Abstract
Histone deacetylases are intimately involved in the transcriptional regulation of genes, and they are high priority drug targets for cancer therapy. Due to prevalence of several sulfhydryl groups on the surface of histone deacetylase 8, we explored the possibility of its binding to colloidal gold nanoparticles by determining its potentials to inhibit the flocculation as well as retaining the enzyme activity. It was observed that although both these processes conformed to the binding affinity of the gold-histone deacetylase 8 conjugate as being equal to 15–20 nM, only 30% of the nanoparticle-bound enzyme exhibited the enzymatic activity. In the light of the structural features of histone deacetylase 8, we propose that the enzyme interacts with the gold nanoparticles via the surface exposed thiol groups, and such interaction occurs in two alternative modes. Whereas the enzyme bound via mode-1 is catalytically inactive (presumably due to the orientation of the enzyme’s active site toward the gold nanoparticle surface), and it prevents the flocculation of the nanoparticles, the enzyme bound via mode-2 shows the full catalytic activity (as its active site is believed to be oriented away from the nanoparticle surface). Although the histone deacetylase 8 bound to AuNP via mode-2 exhibits the same inhibitory potency against Trichostatin A as the free enzyme, the former is more susceptible to thermal denaturation. The potential of potent interaction between gold nanoparticles and histone deacetylase 8 via alternative modes may find diagnostic and/or therapeutic applications for different forms of cancers.
Keywords: Histone Deacetylase, Gold Nanoparticles, Binding, Cancer
1. Introduction
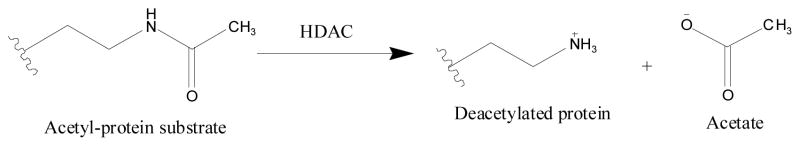
Histone Deacetylases (HDACs) are a family of hydrolases, which catalyze the deacetylation of ε-acetyl lysine residues of histones, and serve as an important mechanistic role in regulating gene transcription 1, 2 (Scheme 1).
Scheme 1.
Aside from histones, this family of enzymes cleaves numerous non-histone substrates and plays key roles in cell cycle control, apoptosis and transcriptional regulation 3. HDACs have been found to be over-expressed in different tumor cells4. Due to their strong influence in regulation of gene expression, these enzymes have been linked to both cancer initiation and progression and thus have great therapeutic potential 5. Toward this goal, several HDAC inhibitors have been synthesized and reported to affect cell growth, differentiation and apoptosis in malignant cell 6, 7. Recently, an HDAC8 inhibitor, suberoylanilide hydroxamic acid (SAHA), has been approved by FDA for the treatment of cutaneous T-cell lymphoma, and several other inhibitors are in clinical trials for treatment of various forms of cancer 8.
Based on phylogeny, human HDACs have been classified into three classes 9. HDAC 1–3 and 8 belong to Class I whereas HDAC 4–7, 9 and 10 belong to class II. Class III HDACs, also known as Sirtuins, are unrelated enzymes since they require NAD+ for deacetylase activity. With the exception of HDAC8, most HDACs exist as high molecular weight multi-protein complexes and are found to be catalytically inactive when expressed in heterologous host 10, 11. Of the different HDACs, HDAC8 is the only enzyme which is efficiently expressed in E.Coli host cells 12, and thus has been widely studied in different laboratories, including our own lab, from structural-functional and mechanistic point of views. 8, 9, 13
The X-crystallographic data of HDAC8 shows the presence of ten cysteine residues and nine methionine residues of which three cysteine and four methionine residues are exposed on the protein surface and others are partially buried in the protein mass13. To our interest, whereas Cys 275 & Cys 352 are confined toward the active site region, Cys 125 is present on the opposite end of the protein surface. In addition there are four surface exposed methionine residues in HDAC8 of which Met 27, Met 64 and Met 130 are present on the opposite surface on the enzyme. Due to the presence of surface exposed Cys and Met residues in HDAC8, it had potential to interact with colloidal gold nanoparticles (AuNP), and our interest in this enzyme partly emerged due to this fact.
In this paper, we demonstrate a fairly strong interaction of HDAC8 with AuNP, and that such interaction exposes the enzyme in two different orientations. The mechanistic features intrinsic to such interactions are highlighted in the following sections.
