nov;141(3):937–943. doi: 10.1093/genetics/141.3.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U. W., Detmers P. A., Hwang C. Activation for cell fusion in Chlamydomonas: analysis of wild-type gametes and nonfusing mutants. J Cell Biol. 1982 Feb;92(2):378–386. doi: 10.1083/jcb.92.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U. W., Hwang C., Martin H. Isolation and genetic analysis of mutant strains of Chlamydomonas reinhardi defective in gametic differentiation. Genetics. 1976 Feb;82(2):169–186. doi: 10.1093/genetics/82.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U. W., Jurivich D. Tipping and mating-structure activation induced in Chlamydomonas gametes by flagellar membrane antisera. J Cell Biol. 1978 Dec;79(3):680–693. doi: 10.1083/jcb.79.3.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmann A., Sumper M. An inducible arylsulfatase of Volvox carteri with properties suitable for a reporter-gene system. Purification, characterization and molecular cloning. Eur J Biochem. 1994 Apr 1;221(1):143–150. doi: 10.1111/j.1432-1033.1994.tb18723.x. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. A regulatory hierarchy for cell specialization in yeast. Nature. 1989 Dec 14;342(6251):749–757. doi: 10.1038/342749a0. [DOI] [PubMed] [Google Scholar]
- Kindle K. L. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Fukuzawa H., Shimada T., Saito T., Matsuda Y. Primary structure and expression of a gamete lytic enzyme in Chlamydomonas reinhardtii: similarity of functional domains to matrix metalloproteases. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4693–4697. doi: 10.1073/pnas.89.10.4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk M. M., Kirk D. L. Translational regulation of protein synthesis, in response to light, at a critical stage of Volvox development. Cell. 1985 Jun;41(2):419–428. doi: 10.1016/s0092-8674(85)80015-5. [DOI] [PubMed] [Google Scholar]
- Larson A., Kirk M. M., Kirk D. L. Molecular phylogeny of the volvocine flagellates. Mol Biol Evol. 1992 Jan;9(1):85–105. doi: 10.1093/oxfordjournals.molbev.a040710. [DOI] [PubMed] [Google Scholar]
- LeDizet M., Piperno G. The light chain p28 associates with a subset of inner dynein arm heavy chains in Chlamydomonas axonemes. Mol Biol Cell. 1995 Jun;6(6):697–711. doi: 10.1091/mbc.6.6.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipke P. N., Kurjan J. Sexual agglutination in budding yeasts: structure, function, and regulation of adhesion glycoproteins. Microbiol Rev. 1992 Mar;56(1):180–194. doi: 10.1128/mr.56.1.180-194.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M., Gillespie J. H. Codon usage divergence of homologous vertebrate genes and codon usage clock. J Mol Evol. 1991 Jan;32(1):6–15. doi: 10.1007/BF02099923. [DOI] [PubMed] [Google Scholar]
- Martin N. C., Goodenough U. W. Gametic differentiation in Chlamydomonas reinhardtii. I. Production of gametes and their fine structure. J Cell Biol. 1975 Dec;67(3):587–605. doi: 10.1083/jcb.67.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesland D. A., Hoffman J. L., Caligor E., Goodenough U. W. Flagellar tip activation stimulated by membrane adhesions in Chlamydomonas gametes. J Cell Biol. 1980 Mar;84(3):599–617. doi: 10.1083/jcb.84.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz E. C., Palumbi S. R. Positive selection and sequence rearrangements generate extensive polymorphism in the gamete recognition protein bindin. Mol Biol Evol. 1996 Feb;13(2):397–406. doi: 10.1093/oxfordjournals.molbev.a025598. [DOI] [PubMed] [Google Scholar]
