Abstract
Single nucleotide polymorphism (SNP) genotyping has emerged as a technology to incorporate copy-number variants (CNVs) into genetic analyses of human traits. However, the extent to which SNP platforms accurately capture CNVs remains unclear. Using independent, sequence-based CNV maps, we find that commonly used SNP platforms have limited or no probe coverage for a large fraction of CNVs. Despite this, in nine samples we inferred 368 CNVs using Illumina SNP genotyping data and experimentally validated over two-thirds of these. We also developed a method (SCIMM) to robustly genotype deletions using as few as two SNP probes. We find that HapMap SNPs are strongly correlated with 82% of common deletions, but the newest SNP platforms effectively tag about 50%. We conclude that currently available genome-wide SNP assays can capture CNVs accurately, but improvements in array designs, particularly in duplicated sequences, are necessary to facilitate more comprehensive analyses of genomic variation.
Introduction
Copy-number variants (CNVs) occur commonly in the human genome1-4, often affect genes, contribute to genomic evolution and genetic diversity (for a review, see 5), and influence a number of human traits 6-10. Considering this observations, it is likely that future genetic studies would benefit from analyzing CNVs in addition to SNPs11. However, relative to SNPs, CNVs are a priori likely to have larger phenotypic effects5, and the mutation rate generating them in some regions of the genome is substantially higher12. Consequently, an important technological goal is the development of a platform capable of discovering rare CNVs in addition to genotyping common variants, which are related but distinct challenges. One promising solution is to leverage commercially available genome-wide SNP platforms, which have been and will continue to be widely applied in association studies 13. These assays can indirectly interrogate CNVs via linkage disequilibrium (LD) 3,14-16 and directly quantify copy-number for some variants17-20.
Due to an absence of high-resolution, independently generated maps of variation, the extent to which commercial SNP platforms accurately capture CNVs remains largely unknown. To address this, we leverage genome-wide fosmid end sequence pair (ESP) maps recently developed for nine humans2,4. We find that even newer platforms miss a large fraction of the CNVs present in any given individual. However, using a Hidden Markov Model (HMM) approach we show that many CNVs can be discovered within a given sample and systematically validated. We also develop a novel algorithm, known as SNP-Conditional Mixture Modeling (SCIMM), to robustly genotype common variants directly in large collections of individuals and evaluate their correlations with neighboring SNPs. Our results have implications for retrospective analysis of existing genome-wide SNP data as well as future assay designs.
Results
Probe coverage and SNP-platform comparisons
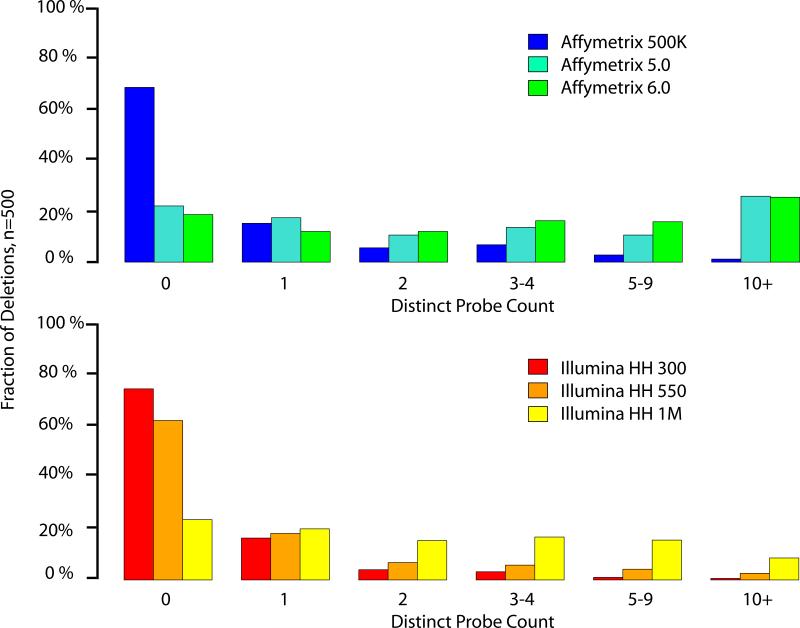
We first assessed the probe coverage for commonly used SNP arrays within variants identified systematically in nine human genomes by fosmid ESP mapping and validated by orthogonal approaches4. Using breakpoints inferred from high-density oligonucleotide array-comparative genomic hybridization (CGH) experiments for 500 deletions larger than 1 kb, we find that older genome-wide platforms (Illumina HumanHap 300 and Affymetrix 500K) lack probes within ∼75% of deletions, and fewer than 20% harbor multiple probes (Fig. 1). Newer arrays (Illumina Human 1M and Affymetrix 6.0) show improved coverage, but ∼20% of deletions harbor zero probes and most span fewer than 5. We obtain similar results when we consider deletions annotated by complete fosmid sequencing and alignment to the reference assembly, with ∼30% missed even on newer platforms (Supplementary Table 1).
