Abstract
The chorioallantoic placenta is a shared derived feature of “placental” mammals essential for the success of eutherian reproduction. Identifying the genes involved in the emergence of the placenta may provide clues for understanding the biology of this organ. Here we identify among 4960 single copy genes in mammals, 222 that show high expression levels in human placentas at term. Further, we present evidence that 94 of these 222 genes evolved adaptively during human evolutionary history since the time of the last common ancestor of eutherian mammals. Remarkably, the majority of positive selection occurred on the eutherian stem lineage suggesting that ancient adaptations have been retained in the human placenta. Of these positively selected genes, 28 have been shown to play a role in human pregnancy and placental biology, and at least 26 have important pregnancy-related phenotypes in mice. Adaptations in genes highly expressed in human placenta are attractive candidates for functional and clinical studies.
Keywords: evolution, positive selection, gene expression, Eutheria, mouse gene phenotypes, pregnancy, comparative genomics
Introduction
The placenta is the first organ to be differentiated during development in eutherian mammals, and during its life it performs functions analogous to those of lung, intestine, kidney, liver, ovary, pituitary, and hypothalamus [1–3]. Failures in placentation and pathology of the placenta often result in human reproductive disorders and pregnancy complications including spontaneous abortion [4], pre-eclampsia [5], and intrauterine growth restriction [6]. Because placental anatomy is so variable, much research has focused on the evolution, development, and pathology of the placenta within eutherian mammals. Here we ask what genes may have been involved in the emergence of the chorioallantoic placenta and their subsequent evolutionary modifications during human descent.
Studies of gene expression in the placenta are one of the key sources for discovering the genes controlling placental development [7]. One of the significant differences between the choriovitelline marsupial placenta and the chorioallantoic eutherian placenta is extravillous trophoblast invasion of the placenta into the eutherian uterus [8]. Recently [9], we suggested that many key features of the human placenta (e.g. hemochorial interface, discoid shape) represent the ancestral character state among placental mammals, and these features have been maintained throughout human descent since the time of the last common ancestor (LCA) of placental mammals. Furthermore, there is evidence that many genes expressed in the placenta originated well before the emergence of the organ [10]. In addition to these ancient genes, subsequent gene duplication and divergence have likely contributed to changes in placental morphology during mammalian evolution [10, 11]. For example, adaptive evolution has been detected in pregnancy-associated glycoproteins (PAG) [12], and many placenta-specific genes are uniquely found in specific mammalian clades [11].
Considerable variation in traits involved in placentation exists among placental mammals. During the evolution of extant placental mammals four major clades have emerged, although the relationship among them remains controversial [13, 14]. Because of the placenta’s importance in reproductive success, it is likely that intense selective pressure has shaped changes in placental anatomy and function during mammalian cladogenesis. Investigations of genes involved in the biology of the placenta are crucially important for understanding both placenta-related diseases and the evolutionary history of variation in the anatomy and physiology of the mammalian placenta. While much genomic research has examined human placental expression data to identify placenta related genes (e.g. [7]), this approach is limited by the fact that > 15,000 genes are expressed in the human term placenta [15]. Mouse mid-gestation placenta and embryo expression profiles have also identified thousands of expressed genes [16]. Thus, the sheer number of expressed genes makes it a challenge to identify the genes most important in placental biology based on expression data alone.
In the present study, we integrated DNA sequence and gene expression data using evolutionary analysis methods to implicate single copy genes involved in placental evolution. Specifically, we tested whether adaptations in single copy genes highly expressed in the term human placenta occurred predominantly in earlier periods of human evolution, defined here as prior to the divergence of the human and chimpanzee lineages [7, 9]. Single copy genes were chosen because techniques for their analysis are exceptionally robust (see Materials and Methods). Highly expressed genes were chosen based on the assumption that genes expressed at high levels in a given tissue likely contribute to the development and function of that tissue [17, 18]. Our expectation was that a proportionally greater number of genes highly expressed in the human placenta would show evidence of adaptive evolution during the emergence of the chorioallantoic placenta on the eutherian stem lineage (i.e., after the divergence from marsupials but before the last common ancestor of extant placental mammals; Fig. 1) than would be found in subsequent descendant lineages leading to humans. Our rationale for this prediction is that many anatomical features of the human placenta are phylogenetically ancient. Nevertheless, in subsequent human evolutionary history, different genes might then have undergone adaptations associated with the modifications of placental functions and/or phenotypes that distinguish the human placenta from placentae of other mammals.
Fig. 1. Phylogenetic relationships among study species.
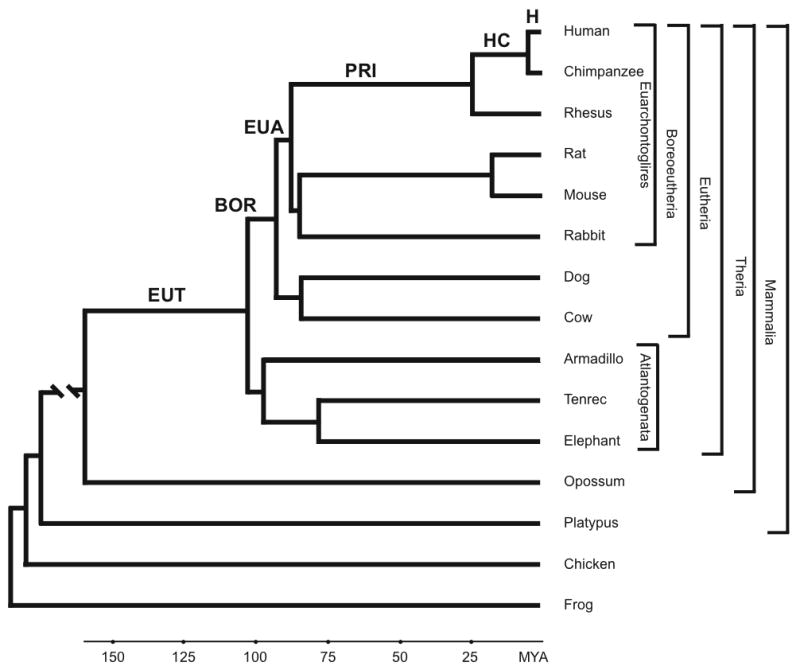
The phylogenetic relationships and divergence time estimates among eutherian mammals are from the literature [13, 14]. Branch lengths outside Theria are not drawn to scale. Stem lineages that were tested for positive selection among the set of placentally highly expressed protein coding genes during human descent from the LCA of therian mammals are indicated and abbreviated as follows: Eutheria (EUT), Boreoeutheria (BOR), Euarchontoglires (EUA), primates (PRI), Homo-Pan group (HC), and human (H). Superordinal clades discussed in the text are also indicated.