2. MATERIALS AND METHODS
2.1 Materials
Gold colloid solution of 10 nm diameter and Trichostatin A (TSA) were purchased from Sigma (Saint Louis, MO). HDAC assay kit (Fluor de Lys™ assay system) was purchased from BIOMOL International L.P. (Plymouth Meeting, PA), and the enzyme was assayed as described by the supplier. Bradford dye reagent was purchased from Bio-Rad Laboratories Inc. (Hercules, CA).
2.2 Methods
2.2.1 HDAC8 expression and purification
Human HDAC8 was cloned, expressed and purified in our laboratory according to Hu et al. 12 A detailed account of above protocols will be published elsewhere. The concentration of the purified HDAC8 was determined by the Bradford method 14.
2.2.2 Stabilization of AuNP by HDAC8
The minimum amount of HDAC8 required to stabilize AuNP (against flocculation) was determined according to Geoghegan and Ackerman 15. A series of HDAC8 solutions of increasing concentration was prepared and 10 μl of each solution was added to 90 μl AuNP solution in a 96 well microtiter plate and mixed. After 5 minutes, 10 μl of 10% NaCl was added, mixed and allowed to stand for 5 minutes. Following this, absorbance of the mixture was recorded at 580 nm and plotted as a function of HDAC8 concentration. The data was analyzed by non-linear regression using Grafit (Erithacus Software Inc. UK) software with a modified quadratic function as described by Qin and Srivastava 16.
2.2.3 Preparation of AuNP-HDAC8 conjugate
HDAC8 solution (10 μl, 8 μM) was added to 90 μl of colloidal gold solution and incubated on ice for 5 minutes with intermittent shaking. A 10 μl solution of 0.1% PEG (M.W. 16,000) was added to prevent the aggregation of AuNP 17. The solution was centrifuged at 20,000 rpm for 20 min at 4°C, and the supernatant was separated. Both supernatant and pellet were assayed for HDAC8 activity to confirm the presence of active enzyme and the amount of AuNP bound HDAC8 was quantitated.
2.2.4 Binding of enzymatically active HDAC8 to AuNP
The binding constant and stoichiometry of AuNP-HDAC8 conjugate was determined by measuring the increase in the enzyme activity upon titration of the enzyme to AuNP, essentially as employed in the preparation of the bio-conjugate. Solutions of increasing concentration of HDAC8 were prepared and added to 200μl AuNP on ice. After 5–10 minutes, 4 μl of 1% PEG was added and the solutions were centrifuged at 20,000 rpm for 20 min at 4°C. The supernatant was separated and both pellet and supernatant were assayed for HDAC8 activity. The enzyme activity was plotted against HDAC8 concentration and the data was analyzed as described by earlier to obtain the stoichiometry and binding constant.
2.2.5 Inhibition of free and AuNP-conjugated HDAC8 by TSA
AuNP-HDAC8 conjugate was prepared using 3 μM HDAC8 as described in the previous section and all pellets were pooled together. HDAC8 concentration in the resulting conjugate was estimated by the Bradford method. Two sets of assays were carried out (i.e., for free HDAC8 and AuNP-HDAC8 conjugate), each containing a series of increasing concentrations of TSA in the assay mixture. Enzyme activity was plotted as a function of TSA concentration and the data were analyzed using quadratic function for the competitive inhibition model as described by Banerjee et al. 18.
2.2.6 Thermal stability of free and AuNP-HDAC8 conjugate
Effect of AuNP on the stability of HDAC8 was probed by measuring the rate of inactivation of free versus AuNP-bound HDAC8 at 40°C. AuNP-HDAC8 conjugate was prepared using 3 μM HDAC8 and centrifuged as described earlier. All AuNP-HDAC8 conjugate pellets were pooled together and divided into 20 μl aliquots in microcentrifuge tubes. These aliquots were incubated at 40°C for varying periods of time using a water bath and immediately transferred on to ice after incubation. Aliquots of free HDAC8 were similarly treated. All aliquots were then assayed for HDAC8 activity, and the enzyme activity was plotted as a function of incubation time. The data were analyzed by exponential function and the rate constants were calculated for each set using Grafit software.