Figure 1.
Probe coverage histogram for 500 non-redundant deletion events greater than 1 kb in size discovered in nine human samples by fosmid ESP placements and refined using oligonucleotide array-CGH experiments4. We analyzed three array platforms available from Affymetrix (top) and Illumina (bottom). ‘Distinct’ probes correspond to each distinct location in the genome (hg17) physically represented on the array and internal to the annotated deletion breakpoints (physically redundant probes for a given location are not counted).
CNV discovery and validation
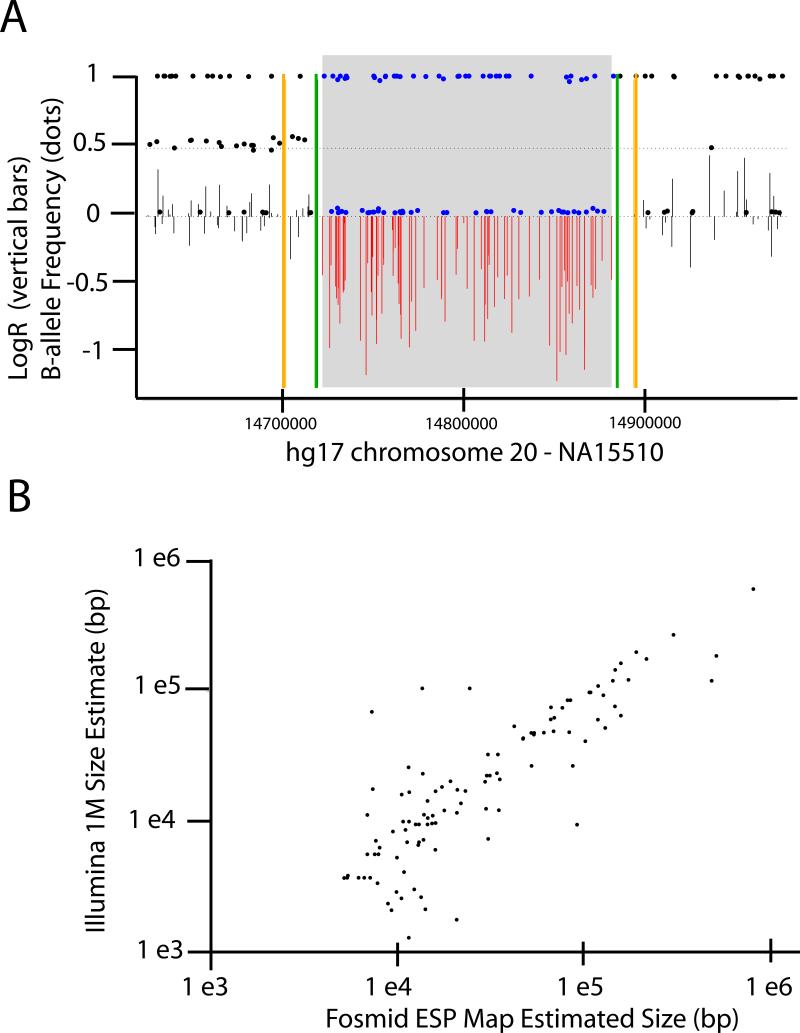
To discover CNVs within a given sample using Illumina Infinium 17 data, we applied a simple HMM-based approach using HMMSeg21 (see Methods). The procedure simultaneously analyzes both the normalized total intensity (‘LogR Ratio’) and allelic intensity ratios (‘B-allele Frequency’)17 to detect regions of homozygous deletion, hemizygous deletion, or amplification. In the nine samples for which a fosmid library is available4 we identified a total of 368 events greater than 1 kb in length (258 deletions,110 amplifications; Supplementary Table 2). We find that 116 of 258 (∼45%) predicted deletions overlap a deletion discovered by fosmid ESP mapping, with a strong correlation in estimated sizes (R2 = 0.79; Fig. 2). We observe that a substantial albeit smaller fraction of the inferred amplifications map to previously defined insertion events (15 of 110 amplifications). The vast majority of the non-validated amplifications are large (81% are > 40 kb) and heavily enriched for segmental duplications in the reference assembly (72% of events, 70% of nucleotides), reflecting the known enrichment for CNVs in duplication-rich regions of the reference assembly1,3,22. ESP mapping, however, has reduced sensitivity to both large insertions and variants within duplication-rich sequence2; thus, the lower rate of validation for amplification events is expected.
Figure 2.