- Quarmby L. M., Hartzell H. C. Two distinct, calcium-mediated, signal transduction pathways can trigger deflagellation in Chlamydomonas reinhardtii. J Cell Biol. 1994 Mar;124(5):807–815. doi: 10.1083/jcb.124.5.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAGER R., GRANICK S. Nutritional studies with Chlamydomonas reinhardi. Ann N Y Acad Sci. 1953 Oct 14;56(5):831–838. doi: 10.1111/j.1749-6632.1953.tb30261.x. [DOI] [PubMed] [Google Scholar]
- Schmitt R., Fabry S., Kirk D. L. In search of molecular origins of cellular differentiation in Volvox and its relatives. Int Rev Cytol. 1992;139:189–265. doi: 10.1016/s0074-7696(08)61413-8. [DOI] [PubMed] [Google Scholar]
- Schnell R. A., Lefebvre P. A. Isolation of the Chlamydomonas regulatory gene NIT2 by transposon tagging. Genetics. 1993 Jul;134(3):737–747. doi: 10.1093/genetics/134.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal R. A., Huang B., Ramanis Z., Luck D. J. Mutant strains of Chlamydomonas reinhardtii that move backwards only. J Cell Biol. 1984 Jun;98(6):2026–2034. doi: 10.1083/jcb.98.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellig T., Bölker M., Lottspeich F., Frank R. W., Kahmann R. Pheromones trigger filamentous growth in Ustilago maydis. EMBO J. 1994 Apr 1;13(7):1620–1627. doi: 10.1002/j.1460-2075.1994.tb06425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson W. J., Vacquier V. D. Extraordinary divergence and positive Darwinian selection in a fusagenic protein coating the acrosomal process of abalone spermatozoa. Proc Natl Acad Sci U S A. 1995 May 23;92(11):4957–4961. doi: 10.1073/pnas.92.11.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker P. K., Lundrigan B. L. Rapid evolution of the sex determining locus in Old World mice and rats. Nature. 1993 Aug 19;364(6439):715–717. doi: 10.1038/364715a0. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks D. P., Beerman N., Griffith O. M. A small-scale five-hour procedure for isolating multiple samples of CsCl-purified DNA: application to isolations from mammalian, insect, higher plant, algal, yeast, and bacterial sources. Anal Biochem. 1986 Feb 1;152(2):376–385. doi: 10.1016/0003-2697(86)90423-9. [DOI] [PubMed] [Google Scholar]
- Wendland J., Vaillancourt L. J., Hegner J., Lengeler K. B., Laddison K. J., Specht C. A., Raper C. A., Kothe E. The mating-type locus B alpha 1 of Schizophyllum commune contains a pheromone receptor gene and putative pheromone genes. EMBO J. 1995 Nov 1;14(21):5271–5278. doi: 10.1002/j.1460-2075.1995.tb00211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield L. S., Lovell-Badge R., Goodfellow P. N. Rapid sequence evolution of the mammalian sex-determining gene SRY. Nature. 1993 Aug 19;364(6439):713–715. doi: 10.1038/364713a0. [DOI] [PubMed] [Google Scholar]
- Witman G. B., Carlson K., Berliner J., Rosenbaum J. L. Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J Cell Biol. 1972 Sep;54(3):507–539. doi: 10.1083/jcb.54.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfsberg T. G., Straight P. D., Gerena R. L., Huovila A. P., Primakoff P., Myles D. G., White J. M. ADAM, a widely distributed and developmentally regulated gene family encoding membrane proteins with a disintegrin and metalloprotease domain. Dev Biol. 1995 May;169(1):378–383. doi: 10.1006/dbio.1995.1152. [DOI] [PubMed] [Google Scholar]
- Youngblom J., Schloss J. A., Silflow C. D. The two beta-tubulin genes of Chlamydomonas reinhardtii code for identical proteins. Mol Cell Biol. 1984 Dec;4(12):2686–2696. doi: 10.1128/mcb.4.12.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hostos E. L., Schilling J., Grossman A. R. Structure and expression of the gene encoding the periplasmic arylsulfatase of Chlamydomonas reinhardtii. Mol Gen Genet. 1989 Aug;218(2):229–239. doi: 10.1007/BF00331273. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]