3. RESULTS
To ascertain whether HDAC8 was able to bind and stabilize colloidal gold nanoparticles (AuNP), we added HDAC8 to the nanoparticle solution and monitored it’s flocculation by NaCl. The latter was ascertained by monitoring change in the color of AuNP solution from red to blue (absorption maximum = 520 nm). While the control AuNP solution turned blue on addition of NaCl, no change was seen in the gold solution treated with HDAC8. With this observation we then proceeded to determine the binding isotherm for the interaction of HDAC8 to AuNP by following the signal (A580) as a function of increasing amount of the enzyme.
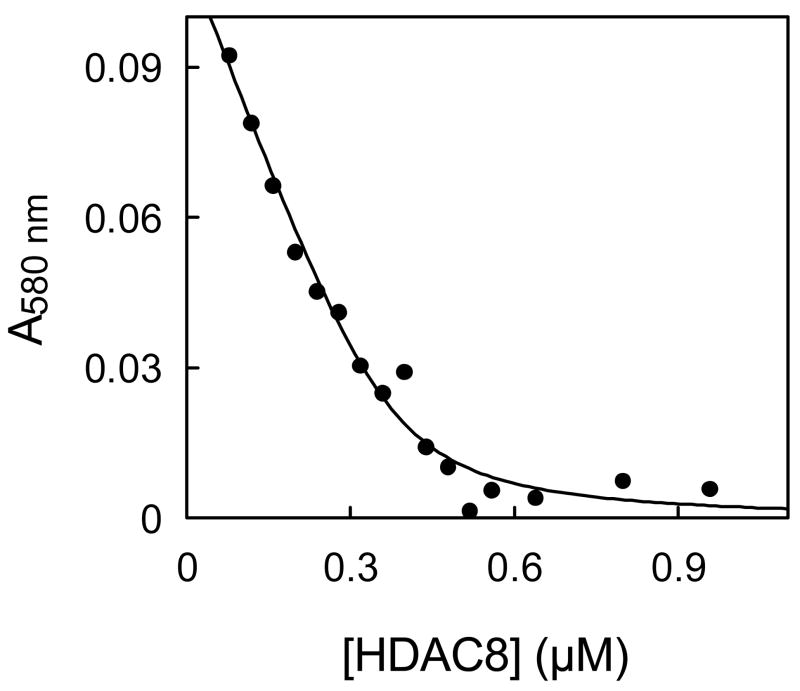
In a typical experiment, we added increasing amount of HDAC8 to AuNP solution and observed the stabilization of the resulting solution by measuring the absorbance of the solution at 580 nm after adding 250 mM NaCl. Flocculation of the nanoparticles results in broadening as well as red shift of the AuNP absorption spectrum. Whereas AuNP solution shows very little absorbance at 580 nm, its flocculated counterpart shows high absorption at this wavelength. Hence the decrease in absorbance at 580 nm was observed with increasing HDAC8 concentration (Fig. 1) indicating an increase in stabilization of AuNP in the presence of HDAC8. The sharp inflection of the A580 versus [HDAC8] plot revealed that the minimum enzyme concentration required to stabilize 10 nm AuNP was 0.5 μM. Since the concentration of AuNP was ~ 10 nM, it appeared that about 50 HDAC8 molecules are required per AuNP to stabilize the colloid AuNP solution. The analysis of data yielded the binding constant of the AuNP-enzyme conjugate to be 20 nM suggesting a very strong interaction between AuNP and HDAC8, and we believe this interaction is mediated via surface exposed thiol groups of the enzyme and the nanoparticle.
Figure 1.
Stabilization of AuNP by HDAC8. The concentration of HDAC8 required to prevent flocculation of AuNP (10 nM) by 250mM NaCl was monitored via measuring the absorption changes at 580 nm. The solid smooth curve represents the best fit of the titration data for the Kd value of 20 nM and the stoichiometry of 45 HDAC8/AuNP
To ascertain whether HDAC8 retained activity on binding to AuNP, we prepared AuNP-HDAC8 conjugate by adding excess HDAC8 to AuNP solution, and centrifuged the mixture to separate the unbound enzyme. The AuNP-HDAC8 conjugate formed a compact pellet on centrifugation which could be separated by aspirating the supernatant. Both pellet and supernatant were assayed for the HDAC8 activity. The experimental data revealed that no measurable enzyme activity was present in the supernatant, but only 30% of the enzyme activity was present in the pellet fraction. The lack of the enzyme activity in the supernatant fraction was further confirmed by the absence of any detectable protein by the Bradford method. Clearly, about 70% of HDAC8 lost activity upon binding with AuNP. We believe the inactive form of the AuNP-bound HDAC8 is due to the orientation of the enzyme’s active site pocket toward the surface of the nanoparticle (i.e., binding via mode-1; see Discussion).