Deletion predictions validated by fosmid ESP placement data. a. Example of a deletion event inferred using Illumina Human 1M data. Intensity data for all probes in the indicated genomic interval (X-axis) for sample NA15510 (aka ‘G248’) are plotted. ‘LogR Ratio’ and ‘B-allele Frequency’ 17 are plotted as vertical bars and filled dots, respectively. The gray box indicates the deletion span inferred by segmentation of the SNP data; probes internal to this box are colored red (‘LogR Ratio’) or blue (‘B-allele Frequency’). Green vertical bars indicate the deletion borders defined by complete fosmid re-sequencing. b. Correlation in size estimates between deletions inferred from SNP genotyping data (Y-axis) that overlap deletions annotated by fosmid ESP mapping (X-axis). Both axes are log-scaled.
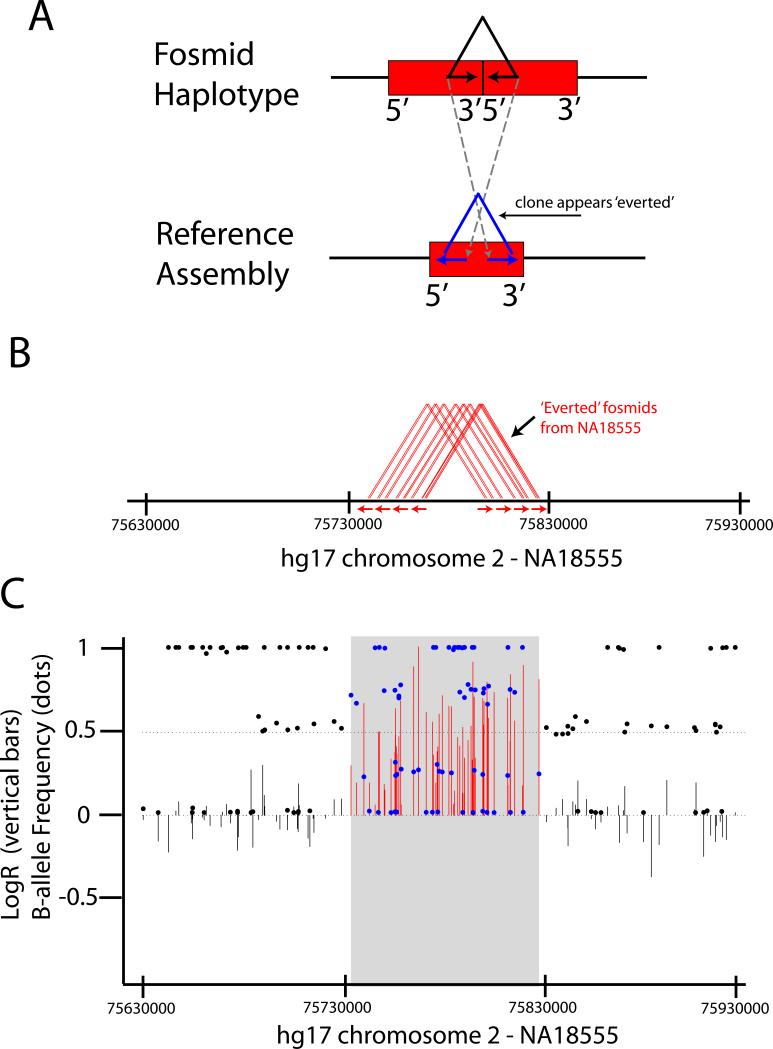
We subsequently leveraged the entire fosmid ESP maps available for these samples (∼900,000 clones per individual; http://hgsv.washington.edu) to determine if the non-validated predictions are false positives or previously missed variants. Many of the deletions inferred here were below the size thresholds used previously4, and by relaxing thresholds we find support for 18 (∼7%) additional deletions (Supplementary Table 3). We also sought to overcome the inability to validate large amplifications through fosmid ESP placements using two approaches. First, we hypothesized that if a sequence unique in the reference assembly is tandemly duplicated in a given sample, then a clone that spans the duplication breakpoint will align to the reference genome such that the reads orient away from the center of the clone (Fig. 3). We find that an additional 18 (16%) amplifications inferred using SNP data overlap a cluster of such clones. Second, we considered variants annotated within eight of these nine samples by a combination of SNP array and BAC-CGH analysis3, the latter of which has better power to detect large variants in duplication-rich regions. We find that an additional 25 (23%) inferred amplification events overlap a previously defined ‘gain’ within the same sample. Combining these and other analyses of fosmid ESP placements (Supplementary Table 4,5, Supplementary Figure 1), we conclude that at least 67% and 64% of the inferred deletion and amplification events, respectively, correspond to experimentally validated variants. The actual true positive rate is likely higher than this (Supplementary Table 4).