While this scenario has an intuitive appeal and genes important in placental development and function have been shown to evolve adaptively in mammalian lineages [19, 20], no statistical evidence on a genome-wide scale has yet been offered in support of the idea that positive selection on genes highly expressed in the human placenta occurred during the emergence of this vital organ. Our findings provide statistical support for the proposal that single copy genes important in human placentation underwent a burst of adaptive change in the more ancient lineages within the mammalian clade, in particular the eutherian stem lineage. This study illustrates the power that evolutionary and comparative genomics methods have to categorize the genes initially involved in the emergence of novel organs. In addition, it demonstrates the utility of an evolutionary approach in identifying candidate genes involved in organ-specific disorders.
Results
Identifying single copy and highly expressed genes in human placenta
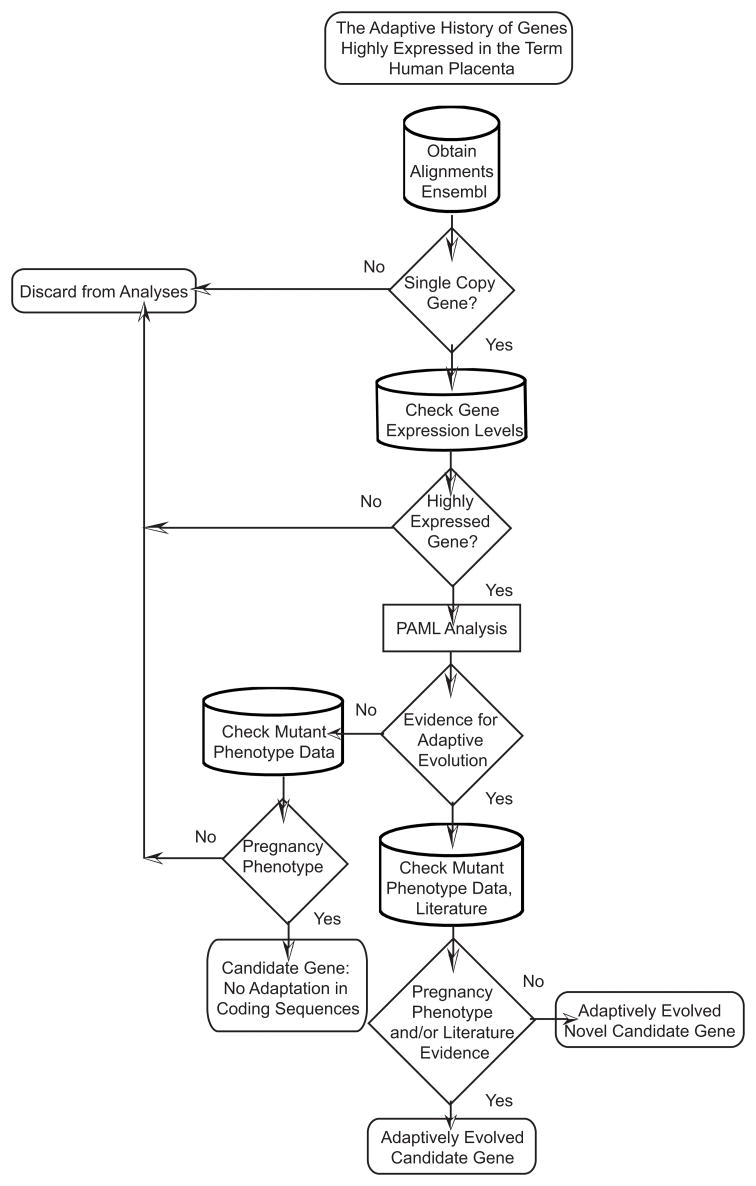
In total we identified 4960 single-copy genes within Eutheria that met with our strict sequence quality control rules (see Materials and Methods). We then intersected the single copy genes with microarray data from 44,775 human transcripts in 79 tissues/cell lines [15]. Those single copy genes that had placental expression values = 3 times the median value among all tissues/cell lines were included in further evolutionary analysis. With this consideration in mind, we identified a total of 243 single copy genes in Eutheria that were also highly expressed in the term human placenta. To conduct evolutionary analysis on the stem of Eutheria, we required opossum orthologs as an outgroup. Two hundred and twenty-two of the 243 genes met this criterion. The complete workflow of this study is presented in Fig. 2.
Fig. 2. Workflow for this study.
This workflow includes the relationship among the single copy gene inference, MGI database searching, literature searching, gene expression and evolutionary analyses.
Evolutionary analysis identifies ancient adaptively evolved genes
The ratio ω = dN/dS is a commonly used metric for testing the rate of protein sequence evolution and is also a robust analytical method for testing positive selection. In order to detect adaptive evolution during human ancestry, we used a statistical method to infer lineage-specific ω values [21] for all the eutherian lineages leading to human terminal (Fig. 1). By using this method, we identified candidate genes showing the signature of positive selection that may have contributed to the evolution of the placenta during the transformation of the ancient placenta to its current form in humans. Likelihood ratio tests (LRTs) revealed that a total of 94 unique genes showed this signature on one or more lineages during eutherian descent (Table1 and Table S1 show LRT parameters; Table S2 lists positively selected sites (PSS). While these positively selected genes (PSGs) represent a large proportion (42.3%) of all single copy, highly expressed genes, even more remarkable is the proportion of PSGs whose signature of selection occurs on the eutherian stem lineage: 62 of the 94 PSGs, or 66%, show this pattern.
Table 1.
Summary of the number of PSGs and their expression level on the different branches
| Branches tested for positive selection* | |||||||
|---|---|---|---|---|---|---|---|
| Expression(X)# | EUT | BOR | EUA | PRI | HC | HUM | Total PSG |
| 3 ≤ X ≤ 5 | 24 | 0 | 6 | 5 | 3 | 3 | 40 |
| 5<X ≤ 10 | 16 | 0 | 4 | 8 | 5 | 3 | 36 |
| X>10 | 22 | 1 | 10 | 5 | 3 | 5 | 46 |
| Total PSGs | 62 | 1 | 20 | 18 | 11 | 11 | 123 |
| Total Analyzed | 222 | 43 | 183 | 219 | 220 | 222 | |
The number of PSGs on individual branches after Bonferroni correction. EUT: Eutheria; BOR: Boreoeutheria; EUA: Euarchontoglires; PRI: primates; HC: human-chimpanzee group; HUM: human terminal (also see Figure 1); BOR running limited by poor genome sequences.
Expression (X): number shown reflects the (expression value)/(median value) for PSGs highly expressed in the human placenta;
The number of PSGs detected on the eutherian stem lineage was significantly different when compared sequentially to the number of PSGs detected on each descendant branch of human ancestry, even when accounting for multiple corrections (two-tail Fisher’s exact test, p<0.01 in all cases) (Table 1). Among the 62 PSGs showing strong evidence for positive selection on the eutherian stem, 50 were selected only on that lineage. In contrast to this pattern of selection on a single branch, 44 of the 94 PSGs showed evidence of positive selection on two or more of the tested lineages. These genes exhibited one of two modes of selection during human descent from the LCA of therian mammals: continuous or interval selection. Eighteen genes were selected on at least two adjacent lineages (continuous selection) and twenty-six genes exhibited interval selection such that the initial lineage in which positive selection was detected was followed by either purifying selection or neutral evolution, and subsequently positively selected on a descendent lineage (Table S1).
