Abstract
Phosphatase of regenerating liver-1 (PRL-1) belongs to a unique subfamily of protein tyrosine phosphatases (PTPases) associated with oncogenic and metastatic phenotypes. While considerable evidence exists to supports a role for PRL-1 in promoting proliferation, the biological regulators and effectors of PRL-1 activity remain unknown. PRL-1 activity is inhibited by disulfide bond formation at the active site in vitro, suggesting PRL-1 may be susceptible to redox regulation in vivo. Because PRL-1 has been observed to localize to several different subcellular locations and cellular redox conditions vary with tissue type, age, stage of cell cycle and subcellular location, we determined the reduction potential of the active site disulfide bond that controls phosphatase activity to better understand the function of PRL-1 in various cellular environments. We used high-resolution solution NMR spectroscopy to measure the potential and found it to be −364.3 ± 1.5 mV. Because normal cellular environments range from −170 to −320 mV, we concluded that nascent PRL-1 would be primarily oxidized inside cells. Our studies show that a significant conformational change accompanies activation, suggesting a post-translational modification may alter the reduction potential, conferring activity. We further demonstrate that alteration of the C-terminus renders the protein reduced and active in vitro, implying the C-terminus is an important regulator of PRL-1 function. These data provide a basis for understanding how subcellular localization regulates the activity of PRL-1 and, with further investigation, may help reveal how PRL-1 promotes unique outcomes in different cellular systems, including proliferation in both normal and diseased states.
Phosphatase of Regenerating Liver (PRL) enzymes are a unique subfamily of protein tyrosine phosphatases (PTPases) and play an important role in maintaining appropriate tyrosine phosphorylation levels in the cell during development and tissue regeneration (1). This subfamily includes three homologous members (PRL-1, −2 and −3) that share a high degree of amino acid sequence identity (>75%). PRL-1 (20 kDa) was first discovered in regenerating liver following partial hepatectomy as the product of an early-immediate response gene (2) and has since been implicated in the repair of other tissues including neurons, oligodendrocytes and the cerebral cortex in response to transient forebrain ischemia (3). PRLs are normally expressed at low levels in most mature cells, while higher expression levels are observed during embryonic development (4) and during cellular proliferation (5–9) or differentiation (5, 10, 11) depending on the cell type. Recently, PRL enzymes have gained considerable interest because each of the three isoforms can aggressively promote cancer (12, 13). Each isoform is expressed at high concentrations in various cancer tissues and promotes cellular transformation and tumorigenesis (14–22). A substantial amount of research is available that shows PRL-1 (8, 16, 17, 23–26) or PRL-3 (18, 27–32) expression promotes cellular motility, invasiveness, and metastasis. Furthermore, it has been shown that during these cellular processes, PRL-1’s critical active site cysteine (C104) is required to facilitate proliferation, motility and metastasis (7–9, 26, 30, 33, 34). Studies examined the inactive C104S/A mutant and concluded that PTP activity is responsible for the observed outcomes. Although phenotypic responses to PRL-1 expression and activity have been wellestablished, the exact biochemical role that PRL-1 plays in these processes has yet to be described.
Like all PTPases, PRL enzymes accomplish the removal of phosphate from their phosphotyrosine substrate via their CX5R active site motif (35). In this motif, the conserved cysteine initiates the catalytic reaction by attacking the phosphorous on the incoming substrate, while the phosphate binding loop (P-loop) and conserved arginine residue bind and position the phosphate group (36). Unlike most cysteine residues in proteins that have pKa values near 8, the PTPase catalytic cysteine is typically deprotonated (pKa≈5) due to the partially positive microenvironment of the active site, making it extremely reactive at physiological pH (1). The exceptional reactivity of the thiol group required for enzyme activity also renders the catalytic cysteine highly susceptible to oxidation (1, 36). As established for PTP-1B and the dualspecificity phosphatase PTEN, oxidation by singlet oxygen and hydroxyl radical leads to disulfide bond formation at the nucleophilic cysteine, inactivating the enzyme (37, 38). In PRL-1, oxidation at the nucleophilic C104 abolishes PTPase activity by promoting disulfide bond formation with C49, a spatially proximal but sequentially distal cysteine residue (36). Disulfide bond formation is reversible in vitro and in vivo and reduction is accomplished via reducing agents present in solution. To restore enzymatic function, reduction at the active site must occur in order to expose the nucleophilic thiol. Reduced forms of PRL enzymes have been isolated and characterized by NMR and X-ray crystallography (36, 39–41). In vitro activity assays and structure analyses are often performed in the presence of high concentrations of reducing agents to prevent inactivation. Despite this precaution, crystallization of PRL-1 produced a structure of the inactive oxidized form. No other PTPase structure has been reported in the inactive state and many remain active in the absence of reducing agents, suggesting that PRL-1 may be more prone to oxidation than other family members.
Disulfide bond formation in vivo depends on the presence of reactive oxidants as well as the redox environment of the cell, which is primarily determined by the amount of glutathione and the ratio of its reduced to oxidized forms [GSH:GSSG] (42). The reduction potential and capacity of a cell vary with compartment, stage of cell cycle, tissue type, age and health. Interestingly, reports describing PRL-1’s subcellular location have been quite varied. Localization of PRL-1 in the cell appears to be influenced by a number of factors, including stage of cell cycle, tissue type and modification at the C-terminus (2, 6, 9, 43). The specific roles that PRL-1 plays at each of these locations has yet to be elucidated, but based on the data presented here, the redox environment and modification state of PRL-1 are likely to regulate the function of this enzyme in vivo. In the present study, we used high-resolution NMR to determine the reduction potential of the active site disulfide bond in full-length PRL-1. Determining the reduction potential of the disulfide bond involving the catalytic cysteine provides a basis for understanding how subcellular localization may regulate the phosphatase activity of PRL-1 in different types of cellular environments.