Figure 3.
Amplification events validated by clusters of ‘everted’ fosmid ESP placements. a. Consider a block of sequence (red bar) that is unique in the reference assembly (bottom portion) but tandemly duplicated in the haplotype of interest (top portion). In principle, for clones that span the breakpoint of this duplication, the end sequences will be inverted when aligned to the reference assembly such that they are oriented away from the center of the clone. Note that this should occur at all such duplication breakpoints, regardless of duplication size. b. We identified 233 sites in the nine fosmid libraries harboring multiple overlapping ‘everted’ clones, one of which is shown here. This site is supported by eight distinct clones (red triangles), each of which has reads oriented outwards as indicated by the underlying red arrows. c. Illumina Human 1M data for the same region in the same sample (X-axes are identical) is shown, with LogR and B-allele Frequency plotted as vertical bars and dots, respectively (similar to Figure 2A). The gray box corresponds to the duplication interval inferred by segmentation of the SNP data.
Genome-wide CNV discovery must be conducted per-sample to detect rare events and must also account for the low prior probability that any given probe is inside a CNV. Therefore, high specificity was our primary goal. However, we also considered the extent to which known variants in these samples had been missed. Restricting our analysis to defined ‘detectable’ (i.e. spanning enough probes) deletions, we find that our sensitivity is ∼47% (7/12 sequence-defined deletions, 30/66 CGH-defined deletions). We note that many of the deletions that were missed correspond to duplication-rich sites: ∼67% of the nucleotides within the false negatives are within a segmental duplication, representing a 13-fold enrichment over the genomic average 23. Thus, most of the missing deletions correspond to sequences present in multiple copies in the reference assembly.