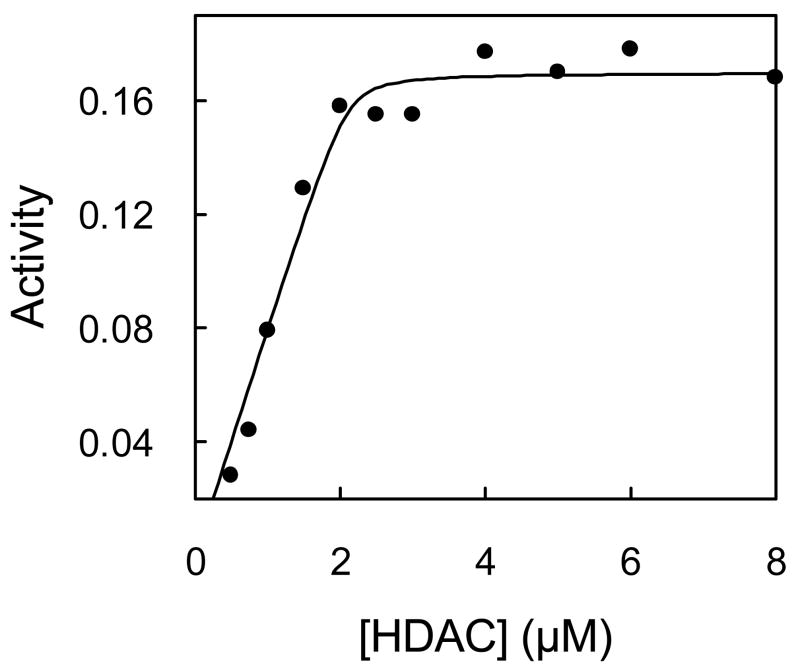
To estimate the capacity of AuNP for the active form of HDAC8, we prepared AuNP-HDAC8 conjugate with increasing concentrations of the enzyme and measured the activity of the bound enzyme preparations. As shown in Fig. 2, the activity of the enzyme increased as a function of HDAC8 concentration, and attained saturation at ~ 2μM of enzyme. At concentrations above 2 μM, the HDAC8 activity started appearing in the supernatant fraction as well, and it increased linearly as a function of the enzyme concentration. The non-linear regression analysis of the data gave a binding constant of the active form of the AuNP-HDAC8 conjugate as being equal to 15 nM and a stoichiometry of 40 active HDAC8 per AuNP.
Figure 2.
Binding Isotherm of the active form of HDAC8 to AuNP. AuNP-HDAC8 conjugates were prepared with increasing HDAC8 concentrations and activity of the conjugate was plotted as a function of the enzyme concentration. The line is the best fit of the data for the Kd value of 15 nM and the stoichiometry of 42 HDAC8/AuNP.
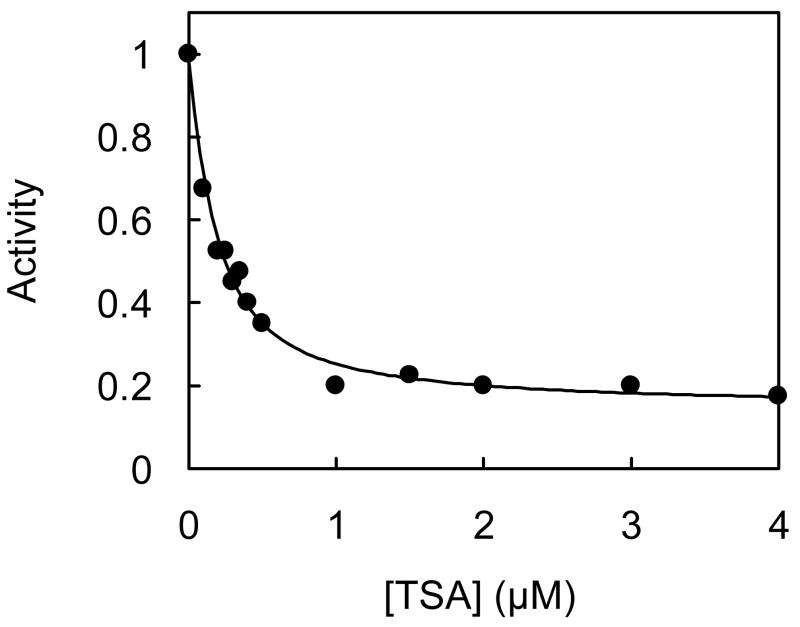
We determined the effect of AuNP-conjugation of HDAC8 on inhibition by a highly potent inhibitor, TSA. To ascertain this, we prepared AuNP-HDAC8 conjugate and assayed its enzyme activity in the presence of varying concentrations of TSA. Similar assays were performed with free HDAC8. The data was analyzed by competitive inhibition model using the non-linear regression analysis (Fig. 3). The experimental data revealed that both free and AuNP-conjugated HDAC8 showed similar Ki values of ~130 nM. Hence, the HDAC8 bound in the active form (i.e., via mode-2; see Discussion) to AuNP has the identical conformational state (as regards to its ability to interact with TSA) as the free enzyme.