In order to test the relationship between tissue-specificity of placentally expressed genes and positive selection, we compared the tissue specificity index τ between PSGs and non-PSGs. A larger τ value indicates higher tissue specificity for a given gene. Results of this test showed that mean τ values for PSGs and non-PSGs are similar (τ̄PS = 0.8810 ± 0.1125, τ̄non − PS = 0.8792 ± 0.0981, p=0.9036, two-tail t-test). Therefore, tissue specificity alone is not an indicator of adaptive potential at coding sequences. The tissue specificity index for every tested gene is shown in supplementary data (Table S1).
Functional annotation analysis of the PSGs
To identify functional groups of genes with over representation of PSGs, we used the DAVID [22] classification tool. The highest enrichment score for the complete list of PSGs is the signal group (enrichment score=14.42, Table 2). Other top scores for the total list of PSGs are extracellular matrix group and collagen group. In examinations of PSGs on individual branches, the annotations signal and/or glycoprotein have the highest enrichment scores for four of the five analyzed lineages (i.e., all but the human-chimpanzee stem). Analysis of enrichment scores shown in Table 2 indicates a significant difference among branches (Kruskal-Wallis statistic=12.23; p=0.016). However, the statistical difference is due exclusively to the comparison of enrichment scores from two lineages - the eutherian stem and human terminal lineages (Dunn’s multiple comparison test, p<0.05). Indeed, post hoc tests comparing the eutherian stem and human-chimpanzee stem lineage, which has the same number of PSGs (n=11) as the human terminal, failed to detect a similar significant difference. These results further emphasize the ancient nature of adaptations relevant to human placental biology.
Table 2.
Functional annotation clustering of genes in total and on different branches
| Class | Period of descent* | # Gene | Cluster | Enrichment Score | # Annotations | # Genes |
|---|---|---|---|---|---|---|
| PSG | Total | 94 | Signal | 14.42 | 4 | 53 |
| Extracellular matrix | 6.23 | 9 | 24 | |||
| Collagen | 4.72 | 11 | 9 | |||
| EUT | 62 | Signal | 9.08 | 4 | 35 | |
| ECM-receptor interaction | 5.54 | 9 | 18 | |||
| Collagen | 4.09 | 15 | 10 | |||
| EUA | 20 | Glycoprotein | 8.75 | 3 | 16 | |
| Focal adhesion | 2.95 | 8 | 5 | |||
| Phosphorylation | 2.39 | 3 | 4 | |||
| PRI | 18 | Glycoprotein | 2.63 | 3 | 12 | |
| Macromolecule biosynthesis | 0.72 | 4 | 3 | |||
| Transmembrane | 0.69 | 14 | 9 | |||
| HC | 11 | Localization | 0.62 | 5 | 4 | |
| Cellular physiological process | 0.57 | 4 | 8 | |||
| Intracellular membrane-bound organelle | 0.55 | 5 | 7 | |||
| HUM | 11 | Signal | 0.6 | 7 | 5 | |
| Metabolism | 0.34 | 3 | 5 | |||
| Intracellular membrane-bound organelle | 0.07 | 4 | 3 | |||
| NMH | 54 | Signal | 2.64 | 4 | 19 | |
| Leucine-rich repeat | 1.64 | 4 | 4 | |||
| Leucine-rich repeat | 1.55 | 9 | 4 | |||
| Non_PSG | 128 | Negative regulation of cellular physiological process | 2.24 | 5 | 20 | |
| Transmembrane | 2 | 6 | 41 | |||
| Fibronectin, type III | 1.82 | 3 | 5 | |||
| Total | 222 | Signal | 14.9 | 4 | 85 | |
| Cell communication | 7.23 | 4 | 21 | |||
| Extracellular matrix | 7.22 | 3 | 19 |
EUT: Eutheria; EUA: Euarchontoglires; PRI: primates; HC: human-chimpanzee group; H: human terminal (also see Figure 1); 94 unique genes are positively selected and some show this signature on >1 lineage. BOR: not counted because of the small number of genes analyzed. NMH: PSGs that have neither (N) mouse phenotypes (M) nor identified roles in human pregnancy (H) by literature searching.
Genes involved in human pregnancy and placentation show evidence of ancient positive selection
In total, 90 of the 94 positively selected genes show evidence of ancient adaptation (i.e. prior to the time of the last common ancestor of humans and chimpanzees). We surmised that if ancient PSGs are really important in the human placenta, these PSGs might be associated with certain placenta or pregnancy-related phenotypes in humans. Indeed, literature searches for PSGs found that at least 30% (28/94) have been shown to be involved in human pregnancy and placentation (Table S3, S4). Notably, all of these 28 genes are also ancient PSGs. This finding leads us to propose that among the single copy genes we studied, ancient PSGs play a more prominent role in human pregnancy or pregnancy-related phenotypes than genes with evidence for selection on the human terminal lineage (p<0.01). Moreover, 22 genes show evidence of positive selection on only one lineage (nEUT=13, nEUA =4, nOthers=5) and are associated with pregnancy-related phenotypes (Table S3, S4). While these findings highlight the potential importance of ancient adapatation, we note that a considerable proportion of the newly identified PSGs (56.4%=53/94 of all PSGs; 55.5%=50/90 of ancient PSGs; Fig. 3, Table S4) have not yet been studied in the context of the placenta or pregnancy-related phenotypes.
Fig. 3. Relationship among orthologs, expression data, positively selected genes (PSGs), and human and mouse pregnancy-related phenotypes.