Targeted genotyping
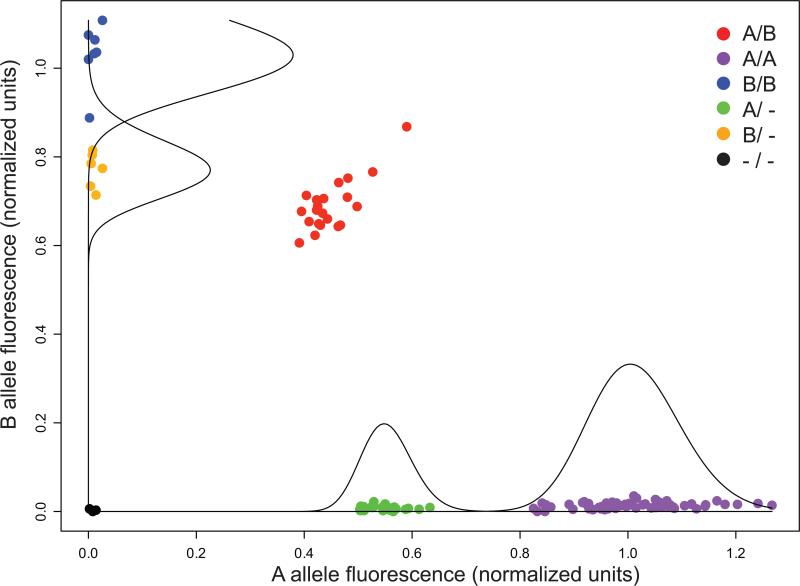
In contrast to discovery, targeted genotyping can leverage the knowledge that a CNV exists at a particular location and, for common variants, borrow information across samples, reducing the number of probes required for analysis. We therefore implemented a strategy, denoted SCIMM (SNP-Conditional Mixture Modeling), for genotyping polymorphic insertion/deletion variants spanning as few as 2 probes. SCIMM employs mixture-likelihood based clustering24, motivated by the observation that hemizygous or homozygously deleted samples often manifest as distinct clusters in the fluorescence intensity data for SNP probes inside common deletions (Fig. 4; see Methods). A second algorithm, SCIMM-Search, identifies copy-number informative probes within known deletions (Supplementary Methods).
Figure 4.
Example of fluorescence intensity measurements for each of 126 samples for a single SNP probe (rs10076425). These data are used by SCIMM (SNP-Conditional Mixture Modeling) to automatically determine insertion/deletion genotypes (see Methods). The genotype for each sample is denoted by color as indicated in the legend. Mixture component distributions are represented by superimposed curves on both the X and Y-axes. In this case, insertion/deletion status is computed by analyzing this probe in conjunction with four additional probes within a deleted region at chr4:10070382−10076653 (Supplementary Data S3).
To validate this approach, we analyzed data generated by the Illumina Human 1M assay for 126 samples (125 HapMap samples plus NA15510; see Methods), including 28 parent-child trios. We compared insertion/deletion genotypes produced by our algorithm for 18 common, autosomal deletions that have been independently genotyped using quantitative PCR and GoldenGate fluorescence data4,25 (Supplementary Table 6,7). SCIMM-Search identified informative probe sets for 13 of these sites. SCIMM generated genotypes with correlation (r2) to the reference genotypes exceeding 80% at all sites and > 97% genotype concordance at 12 sites (Supplementary Table 6).
We subsequently applied SCIMM-Search to 252 non-overlapping, independently defined autosomal deletions spanning two or more probes on the Illumina Human 1M array, identifying informative probe sets for 136 of these sites (Supplementary Fig. 2; Supplementary Table 8). Of the 130 sites passing subsequent manual review, 126 are polymorphic (allele frequency >1%), with only six Mendelian inconsistencies across the 3,640 trio offspring genotypes (Supplementary Tables 9,10). We also applied SCIMM to data produced for 120 HapMap samples using the Illumina HumanHap 550 assay. We find that deletions spanned by 2 or more HumanHap 550 probes yield highly concordant genotypes (99.8% identical). This demonstrates high technical reproducibility and the applicability of reduced probe sets to lower-density data. However, single probe genotypes were more prone to discordancy (Supplementary Fig. 3), indicating that multiple probes are required for accurate genotypes.