Figure 3.
Inhibition of AuNP-HDAC8 by TSA. AuNP-HDAC8 conjugate was prepared with 3 μM HDAC8 and assayed for enzyme activity in the presence of varying concentrations of TSA. Activity was plotted as a function of TSA concentration. The fitted curve represents the best fit of the data for competitive inhibition model with Ki = 127 ± 18 nM. Ki for inhibition of free HDAC was similarly determined to be 131 ± 15 nM, in agreement with the reported value 19.
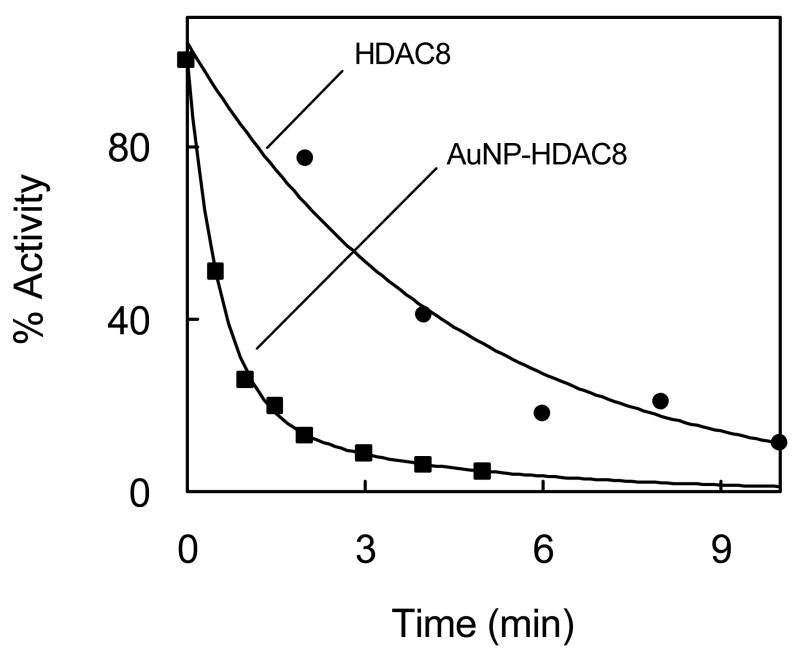
We investigated the effect of AuNP on stabilization/destabilization of HDAC8. This was ascertained by the thermal denaturation of HDAC8 at 40°C in the absence and presence of AuNP. AuNP-HDAC8 conjugate was prepared using saturating concentration of HDAC8 and separated from any unbound HDAC8 by centrifugation. The pellet containing AuNP-HDAC8 conjugate, as well as free HDAC8, were incubated at 40°C for varying periods of time, and then assayed for the enzyme activity. Free and AuNP-conjugated HDAC8 without incubating at high temperature were used as controls. Fig. 4 shows the time dependant loss in activity of HDAC8 under the above conditions. Note that AuNP conjugated HDAC8 is seen to lose activity at a much faster rate than free HDAC8. The data were analyzed by single and biphasic kinetic equations. The analytical outcome revealed that the free HDAC8 lost the enzyme activity with a single exponential decay rate of 0.2/min, whereas the AuNP-HDAC8 lost the enzyme activity in a double exponential manner with fast and slow rate constants of 1.2/min and 0.3/min respectively. We believe the biphasic profile of AuNP bound HDAC8 is due to microheterogeneity in the enzyme population.
Figure 4.