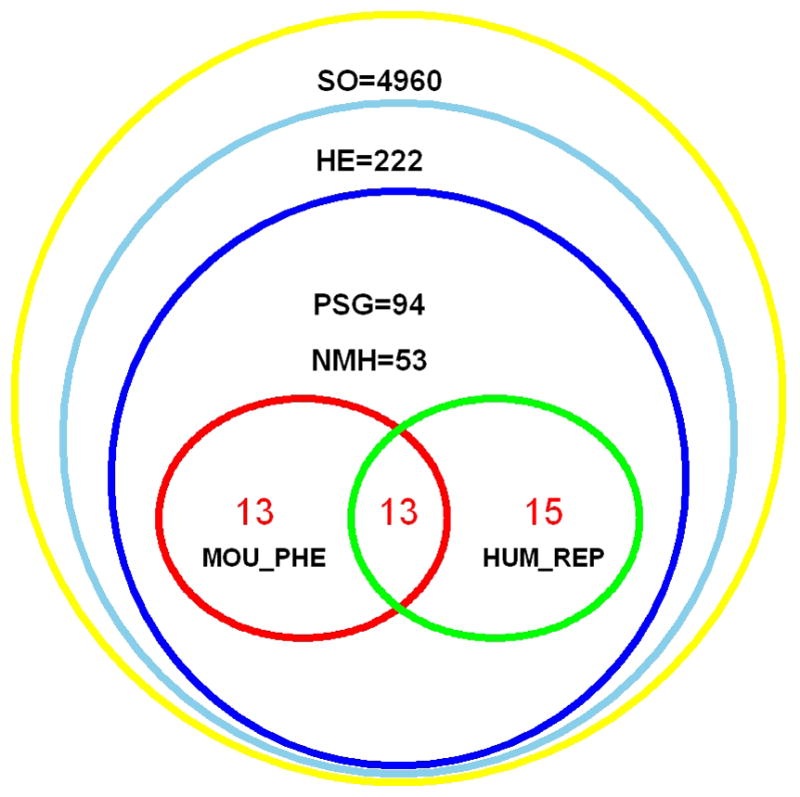
SO = single copy ortholog; HE = highly expressed gene in placenta; MOU_PHE = PSG with pregnancy-related mouse gene phenotypes; HUM_REP = PSGs with human pregnancy-related phenotypes. NMH: PSGs that have neither (N) mouse (M) nor known human pregnancy-related phenotypes (H) by literature searching.
Adaptive evolution of genes associated with pregnancy-related phenotypes in mouse models
To further identify placental and/or pregnancy-related phenotypes for these PSGs, we investigated the prevalence of PSGs in the mouse knock-out gene database [23]. Twenty-six PSGs have pregnancy-related phenotype records in the mouse gene phenotype database (MGI); furthermore, 13 of these 26 genes also have been shown to be involved in human pregnancy and placentation. Among the 26 PSGs with MGI pregnancy-related records, mutations in 18 of the PSGs in the MGI database are lethal (Table S4). MGI database searches identified 20 non-PSGs with pregnancy-related records. There is a greater proportion of PSGs (26/94=27.66%) with pregnancy-related phenotypes than non-PSGs (20/128=15.62%); however, the difference between the two groups is not significant (one tail Fisher’s exact test, p=0.055). Fifteen PSGs with known human placental and pregnancy-related phenotypes lack MGI records (Table S4, Fig. 3). Fifty-three PSGs lack both mouse mutant phenotype records and evidence of human placental and/or pregnancy-related phenotypes (Fig. 3). An additional 19 genes are related to innate immune function (10 of which also have MGI records or human placenta related records) and we propose these are further candidate genes that may have important roles in placentation and/or pregnancy (see Discussion).
Discussion
The human placenta in its current form is the product of an evolutionary history that predates the emergence of its chorioallantoic form. Indeed, recent work suggests that many genes expressed in the placenta originated well before the emergence of this organ [10]. Nevertheless, here we have hypothesized that genes important to human placentation underwent a burst of adaptive change with the emergence of a novel form of this organ. One of the significant differences between the choriovitelline marsupial placenta and the chorioallantoic eutherian placenta is extravillous trophoblast invasion of the placenta into the eutherian uterus [8]. Finding that adaptations in genes associated with trophoblast invasion occurred on the eutherian stem lineage suggests that the evolution of the invasive trophoblast also occurred on the eutherian stem lineage. Recent evidence suggests that the placenta of the last common ancestor of extant eutherians was invasive [9, 24, 25]. Phylogenetic reconstructions demonstrate that the placenta of the ancestral placental mammal had a hemochorial placental interface with a discoid shape and a labyrinthine interdigitation [9, 26]. Humans maintain this anciently evolved, invasive, hemochorial placenta. Thus, it is necessary to identify the selective pressure(s) that favored the evolution of this highly invasive hemochorial placentation.
Analyses of single copy gene sequences and gene expression
Our current study focused on highly expressed, single copy genes in the term human placenta. The potentially large sequence divergence among orthologs from distantly related vertebrate genomes is a possible pitfall for positive selection analyses. This is one reason why we used the relatively conserved single copy genes rather than members of multi-gene families as inferred by the Ensembl comparative pipeline. If the analyzed sequences have diverged too much, the pipeline will fail to cluster the sequences together. However, as all algorithms face difficulties in some cases, there is still the possibility that sequence divergence may have, in some cases, affected our alignments. We therefore manually checked the alignments to ensure the alignment quality was acceptable (see supplemental data for aligned translated protein sequences and aligned DNA sequences based on translated protein sequences).
It is possible that some genes with relatively low expression levels in the placenta (e.g. transcription factors) are also important in placental biology but were not detected by our methodology. The role played by modestly expressed genes in the placenta needs to be investigated in further research. Moreover, our study neglects many aspects of adaptations that may be associated with pregnancy. For example, neutral changes that occurred early in mammalian and vertebrate evolution may not have been subject to selective forces until the placenta emerged. Our study design would not have identified such changes. Functionally important, but non-PSGs may also have undergone adaptations in their regulatory sequences. Our study design would not have detected these changes. An additional possible caveat to consider is that the PSGs identified in our study may not be highly expressed in all mammalian placentas, making it difficult to extrapolate whether PSGs highly expressed in human term placenta are relevant to placental biology in other mammalian clades and stages of pregnancy. Recently, a study by Knox and Baker [10] showed that highly expressed genes were shared by both human and mouse placentas and that many of these genes have origins that are more ancient than the origin of the placenta. Thus, ancient genes that were co-opted during placental evolution in part directed the development and functions of the placenta [10]. However, that study also noted that both mouse and human late gestation placentas express many clade specific gene duplicates [10]. Our results also demonstrate that genes highly expressed in the human placenta have undergone positive selection after the emergence of the Euarchontoglires clade (Table 1). These more recent PSGs might be associated with changes in the morphology of primate placentas such as the emergence of villous interdigitation and an increase in the depth of trophoblast invasion. Whether the substitutions observed in the PSGs played a role in these morphological changes awaits further investigation.
Most adaptations occurred on the eutherian stem lineage
Our study takes an important first step towards the goal of understanding the selective pressures favoring human placentation. We demonstrate that single copy genes highly expressed in human placenta show significant evidence of ancient adaptations that occurred prior to the divergence of the human-chimpanzee group (Table 1). Specifically, numbers of PSGs on the eutherian stem lineage are significantly larger than on any other descendant branches (two tail Fisher exact test, p<0.01, Table 1).
We used a previously published set of housekeeping genes [27] in order to test whether the highly expressed PSGs we identified reflected an increase above the expected amount of selection on the placental stem lineage. Using this approach, we obtained multiple sequence alignments of 282 human housekeeping genes with our online tool OCPAT [28] as a comparison set to the highly-placentally-expressed genes (Table S6). Branch site tests identified 14 housekeeping PSGs after correction. The ratio of PSGs highly expressed in the placenta and positively selected on the stem eutherian lineage (62/222=28%) is significantly larger (Two tail Fisher’s exact test p<0.01) than for the housekeeping genes (14/282 = 4.96%).