Previous studies have evaluated LD between SNPs and insertion/deletion polymorphisms, suggesting that associations between CNVs and phenotypes may be detected through correlation with SNP genotypes3,15,16. We searched for tagSNPs for each of the successfully genotyped deletion polymorphisms (126 total) using SNP genotype data from Phase II HapMap data 26 and from four genome-wide SNP data sets (Table 1). We found that 82% (69/84) of the common deletions (worldwide frequency > 5%) were strongly correlated to a HapMap SNP (worldwide r2 > 0.8); in contrast, each high-density genome-wide SNP platform effectively tagged only about half (48%-54%) of the common deletions (Supplementary Table 8) by the same criterion.
Table 1.
Pairwise correlation estimates between SNP and deletion genotypes.
| SNP Data Set | Fraction of sites with at least one tagSNP, r2>0.8 | Fraction of sites with at least one tagSNP, r2>0.7 | Mean (max r2), all sites |
|---|---|---|---|
| HapMap 2.0 | 82% | 88% | 0.88 |
| Illumina Human 1M | 54% | 70% | 0.77 |
| Affymetrix 6.0 | 51% | 65% | 0.73 |
| Illumina HumanHap 650Y | 48% | 64% | 0.74 |
| Illumina HumanHap 550 | 48% | 61% | 0.71 |
Discussion
While it has previously been shown that SNP-array data can be used to infer the presence of intermediate size CNVs17-20, the reliability of the resulting annotations has never been systematically validated. Exploiting data from the Illumina Human 1M BeadArray, we accurately predicted the identity and size of 368 intermediate-size CNVs in nine samples, at least two-thirds of which were validated with independent experimental data. Our validation expands previous uses of fosmid ESP mapping information and includes a novel technique to confirm the presence of large duplication events. This technique circumvents a previously recognized limitation of the approach (inability to identify large insertions) and may prove useful in future applications of high-throughput clone ESP data27,28.
We also developed a novel genotyping algorithm that accurately infers genotypes for polymorphic deletions using as few as two probes. We found that 82% of the genotyped sites can be tagged by SNPs near the CNV; however, the best available platforms only tag ∼50% (Table 1). We note that the set of deletion events that we successfully genotyped is not a random sample. In particular, segmental duplications in the reference assembly are under-represented on all genome-wide SNP platforms (not shown), and even when probes are present, cross-hybridization of paralogous sequences can confound deletion genotyping (Supplementary Methods). However, the mutation rate for events in and around clusters of segmental duplications is substantially higher than the background mutation rate8,12. Thus, the regions that might be prone to recurrent deletion generation, and would in turn not be in strong LD with neighboring variants, are probably enriched in the set of events for which we did not obtain genotypes. Our estimate that 18% of common deletion variants are not strongly correlated with any known SNP is thus likely to be a lower-bound estimate, and underscores the need for independent experimental information to evaluate CNV detection 11,27.
Direct interrogation of copy number variation by genome-wide SNP platforms is limited by probe coverage. Even on the newest genome-wide SNP platforms, at least 20% of all intermediate-size deletion events span zero, and most fewer than 5, probes (Fig. 1). Older array designs have particularly poor coverage and retrospective mining of these data are likely to be of limited utility. This finding is in contrast to the higher coverage estimates one would obtain with lower resolution, and generally inflated, CNV annotations, such as those derived from BAC-CGH experiments 4,29 (Supplementary Figure 4). We note that differences in assay chemistry, probe specificity, and physical redundancy strongly influence dynamic range; thus, probe count alone does not provide sufficient information for comparison of different platforms.