Thermal denaturation of free and AuNP-bound HDAC8. AuNP-HDAC8 conjugate was prepared as described in the legend of Fig 3. The time dependant change in free and AuNP-bound enzyme activity was determined upon incubation at 40°C. The fitted lines represent the best fit for exponential decay. Data for free HDAC8 gave single exponential decay with a rate constant of 0.2 ± 0.02 min−1 while AuNP-HDAC8 gave bi-exponential decay with rate constants = 1.2 ± 0.2 and 0.3 ± 0.1 min−1 respectively.
Since the HDAC8 assay system involved the use of Trypsin as a coupling enzyme, it appeared plausible that HDAC8 in the AuNP-conjugate was unfolding at 40°C and was slow in reverting back to its folded state, resulting in a facile degradation of the enzyme by Trypsin. To probe this, the above experiment was repeated, and after the heat treatment, the aliquots were kept on ice for 30 minutes before assaying for enzyme activity. This experimental approach produced the same results as the original experiment.
4. Discussion
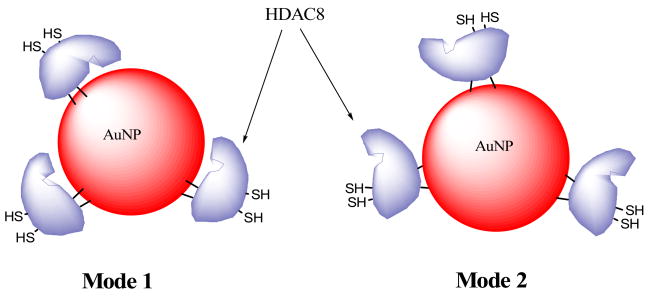
We report herein, for the first time, that HDAC8 strongly interacts with AuNP, and such interaction involves orientation of the enzyme in at least two alternative modes. In both these modes, the enzyme interacts with AuNP via thiol/thioether groups. In one mode (mode-1), the active site of HDAC8 appears to be juxtaposed towards the surface of AuNP and thus does not exhibit catalytic activity. Whereas in the other mode (mode-2), the active site of the enzyme is fully exposed to the exterior environment. These alternative modes of AuNP interaction are shown in the cartoon of figure 5.
Figure 5.
Cartoon showing the alternative binding modes of HDAC8 to AuNP. Note that in binding mode-1, the enzyme’s active site is masked due to its orientation toward the nanoparticle, and thus does not show activity. In binding mode-2, the active site is exposed to the exterior environment, yielding the enzyme activity as well as showing similar inhibitory potential against TSA as the free enzyme.
We were able to stabilize AuNP by HDAC8 and prevent flocculation of the AuNP on addition of NaCl. We did not have to remove salt and buffer ions from the protein preparation in order to stabilize AuNP with HDAC8 15. This is presumably due to a very strong (thiol mediated) interaction between AuNP and HDAC8. The strong binding was further evident, as the addition of reducing agent or excess BSA was unable to displace HDAC8 from the AuNP (data not shown), which is in accord with the data of Sasaki et al.20. The binding isotherm of HDAC8 with AuNP yielded the Kd value of 20 nM, suggesting that the stabilization of AuNP by HDAC8 was mediated via strong Au-S interaction with a ΔG° value of about 40 kcal/mol. Both thiols as well as thio-ethers are known to bind strongly to gold surface 21, 22. The presence of several exposed cysteine thiols as well as methionine thio-ether moieties on the surface of the protein make it a particularly attractive candidate for strong binding of HDAC8 to AuNP. However, these exposed residues are uniquely separated on two faces of HDAC8, with Cys 275 & Cys 352 located on the active site face and Cys 125, Met 27, Met 64 and Met 130 located on the opposite face of the protein. This coupled with the fact that about 70% of AuNP bound HDAC8 does not show enzyme activity (although it stabilizes AuNP against NaCl assisted flocculation) and only 30% of AuNP bound HDAC8 remains catalytically active, it is conceivable that HDAC8 binds to AuNP via two alternative modes. Whereas in mode-1, the enzyme’s active site pocket is oriented toward the surface of AuNP such that the accessibility of the substrate is impaired, in mode-2 the active site pocket is fully exposed to the exterior solvent environment and thus the enzyme is not only catalytically active but it is inhibited by TSA with equal potency as the free enzyme (Fig. 5). However, at this time we cannot exclude the possibility that part of HDAC8 bound to AuNP is denatured and the latter form of the enzyme (in binding mode 1) is responsible for preventing the flocculation phenomenon.