We considered the possibility that the increased number of PSGs on the eutherian stem may be an artifact because the eutherian stem lineage represents a relatively long period of evolutionary time. Careful examination of Fig. 1 reveals that the branch lengths of the stem eutherian and stem primate branches are similar. Divergence time estimates show that the stem primate branch (63 million years) is longer than the stem eutherian branch (55 million years) [29]. If branch length were the cause of the high number of PSGs, we would expect a similar number of PSGs on these two branches. Instead, the primate stem has only 18 PSGs while the eutherian stem has 62. A Fisher’s exact test shows that significantly more genes were positively selected on the stem eutherian lineage compared to the stem primate branch (p<0.01; Table 1). In order to further confirm the branch-site model results we also conducted site model analysis for each gene [30]. More than 50% (50 PSGs, 53.20%) of the genes that passed the branch-site model A test also passed the site model test (Table S2).
Most comparative genomic studies seeking to uncover the genetic underpinnings of the human phenotype have focused primarily on recent human adaptations. Our results emphasize that if we limit tests for selection to the human terminal lineage, a large portion of genes that are anciently adaptively evolved and potentially crucial for human placentation would be missed. Similarly, another recent genome scan searching for adaptation in humans also showed that genes showing more ancient selection on the ape stem lineage are associated with human autoimmune and aging-related diseases [31].
Functional annotation and candidate genes for future research
Because we examined single copy genes that were highly expressed in human term placenta, our initial set of 222 genes could shed light on the importance of both PSGs and non-PSGs in human placental biology. DAVID analysis of the PSGs and non-PSGs found distinct functional differences between the two sets of genes (Table 2). For example, the first cluster determined from the total list of PSGs has “signal” as its primary annotation. Genetic studies have demonstrated the crucial importance of signaling interactions among the mother, the placenta, and the embryo/fetus [32]. Therefore, further research might be warranted on the positively selected signaling related genes. In contrast, the most enriched functional annotation cluster among non-PSGs is negative regulation of cellular physiological process. Genes in this cluster include distal-less homeobox 5 (DLX5), notch homolog2 (NOTCH2), and adrenomedullin (ADM). Mutations in genes that show a consistent pattern of purifying selection are also likely to cause dysfunction due to functional constraint, although that functional constraint precedes the emergence of the placenta. As genes often have pleiotropic effects, some genes highly expressed in the human placenta also play important roles in other tissues. For example, KISS1 was first described as a tumor related gene [33, 34] and more recently, it has been described as a potent regulator of the neuroendocrine reproductive axis [35]. Therefore, the adaptive benefit of the substitutions we identified requires further study. Finally, we also consider those genes that show evidence of adaptive evolution, but which have not been studied in the context of placental biology (n=53) as particularly promising candidates for future genetic association and clinical research studies.
Adaptive evolution on the eutherian stem lineage as related to pregnancy
In our analysis, we found that many of the ancient PSGs are associated with pregnancy-related phenotypes in humans and mice (Table S4), suggesting that successful placentation requires the actions of proteins encoded by these PSGs. Examples of genes that show adaptations on the eutherian stem lineage and are also important for placentation include COL4A2 NCOA6, and KISS1. Collagen IV, encoded by COL4A2, is a major constituent of the trophoblast basement membrane [36] and adaptively evolved on the eutherian stem (ω =114.89, p=0.00125, proportion of PSS=0.61%) and human terminal ( ω =999, p=0.000, PSS%=1.12%) lineages. Nuclear receptor coactivator (NCOA6) is essential for embryonic and placental development and evolved adaptively on the eutherian stem lineage (ω =13.686, p=0.00238, PSS%=0.65%). Remarkably, Ncoa6 knockout mice do not survive past embryo day 13.5, and histological examination of their placentas shows abnormal placental morphology and placental development with a greatly reduced spongiotrophoblast layer (Table S4) [37]. KISS-1 metastasis-suppressor (KISS1) also adaptively evolved on the eutherian stem lineage (ω =466.628, p=0.0056, PSS%=8.84%). The placental expression of kisspeptins, products of the KISS1 gene, and their receptor are highest in the first trimester in humans and at day 12.5 in rats [38], and known to regulate trophoblast invasion [39].
Most genes in the human genome have not yet been subject to any functional study, and among those that have, only a subset have been studied in the context of placentation. We therefore suspect that ascertainment bias is a potential problem when comparing the proportion of PSGs to non-PSGs in terms of pregnancy-related phenotypes because of the lack of currently available functional data. Instead, we used the MGI phenotypes records to compare the ratio of the PSGs and non-PSGs related with pregnancy. There is an increasing trend for enrichment of the PSGs with pregnancy (27.66% for PSGs v.s. 15.62% for non-PSGs), even though the increase is not significant (one tail Fisher exact test, p=0.055).
Our finding that 19 innate immune related genes show adaptive evolution on the eutherian stem lineage (Table S1, Table S5) warrants further discussion. This result corroborates experimental evidence that innate immune-related genes are involved in placentation and pregnancy [40, 41]. Since the placenta is a semi-allograft [42, 43], the maternal immune system must have been challenged by the invading placenta throughout human descent from the LCA of extant eutherians. Many studies have found that immune-related genes are highly expressed in the placenta and fetal membranes, and that key immune-regulatory molecules have significant impact on the outcome pregnancy; for example, Toll-like receptors (TLR) [44], and genes expressed in NK cells during pregnancy [45]. Examples of adaptively evolving innate immune-related genes on the eutherian stem lineage include CFB and AXL. Complement factor B (CFB) shows evidence of adaptive evolution on both the eutherian ( ω =24.633, p=0.00414, PSS%=3.54%) and Euarchonotoglires (ω =286.319, p=0.000, PSS%=3.02%) stem lineages. Of note, low concentrations of CFB have been detected in the circulation of patients with recurrent pregnancy loss [46]. AXL, which encodes the AXL receptor tyrosine kinase, also shows evidence of adaptive evolution on the eutherian stem lineage (ω =27.322, p=0.0046, PSS%=1.94%). Importantly, AXL is one of the three TAM (Tyro3, AXL and Mer) receptors that profoundly inhibit both TLR- and cytokine-driven immune responses [47] that are known to have immunoregulatory role in pregnancy [48]. Taken together, these examples suggest that the 19 immune-related PSGs identified in this study are particularly interesting candidates for studies focused on the immune responses at the maternal-fetal interface.