We show that SNP arrays can be used to infer the presence of many individually rare CNVs with reasonable specificity given a considerable probe count, and can furthermore be used to robustly genotype common deletions using as few as two probes. However, when considering balanced events (eg. inversions), novel insertion sequences not represented in the reference assembly4, and the bias against segmental duplications in array designs contrasted with the enrichment for CNVs both within and flanking duplicated sequences1,3,22, we conclude that a large fraction of genomic variation cannot be captured by existing genome-wide SNP platforms. Significant improvements to array designs, perhaps in the form of a targeted CNV genotyping platform, may ultimately be necessary. In any case, it will be important to continue to benchmark such efforts against high-resolution, ultimately sequence-based, maps of variation to accurately assess both successes and failures. These analyses should lay the framework for more comprehensive assessments of human genomic variation.
Methods
Genome-wide SNP genotyping
We obtained SNP genotyping data generated by the Illumina Human 1M and HumanHap 550K platforms directly from Illumina (courtesy of Dan Peiffer, data available via techsupport@illumina.com). The 1M data include within-sample normalized fluorescence (“x” and “y”), between-sample normalized fluorescence (“Log R ratio” and “B-allele frequency”), and SNP calls for 125 HapMap samples (Supplementary Methods), including eight samples for which fosmid libraries have been generated4. We supplemented the genotyping data with one additional sample, NA15510 (also known as ‘G248’), that was previously analyzed by fosmid ESP analysis2. SNP genotyping was done in accordance with manufacturer's protocols17. Note that only 120 HapMap samples (a subset of the 126 described above) were available on the Illumina HumanHap 550K array.
CNV discovery using Illumina Human 1M genotyping data
Large CNV discovery was accomplished by using HMMSeg21, considering both the “LogR Ratio” and “B-allele Frequency” data for each sample simultaneously, based essentially on a previously established approach17. We used a four-state model, one each for null (homozygous deletion), hemizygous deletion, diploid, and amplification. Initial segmentation results were merged and filtered, requiring all variants to be larger than 1 kb in length and to span at least 10 probes for amplifications or hemizygous deletions, or three probes for homozygous deletions. Additional details on data normalization, model specifications, and implementation can be found in Supplementary Methods.
CNV validation using whole-genome fosmid ESP placement analysis
We validated our CNV predictions by comparing their locations and sizes with the locations and sizes of variants that had been previously annotated by analysis of fosmid ESP placements for the same nine individuals; all these data are available in the supplementary information of Kidd et al4 and at http://hgsv.washington.edu. We considered any amount of overlap between the CNV maps as validation, given the restriction that the event is in the same sample and in the same direction (ie. only ESP deletions are used to validate predicted deletions). We performed additional validation by considering all fosmid ESP placement information, borrowing information across samples, and leveraging information on clone placements that were previously excluded as a result of size, alignment score, or other quality-control thresholds (Supplementary Methods).
Probe coverage analysis
Genomic locations of SNP probes were obtained from http://www.affymetrix.com and http://www.illumina.com for the SNP genotyping products provided by the respective companies. We mapped coordinates as appropriate from hg18 to hg17 using the ‘Liftover’ tool at http://genome.ucsc.edu. Locations of CNVs identified through analysis of fosmid ESP mapping were obtained directly from the supplementary data provided in Kidd et al4. An overview of the data sets used here can be found in Supplementary Methods.
Insertion-deletion genotyping
SCIMM is a clustering algorithm which, given a set of probes, produces a classification of each sample as ‘null’, ‘haploid’ or ‘diploid’. Two rounds of mixture-likelihood based clustering implemented by the Expectation Maximization algorithm24 are used; the first operates on per-sample summary intensity values to identify null samples, and the second operates directly on two-channel fluorescence data to classify the remaining samples as either ‘haploid’ or ‘diploid’. SNP genotypes are used for direct inference of copy number (heterozygosity is used as evidence of diploidy) and for model fitting. During the second round of clustering, a single-component (copy-number-invariant) model is also fit to the data to produce a score for the probe set using the Bayesian Information Criterion (BIC)30.
SCIMM-Search is an iterative search algorithm which, given the coordinates of a region spanning a deletion and the identity of a sample known to carry the variant, determines the set of probes used to genotype the deletion variant carried by the reference sample. SCIMM-Search uses the BIC to evaluate alternate probe sets. It is not assumed that all probes within the annotated region are informative for copy number or that informative probes are contiguous. SCIMM-Search allows specification of constraints on genotype consistency between probes and cluster separation for each probe in the probe set. A more comprehensive description along with detailed model specifications and thresholds for both SCIMM and SCIMM-Search are available in Supplementary Methods.