It should be emphasized that the activity of the AuNP bound HDAC8 (in mode 2) exhibits similar Ki value for the inhibitor TSA as that obtained with free HDAC8. Apparently AuNP-bound HDAC8 in the above mode has not undergone discernible conformational changes to alter the binding affinity of the inhibitor. Hence the activity of AuNP-HDAC8 can be taken as a measure of the free enzyme activity and this feature may find application in developing diagnostic protocol of the enzyme via nanoparticles.
Although the AuNP-bound HDAC8 (in mode 2) has similar conformational state as free HDAC8, the former is more susceptible to thermal denaturation than the latter. This is contrary to the effects of AuNPs in stabilizing other enzymes against thermal denaturation 23, 24. A 6 fold higher inactivation rate of AuNP-conjugated HDAC8 as compared to the free HDAC8 clearly suggests that the surface of AuNP helps in destabilization the enzyme (at least at higher temperatures). The unfolding of proteins on binding to surfaces has been well documented in the literature 25. The curvature of the nanoparticle surface in combination with its unusually strong interactions is particularly conducive for conformational change and unfolding of the proteins 26–28. Moreover, initial unfolding of a protein on the AuNP surface can be cooperatively enhanced due to increased electrostatic as well as hydrophobic interactions involving the denatured forms of the enzymes. The latter features are the likely cause for the thermal lability of HDAC8 conjugated to AuNP.
In summary, we have demonstrated that the recombinant form of human HDAC8 strongly interacts with AuNP presumably via the surface exposed thiol groups of the enzyme and such interactions occur through two alternative modes. Since HDAC8 is known to interact with a variety of its protein partners in the physiological milieu, it is likely that the enzyme bound via the above alternative modes would be able to identify its cognate partners (both under in vivo and in vitro conditions) due to the nanoparticle-surface assisted augmentation of biomolecular interactions 29. We are currently in the process of delineating the molecular basis of interaction of HDAC8 to AuNP via the two alternative modes, and having succeeded in this endeavor, we will explore the possibility of employing AuNPs for diagnostics as well as therapeutic applications of various forms of cancers.
Acknowledgments
This work was supported by the National Institute of Health grant CA113746.
Footnotes
This research was supported by the NIH grant CA113746
References
- 1.Wu J, GrunStein M. 25 years after the nucleosome model: chromatin modifications. Trends Biochem Sci. 2000;25:619. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- 2.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 3.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatment for cancer. Nature Rev Cancer. 2006;6:38. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 4.Nakagawa M, Oda Y, Eguchi T, Aishima S, Yao T, Hosoi F, Basaki Y, Ono M, Kuwano M, Tanaka M, Tsuneyoshi M. Expression profile of class I histone deacetylases in human cancer tissues. Oncol Report. 2007;18:769. [PubMed] [Google Scholar]
- 5.Glozak MA, Seto E. Histone deacetylase and cancer. Oncogene. 2007;26:5420. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 6.Gallinari P, Di Marco S, Jones P, Pallaoro M, Steinkuhler C. HDACs, histone deacetylation and gene transcription: from molecular biology to cancer therapeutics. Cell Res. 2007;17:195. doi: 10.1038/sj.cr.7310149. [DOI] [PubMed] [Google Scholar]
- 7.Johnstone RW. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nature Rev Drug Discov. 2002;1:287. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- 8.HDAC inhibitors overcome first hurdle. News feature. Nature Biotechnol. 2007;25:17. doi: 10.1038/nbt0107-17. [DOI] [PubMed] [Google Scholar]
- 9.Gregoretti IV, Lee Y, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Bio. 2004;338:31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Sengupta N, Seto E. Regulation of histone deacetylase activities. J Cell Biochem. 2004;93:57. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- 11.Yang XJ, Seto E. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr Opin Genet Dev. 2003;13:143. doi: 10.1016/s0959-437x(03)00015-7. [DOI] [PubMed] [Google Scholar]