Human and mouse pregnancy-related phenotypes/functions
28 PSGs have human pregnancy-related phenotypes (Table S3) while 26 PSGs have mouse knock-out pregnancy-related phenotypes. Among these 41 PSGs in total, all show ancient signatures of selection (Tables S1 and S3) and about half evolved adaptively on the eutherian stem lineage (18 for human phenotypes/functions, 18 for mouse MGI records). However, there are still considerable numbers of genes that evolved adaptively on other lineages and also appear to be important in the placenta. For example, ADAM12 (a disintegrin and metalloprotease 12) shows evidence of positive selection on the primate stem lineage ( ω =8.155, p=0.00691, PSS%=0.83%). ADAM12 mediates syncytiotrophoblastic shedding of oxytocinase [49] and its reduced maternal serum concentrations were reported in early pregnancies that are complicated with fetal trisomies 18 and 21 [50], and in pregnant women who subsequently developed preeclampsia [51]. FN1 (fibroncetin 1) shows evidence for positive selection on the Euarchontoglires stem lineage (ω =170.096, p=0.0225, PSS%=0.06%). Fibroncetin mediates the attachment of trophoblast to collagen and the extracellular matrix [52], and patients with a positive fibronectin test have been shown to be at higher risk for spontaneous recurrent preterm birth [53]. Evidence from gene expression profiles at the human maternal-fetal interface [54] demonstrated that several of our newly identified ancient PSGs including collagen encoding genes (COL1A2, COL3A1, COL6A1, and COL6A3), the PDZ and LIM domain-containing PDLIM1, and the anti-hemophillic Von Willebrand Factor (VWF), change expression significantly (fold value larger than 1.6) between midgestation and term. Among these genes, one (PDLIM1) shows evidence for selection on the human terminal lineage.
It has recently been reported that phenotypes determined from mouse models may not result in similar phenotypes in human [55]. However, in our study, 11 of the 13 PSGs with known mouse phenotypes but unknown human phenotypes show ancient adaptations before the last common ancestor of the two species; suggesting the possibility that the observed mouse phenotypes in these genes are pertinent to humans.
In conclusion, the current study provides evolutionary genetic evidence that a significantly greater number of adaptive events occurred on the eutherian stem lineage compared with the subsequent descendent lineages in which the human placenta evolved into its current form. The statistical evidence obtained in the present study allows us to propose that the early adaptations in single copy genes highly expressed in term human placentas have been largely maintained during human descent. Moreover, examination of the mouse mutant phenotypes and PubMed literature searches on human pregnancy revealed that a considerable portion of ancient positively selected genes, especially on the stem eutherian lineage, have important roles in mammalian pregnancy and placentation. Other PSGs without functional and/or experimental evidence are attractive and novel candidates for further study of placental biology.
Materials and Methods
Genome data sources
All sequences were obtained from the Ensembl comparative database (v. 47, Oct. 2007). We used two Ensembl comparative homology databases (Ensembl family and Ensembl protein homology, http://www.ensembl.org). The Ensembl family database uses all the available data to cluster genome sequences into protein families and includes many redundant sequences. Conversely, the protein homology database is limited to only the longest transcripts for each gene and relies solely on Ensembl predicted gene datasets (Ensembl help desk). When genome sequence coverage is good, inferences of protein homology are robust. However, it is not possible to accurately infer gene duplication and loss using genomes with coverage lower than 2X [56]. Therefore, we decided to use Ensembl protein homology and only those genomes with fold coverage > 5X (n=11) for initial orthology assessment. Genome coverage, taxon names, and placental interface are listed in Table S7. We used established single copy genes (i.e., 1:1 orthologs) only as determined by Ensembl. The rationale for this choice is that lineage specific gene duplications resulting in sub- and neo-functionalization in new gene copies, along with processes such as gene conversion, complicate interpretations of branch specific adaptive evolution. Instead, we took the conservative approach and examined only those genes with relatively simple evolutionary histories. All gene families with multiple copies in a given genome were excluded from further analysis. In the second step, we included putatively orthologs sequences from the low-fold coverage genomes of rabbit, armadillo, African elephant and tenrec. This approach allowed us to include all four major extant eutherian mammal clades in the analyses. Due to low-quality atlantogenatan (i.e., the elephant, tenrec, armadillo group) sequences some genes were analyzed without inclusion of their orthologs. Finally, we also checked whether the putative orthologs had multiple copies in the armadillo, African elephant or tenrec. If so, these genes were excluded from our datasets.
Human housekeeping genes were used as the control group to compare the ratio of PSGs among genes highly expressed in the placental datasets to a control group. We used a previously published housekeeping gene list [27] and we used our online tool OCPAT [28] to obtain multiple sequence alignments of these genes. Using this tool, we were able to obtain alignments from 282 human housekeeping genes (Table S6).
Sequences and alignment quality control
We used two rules to strike a balance between sequence quality and statistical power. Sequences with a proportion of ‘Ns’ greater than 10% across the length of the gene sequence were removed from analysis. Further, any gene sequence with total length < 300bp was excluded from further analysis. Alignment of multiple DNA sequences based on predicted protein sequences was done using the CLUSTAL W program [57] using Bioperl modules (http://www.bioperl.org).
Evolutionary analysis
The ratio between the relative rate of non-synonymous substitution to the relative rate of synonymous substitution (ω =dN/dS) measures the strength of selection acting on a protein-coding gene. Assuming synonymous mutations are subjected to almost strictly neutral selection, ω <1, ω =1, and ω >1 represents negative selection, neutral evolution, and positive Darwinian selection, respectively. Maximum likelihood analysis of the sequence evolution was performed with CODEML program in the PAML 3.15 software package [58]. The improved branch-site model A (TEST-II) [59] was used to test for positively selected sites along specific lineages. TEST-II is an LRT that compares a null hypothesis with fixed ω2=1 with model A that allows ω2>1 in the foreground lineage. TEST-II is a direct test for positive selection on the foreground lineages and can discriminate relaxed selective constraint from positive selection [21]. A Bayes Empirical Bayes (BEB) method of analysis, introduced to answer the criticisms that branch-sites tests have an excessive false positive rate [60], was used to detect positively selected sites. However, any genome-scale study that surveys for positive selection is susceptible to false positives arising from uncertainty in parameter estimation [61]. When multiple branches on a tree are tested for positive selection using the same data, correction for multiple testing is required. We used the conservative Bonferroni correction procedure for multiple tests as recommended by Yang [61]. Six separate foreground branches were tested for evidence of positive selection: stem of Eutheria, stem of Boreoeutheria, stem of Euarchontoglires, stem of Primates, stem of the human-chimp clade, and the human terminal (Fig. 1) according to a presumed phylogeny [13]. As an additional validation test, Site model (M7 v.s. M8) was used to test selection across the whole tree [30].
In order to reduce the amount of phylogenetic gene tree inference bias in genome level analysis that can be introduced, for example, when either only the gene tree or only the species tree is used, we used a newly developed gene tree construction method called TreeBest [62] to infer individual gene trees. TreeBest was specifically designed for reconciling gene trees with species trees and has been widely used in both the Ensembl comparative and Treefam [62] databases. TreeBest minimizes the number of gene duplications and losses by reconciling an inferred neighbor joining gene tree with a presumed species tree. If the best tree topology is different from user-determined criteria, the gene is discarded for the specific lineage test. We applied an inclusion criterion that required the basic tree topology recovered by TreeBest to follow the species tree such that for each tested branch, the closest outgroup species must be the same as observed in the presumed species tree. For example, in any given TreeBest gene tree, we required the opossum gene as the closest outgroup to eutherian mammals in order to test for selection on the stem eutherian lineage.