Taggability
For each polymorphic insertion/deletion site, we extracted all phase II HapMap SNP genotypes within 200kb of the deletion interval and calculated r2 between each SNP and the SCIMM-generated insertion/deletion samples for 90 unrelated HapMap individuals (Supplementary Data S3). We combined data across populations to obtain a single estimate of correlation (within-population correlation estimates were similar; data not shown) and ignored sites with calls for fewer than 75% of the unrelated samples. We repeated this process for the Affymetrix 6.0, Illumina Human 1M, Illumina 650Y and Illumina HumanHap 550 assays, using SNP genotypes provided by the manufacturers of each assay.
Supplementary Material
Acknowledgments
We thank Dan Peiffer and colleagues at Illumina for sharing Human 1M and HumanHap 550K genotyping data. GMC is supported by a Merck, Jane Coffin Childs Memorial Fund Postdoctoral Fellowship. TRZ acknowledges support from the National Human Genome Research Institute (NHGRI) Interdisciplinary Training in Genomic Sciences grant T32 HG00035. JMK is supported by a NSF graduate fellowship. This work was supported by the National Heart, Lung, and Blood Institute Programs for Genomic Applications grant HL066682 to DAN and NHGRI grant HG004120 to EEE. EEE is an investigator of the Howard Hughes Medical Institute.
References
- 1.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–8. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 2.Tuzun E, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–32. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 3.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper GM, Nickerson DA, Eichler EE. Mutational and selective effects on copy-number variants in the human genome. Nat Genet. 2007;39:S22–9. doi: 10.1038/ng2054. [DOI] [PubMed] [Google Scholar]
- 6.Singleton AB, et al. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez E, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–40. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 8.Sharp AJ, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–42. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 9.Perry GH, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–60. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–43. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 11.Estivill X, Armengol L. Copy number variants and common disorders: filling the gaps and exploring complexity in genome-wide association studies. PLoS Genet. 2007;3:1787–99. doi: 10.1371/journal.pgen.0030190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaffer LG, Lupski JR. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- 13.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 15.Locke DP, et al. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am J Hum Genet. 2006;79:275–90. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCarroll SA, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 17.Peiffer DA, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–48. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komura D, et al. Genome-wide detection of human copy number variations using high-density DNA oligonucleotide arrays. Genome Res. 2006;16:1575–84. doi: 10.1101/gr.5629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colella S, et al. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–25. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–74. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Day N, Hemmaplardh A, Thurman RE, Stamatoyannopoulos JA, Noble WS. Unsupervised segmentation of continuous genomic data. Bioinformatics. 2007;23:1424–6. doi: 10.1093/bioinformatics/btm096. [DOI] [PubMed] [Google Scholar]
- 22.Sharp AJ, et al. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.She X, et al. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 2004;431:927–30. doi: 10.1038/nature03062. [DOI] [PubMed] [Google Scholar]
- 24.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. (Series B).Journal of the Royal Statistical Society. 1977;39:1–38. [Google Scholar]
- 25.Newman TL, et al. High-throughput genotyping of intermediate-size structural variation. Hum Mol Genet. 2006;15:1159–67. doi: 10.1093/hmg/ddl031. [DOI] [PubMed] [Google Scholar]
- 26.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eichler EE, et al. Completing the map of human genetic variation. Nature. 2007;447:161–5. doi: 10.1038/447161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–6. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Smith AJ, et al. Array CGH analysis of copy number variation identifies 1284 new genes variant in healthy white males: implications for association studies of complex diseases. Hum Mol Genet. 2007;16:2783–94. doi: 10.1093/hmg/ddm208. [DOI] [PubMed] [Google Scholar]
- 30.Schwarz G. Estimating the dimension of a model. Annals of Statistics. 1978;6:461–464. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.