- 12.Hu E, Chen Z, Fredrickson T, Zhu Y, Kirkpatrick R, Zhang GF, Johanson K, Sung CM, Liu R, Winkler J. Cloning and characterization of a novel Human class I histone Deacetylase that functions as a transcription repressor. J Biol Chem. 2000;275:15254. doi: 10.1074/jbc.M908988199. [DOI] [PubMed] [Google Scholar]
- 13.Vannini A, Volpari C, Filocamo G, Casavola EC, Brunetti M, Renzoni D, Chakravarty P, Paolini C, Francesco RD, Gallinari P, Steinkiihler C, Marco SD. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc Natl Acad Sci USA. 2004;101:115064. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 15.Geoghegan WD, Ackerman GA. Adsorption of horseradish peroxidase, ovomucoid and anti-immunoglobulin to colloidal gold for the indirect detection of Concanavalin A, wheat germ agglutinin and goat anti-human immunoglobulin G on cell surfaces at the electron microscopic level: A new method, theory and application. J Hist Cyt. 1977;25:1187. doi: 10.1177/25.11.21217. [DOI] [PubMed] [Google Scholar]
- 16.Qin L, Srivastava DK. Energetics of two-step binding of a chromophoric reaction product, trans-3-indoleacryloyl-coa, to medium-chain acyl-coenzyme-a dehydrogenase. Biochemistry. 1998;37:3499. doi: 10.1021/bi9720585. [DOI] [PubMed] [Google Scholar]
- 17.Horisberger M, Rosset J, Bauer H. Colloidal gold granules as markers for cell surface receptors in the scanning electron microscope. Experientia. 1975;31:1147. doi: 10.1007/BF02326761. [DOI] [PubMed] [Google Scholar]
- 18.Banerjee AL, Swanson M, Roy BC, Jia X, Haldar MK, Mallik S, Srivastava DK. Protein surface-assisted enhancement in the binding affinity of an inhibitor for recombinant human carbonic anhydrase-II. J Am Chem Soc. 2004;126:10875. doi: 10.1021/ja047557p. [DOI] [PubMed] [Google Scholar]
- 19.Schultz BE, Misialek S, Wu J, Tang J, Conn MT, Tahilramani R, Wong L. Kinetics and comparative reactivity of human class I and class IIb histone deacetylases. Biochemistry. 2004;43:11083. doi: 10.1021/bi0494471. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki YC, Yasuda K, Suzuki Y, Ishibashi T, Satoh I, Fujiki Y, Ishiwata S. Two dimensional arrangement of a functional protein by cysteine-gold interaction: enzyme activity and characterization of a protein monolayer on a gold substrate. Biophys J. 1997;72:1842. doi: 10.1016/S0006-3495(97)78830-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulman A. Introduction to thin films: From Langmuir-Blodgett films to self assembly. Academic Press; Boston : 1991. [Google Scholar]
- 22.Sun Q, Reddy BV, Marquez M, Jena P, Gonzales C, Wang Q. Theoretical study on gold-coated iron oxide nanostructure: Magnetism and bioselectivity for amino acids. J Phys Chem C. 2007;111:4159. [Google Scholar]
- 23.Li D, He Q, Cui Y, Duan L, Li J. Immobilization of glucose oxidase onto gold nanoparticles with enhanced thermostability. Biochem Biophys Res Comm. 2007;355:488. doi: 10.1016/j.bbrc.2007.01.183. [DOI] [PubMed] [Google Scholar]
- 24.Jiang X, Shang L, Wang Y, Dong S. Cytochrome c superstructure biocomposite nucleated by gold nanoparticle: thermal stability and voltammetric behaviour. 2005;6:3030. doi: 10.1021/bm050345f. [DOI] [PubMed] [Google Scholar]
- 25.Sadana A. Protein Adsorption and Inactivation on Surfaces. Influence of Heterogeneities. Chem Rev. 1992;92:1799. [Google Scholar]
- 26.Mandal HS, Kraatz HB. Effect of the surface Curvature on the Secondary Structure of Peptide Adsorbed on Nanoparticles. J Am Chem Soc. 2007;129:6356. doi: 10.1021/ja0703372. [DOI] [PubMed] [Google Scholar]
- 27.Chah S, Hammond MR, Zare RN. Gold Nanoparticles as a colorimetric Sensor for Protein Conformational Changes. Chem & Biol. 2005;12:323. doi: 10.1016/j.chembiol.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 28.Brewer SH, Glomm WR, Johnson MC, Knag MK, Franzen S. Probing BSA Binding to Citrate-Coated Gold Nanoparticles and Surfaces. Langmuir. 2005;21:9303. doi: 10.1021/la050588t. [DOI] [PubMed] [Google Scholar]
- 29.Lin C, Yeh YC, Yang CY, Chen CL, Chen GF, Chen C, Wu Y. Selective binding of mannose - encapsulated gold nanoparticles to type 1 pili in Escherichia coli. J Am Chem Soc. 2002;124:3508. doi: 10.1021/ja0200903. [DOI] [PubMed] [Google Scholar]