Expression and phenotype data
Gene expression data were obtained from the Human GNF1H, gcRMA indexed by SymAtlas [15]. The annotation files were also downloaded from SymAtlas [15] to map to the expression value to the Ensembl Transcripts IDs. Placental expression data were used to identify those genes highly expressed in the human placenta. Tissue-specific expression patterns were measured as tissue specificity index τ [63]. The formula (1) was used for τ value calculation.
| (1) |
In this formula nH is the number of examined tissues and xi is the expression profile component normalized by the maximal component value. In the current study, we used all 79 tissues/cell lines to calculate the τ value. If a gene was expressed in only one tissue, its τ value would be close to 1. In contrast, if a gene is equally expressed in all tissues, τ =0. If two or more probes mapped to a single gene, we averaged the expression values for each tissue and then calculated the τ value. Highly expressed genes were defined as three times or greater than the median value of the 79 tissues/cell lines.
Because human and mouse placentas are both discoid and hemochorial, mouse mutants can provide important information for human placenta research. Mouse gene mutant phenotype data were obtained from the MGI_4.0 database (http://www.informatics.jax.org/phenotypes.shtml) [23]. In the current research, we used MGI_4.0 database and literature searches to define pregnancy-related phenotypes. Pregnancy-related phenotypes were assumed for a particular PSG if: 1) The mouse phenotype for a certain gene caused significant changes in A) placental/extraembryonic tissue development; B) embryonic/fetal development; C) embryonic/fetal lethality; D) angio-/vasculogenesis; or 2) Current literature searches suggested important roles in human pregnancy and placentation (Table S4).
Functional Annotation Clustering and Tests of Statistical Significance
Functional annotation clusters were determined using the DAVID functional annotation clustering tool [22], with the kappa similarity threshold set to 0.7 and all other options set to default values. The tool calculates an associated enrichment score for each cluster based upon the geometric mean (in negative log scale) of the p values determined for each of its component annotations, as determined via a modified Fisher’s exact test. To compare the tissue specificity for the PSG and the non-PSG both highly expressed in the human placenta, the two tail t-test was used to check possible tissue specificity difference between two groups. Non-parametric ANOVAs and post-hoc tests were performed using GraphPad Prism software (http://www.graphpad.com/prism/Prism.htm). The GO term for immune related genes were annotated by using the GO official website annotated immune system (http://www.geneontology.org/). Numbers of PSGs reported in total and for each lineage have been corrected for multiple testing as recommended by [49].
Supplementary Material
Acknowledgments
This research was supported in part by the Intramural Research program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development/NIH/DHHS. We thank Morris Goodman (Wayne State University) for insightful comments on a draft of this manuscript, and we acknowledge Munirul Islam (Wayne State University) for computational assistance. We thank two anonymous reviewer’s for their valuable insights on this paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luckett WP. Comparative development and evolution of the placenta in primates. In: Luckett WP, editor. Reproductive Biology of the Primates. S. Karger; New York: 1974. pp. 142–234. [PubMed] [Google Scholar]
- 2.Benirschke K, Kaufmann P. Pathology of the human placenta. Spinger; New York: 2000. [Google Scholar]
- 3.Mossman HW. Vertebrate Fetal Membranes. Rutgers University Press; New Brunswick: 1987. [Google Scholar]
- 4.Wells M. The pathology of gestational trophoblastic disease: recent advances. Pathology. 2007;39:88–96. doi: 10.1080/00313020601137367. [DOI] [PubMed] [Google Scholar]
- 5.Jauniaux E, Poston L, Burton GJ. Placental-related diseases of pregnancy: Involvement of oxidative stress and implications in human evolution. Hum Reprod Update. 2006;12:747–755. doi: 10.1093/humupd/dml016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pardi G, Marconi AM, Cetin I. Placental-fetal interrelationship in IUGR fetuses--a review. Placenta. 2002;23(Suppl A):S136–S141. doi: 10.1053/plac.2002.0802. [DOI] [PubMed] [Google Scholar]
- 7.Sood R, Zehnder JL, Druzin ML, Brown PO. Gene expression patterns in human placenta. Proc Natl Acad Sci U S A. 2006;103:5478–5483. doi: 10.1073/pnas.0508035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mess A, Carter AM. Evolution of the placenta during the early radiation of placental mammals. Comp Biochem Physiol A Mol Integr Physiol. 2007;148:769–779. doi: 10.1016/j.cbpa.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 9.Wildman DE, et al. Evolution of the mammalian placenta revealed by phylogenetic analysis. Proc Natl Acad Sci U S A. 2006;103:3203–3208. doi: 10.1073/pnas.0511344103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knox K, Baker JC. Genomic evolution of the placenta using co-option and duplication and divergence. Genome Res. 2008;18:695–705. doi: 10.1101/gr.071407.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rawn SM, Cross JC. The Evolution, Regulation, and Function of Placenta-Specific Genes. Annu Rev Cell Dev Biol. 2008 doi: 10.1146/annurev.cellbio.24.110707.175418. [DOI] [PubMed] [Google Scholar]
- 12.Hughes AL, Green JA, Garbayo JM, Roberts RM. Adaptive diversification within a large family of recently duplicated, placentally expressed genes. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3319–3323. doi: 10.1073/pnas.050002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wildman DE, et al. Genomics, biogeography, and the diversification of placental mammals. Proc Natl Acad Sci U S A. 2007;104:14395–14400. doi: 10.1073/pnas.0704342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Springer MS, et al. The adequacy of morphology for reconstructing the early history of placental mammals. Syst Biol. 2007;56:673–684. doi: 10.1080/10635150701491149. [DOI] [PubMed] [Google Scholar]
- 15.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proceedings of the National Academy of Sciences. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka TS, et al. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc Natl Acad Sci U S A. 2000;97:9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu XJ, Zheng HK, Wang J, Wang W, Su B. Detecting lineage-specific adaptive evolution of brain-expressed genes in human using rhesus macaque as outgroup. Genomics. 2006;88:745–751. doi: 10.1016/j.ygeno.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen R, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CY, et al. The human progesterone receptor shows evidence of adaptive evolution associated with its ability to act as a transcription factor. 2008. pp. 637–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Zhang J. Rapid evolution of primate ESX1, an X-linked placenta- and testis-expressed homeobox gene. Hum Mol Genet. 2007;16:2053–2060. doi: 10.1093/hmg/ddm153. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 22.Huang dW, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elliot MG, Crespi BJ. Placental invasiveness mediates the evolution of hybrid inviability in mammals. Am Nat. 2006;168:114–120. doi: 10.1086/505162. [DOI] [PubMed] [Google Scholar]
- 25.Vogel P. The current molecular phylogeny of Eutherian mammals challenges previous interpretations of placental evolution. Placenta. 2005;26:591–596. doi: 10.1016/j.placenta.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Elliot MG, Crespi BJ. Placental invasiveness mediates the evolution of hybrid inviability in mammals. American Naturalist. 2006;168:114–120. doi: 10.1086/505162. [DOI] [PubMed] [Google Scholar]
- 27.Eisenberg E, Levanon EY. Human housekeeping genes are compact. Trends in Genetics. 2003;19:362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- 28.Liu G, et al. OCPAT: an online codon-preserved alignment tool for evolutionary genomic analysis of protein coding sequences. Source Code Biol Med. 2007;2:5. doi: 10.1186/1751-0473-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hedges SB, Kumar S. Genomic clocks and evolutionary timescales. Trends in Genetics. 2003;19:200–206. doi: 10.1016/S0168-9525(03)00053-2. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uddin M, et al. Distinct genomic signatures of adaptation in pre- and postnatal environments during human evolution. 2008. pp. 3215–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- 33.Stafford LJ, Vaidya KS, Welch DR. Metastasis suppressors genes in cancer. International Journal of Biochemistry & Cell Biology. 2008;40:874–891. doi: 10.1016/j.biocel.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez-Carbayo M, Capodieci P, Cordon-Cardo C. Tumor suppressor role of KiSS-1 in bladder cancer - Loss of KiSS-1 expression is associated with bladder cancer progression and clinical outcome. American Journal of Pathology. 2003;162:609–617. doi: 10.1016/S0002-9440(10)63854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popa SM, Clifton DK, Steiner RA. The role of kisspeptins and GPR54 in the neuroendocrine regulation of reproduction. Annual Review of Physiology. 2008;70:213–238. doi: 10.1146/annurev.physiol.70.113006.100540. [DOI] [PubMed] [Google Scholar]
- 36.Furuhashi N, et al. Serum collagen IV and laminin levels in preeclampsia. Gynecol Obstet Invest. 1994;37:250–253. doi: 10.1159/000292571. [DOI] [PubMed] [Google Scholar]
- 37.Antonson P, et al. Inactivation of the nuclear receptor coactivator RAP250 in mice results in placental vascular dysfunction. Mol Cell Biol. 2003;23:1260–1268. doi: 10.1128/MCB.23.4.1260-1268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terao Y, et al. Expression of KiSS-1, a metastasis suppressor gene, in trophoblast giant cells of the rat placenta. Biochim Biophys Acta. 2004;1678:102–110. doi: 10.1016/j.bbaexp.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Hiden U, Bilban M, Knofler M, Desoye G. Kisspeptins and the placenta: regulation of trophoblast invasion. Rev Endocr Metab Disord. 2007;8:31–39. doi: 10.1007/s11154-007-9030-8. [DOI] [PubMed] [Google Scholar]
- 40.Romero R, et al. Inflammation in preterm and term labour and delivery. Semin Fetal Neonatal Med. 2006;11:317–326. doi: 10.1016/j.siny.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abrahams VM, Mor G. Toll-like receptors and their role in the trophoblast. Placenta. 2005;26:540–547. doi: 10.1016/j.placenta.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 42.Medaware PB. Some immunological and endocrinological problems raised by the eovlution of viviparity in vertebrates, Society for Experimental Biology. Academic Press; New York: 1953. pp. 320–338. [Google Scholar]
- 43.Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol. 2006;6:584–594. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- 44.Kim YM, et al. Toll-like receptor-2 and -4 in the chorioamniotic membranes in spontaneous labor at term and in preterm parturition that are associated with chorioamnionitis. Am J Obstet Gynecol. 2004;191:1346–1355. doi: 10.1016/j.ajog.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 45.Kusakabe K, Naka M, Ito Y, Eid N, Otsuki Y. Regulation of natural-killer cell cytotoxicity and enhancement of complement factors in the spontaneously aborted mouse placenta. Fertil Steril. 2007 doi: 10.1016/j.fertnstert.2007.07.1331. [DOI] [PubMed] [Google Scholar]
- 46.Micheloud D, et al. Hypocomplementemia in the absence of autoantibodies in women with recurrent pregnancy loss. Allergol Immunopathol (Madr ) 2007;35:90–94. doi: 10.1157/13106775. [DOI] [PubMed] [Google Scholar]
- 47.Rothlin CV, Ghosh S, Zuniga EI, Oldstone MB, Lemke G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell. 2007;131:1124–1136. doi: 10.1016/j.cell.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 48.Abrahams VM, et al. A role for TLRs in the regulation of immune cell migration by first trimester trophoblast cells. J Immunol. 2005;175:8096–8104. doi: 10.4049/jimmunol.175.12.8096. [DOI] [PubMed] [Google Scholar]
- 49.Ito N, et al. ADAMs, a disintegrin and metalloproteinases, mediate shedding of oxytocinase. Biochemical and Biophysical Research Communications. 2004;314:1008–1013. doi: 10.1016/j.bbrc.2003.12.183. [DOI] [PubMed] [Google Scholar]
- 50.Spencer K, Cowans NJ. ADAM12 as a marker of trisomy 18 in the first and second trimester of pregnancy. J Matern Fetal Neonatal Med. 2007;20:645–650. doi: 10.1080/14767050701483389. [DOI] [PubMed] [Google Scholar]
- 51.Laigaard J, et al. Reduction of the disintegrin and metalloprotease ADAM12 in preeclampsia. Obstetrics and Gynecology. 2005;106:144–149. doi: 10.1097/01.AOG.0000165829.65319.65. [DOI] [PubMed] [Google Scholar]
- 52.Kao LC, Caltabiano S, Wu S, Strauss JF, Kliman HJ. The Human Villous Cytotrophoblast - Interactions with Extracellular-Matrix Proteins, Endocrine Function, and Cytoplasmic Differentiation in the Absence of Syncytium Formation. Developmental Biology. 1988;130:693–702. doi: 10.1016/0012-1606(88)90361-2. [DOI] [PubMed] [Google Scholar]
- 53.Mazaki-Tovi S, et al. Recurrent preterm birth. Semin Perinatol. 2007;31:142–158. doi: 10.1053/j.semperi.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winn VD, et al. Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology. 2007;148:1059–1079. doi: 10.1210/en.2006-0683. [DOI] [PubMed] [Google Scholar]
- 55.Liao BY, Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0800387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Green P. 2x genomes-does depth matter? Genome Res. 2007;17:1547–1549. doi: 10.1101/gr.7050807. [DOI] [PubMed] [Google Scholar]
- 57.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 59.Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki Y, Nei M. False-positive selection identified by ML-based methods: examples from the Sig1 gene of the diatom Thalassiosira weissflogii and the tax gene of a human T-cell lymphotropic virus. Mol Biol Evol. 2004;21:914–921. doi: 10.1093/molbev/msh098. [DOI] [PubMed] [Google Scholar]
- 61.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 62.Li H, et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–D580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yanai I, et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.