Materials and Methods
Protein Expression and Purification
Wild-type PRL-1 and PRL-1-C170S-C171S were produced as described previously (44). To create the PRL-1-C98A and PRL-1-C104S mutants, the human PRL-1 gene was cloned into the pET-30 Xa/LIC vector (Novagen) and mutated using the PCR-based QuickChange method (Stratagene). The primers used to generate the C98A and C104S mutants, 5’ to 3’, were cgtgaagaacctggtGCTtgtattgctgttcattgc and ggttgtattgctgttcatTCCgttgcaggccttgg, respectively. Capital letters indicate the mutated bases. PCR reactions were treated with DpnI (Promega) for 1.5 hours at 37°C, directly transformed into Novablue GigaSingles competent cells (Novagen) and selected on LB plates with 30 mg/mL kanamycin. Individual colonies were cultured overnight in selective LB at 37°C, and the resultant DNA was purified using the Qiagen MiniPrep System (Stratagene). All mutations were confirmed using bidirectional DNA dye-terminator sequencing (Northwoods DNA, Inc., Bemidjii, MN). All variants of PRL-1 were expressed in BL21(DE3) (Novagen) cells on minimal media and purified as described previously (45). Samples for NMR were produced from cells grown on minimal media containing 15N ammonium chloride (Spectra Stable Isotopes). All solutions used in the purification and analysis of samples involved in this study were sparged with Ar gas to minimize oxidation derived from O2.
Electrospray Ionization Mass Spectrometry
ESI spectra were acquired on a Q-Tof-2 (Micromass Ltd, Manchester UK) Hybrid Mass Spectrometer operated in MS mode and acquiring data with the time of flight analyzer de-tuned to 8000 resolution (FWHH) for sensitivity. For whole protein ESI spectra, purified protein samples were diluted to 1 µg/µL in untreated (“oxidizing”) 20 mM ammonium bicarbonate, pH 8.2, and allowed to incubate at room temperature. Time points were taken on the first, fourth and seventh day post-purification and frozen at −80°C until ready for use. Samples were desalted on a short (3 cm × 1 mm I.D.) reversephase (RP) HPLC column (Hamilton PRP1, Reno, NV). Samples were loaded onto the column from a 1% formic acid solution with protein (5 µg), washed in same solution, and eluted with 90% MeOH/0.5% formic acid directly into the ESI source. The cone voltage was 60 eV, and the voltage on the collision cell was 20 V. Spectra were acquired over the mass range 800 to 3000 u, accumulating data for 5 seconds per cycle.
For tryptic digests, PRL-1-WT and PRL-1-C170S-C171S were diluted to 1 mg/mL in ammonium bicarbonate, pH 8.2, with or without 20 mM β-mercaptoethanol (BME), and trypsin (Promega) was added in a 1:50 ratio protease:protein (w/w) and incubated at 37°C for 16 hrs. To confirm complete digestion, samples at various time points were run on SDS-PAGE and visualized by silver staining. Coupled to MS analysis, capillary HPLC separations were performed using a Zorbax SBC18 RP column (5 cm × 0.32 mm I.D., 3.5 µm bead size, 300 Å pore size) packed by Micro-Tech Scientific (Sunnyvale, CA), with a chromatograph (Waters capLC XL, Milford, MA) that develops gradients at 10 µL/min. A linear gradient of 20 to 80% B was applied over 120 minutes. The solvents were: A) 99% H2O, 1% MeOH, B) 99% MeOH, 1% H2O, and both contained 0.08% formic acid. Argon was admitted to the collision cell at a pressure that attenuates the beam to about 20%. This corresponds to 16 psi on the supply regulator or 5.3 × 10−5 mBar on a penning gauge near the collision cell. The collision cell was operated at 8 V for maximum transmission, and spectra were acquired over the range 250 to 2000 u, accumulating data for 8 seconds per cycle.
SDS-PAGE
Post-purification, all samples were exchanged by dialysis at least 1000 fold into 50 mM sodium phosphate, pH 7.5, with 100 mM NaCl. Protein samples at a final concentration of 1 mg/mL were incubated for 1 hour with varying amounts of reduced or oxidized dithiothreitol (DTT). Oxidized DTT was obtained by incubating H2O2 with DTT at a 3:1 concentration ratio (H2O2:DTT) and was confirmed to be complete by monitoring the absorbance at 283 nm (46, 47). After incubation, non-reducing SDS-PAGE loading buffer was added. Samples were then heated for 6 minutes and finally run on a 12% acrylamide gel. Gels were stained using Coomassie (R-250) and the protein bands were used to assess the relative amounts of reduced or oxidized PRL-1.
Phosphatase Activity Assays
A standard phosphatase activity assay with p-nitrophenyl phosphate (pNPP) was used to determine the relative activity of PRL-1 variants (35). All samples were exchanged at least 1000 fold by dialysis or using Amicon Ultra 10 kDa centrifugal filters (Millapore) into 50 mM HEPES, pH 7.5 before being tested for phosphatase activity. Assay mixtures contained 50 mM HEPES, pH 7.5, 20 mM p-nitrophenyl phosphate (pNPP-freshly prepared) and 0–10 mM reduced DTT. Reactions were initiated by the addition of PRL-1 at a final concentration of 1 mg/mL and monitored for absorption at 405 nm and 37°C using a Cary 100 UV-Vis spectrophotometer with temperature controller. Reactions were run in triplicate and on a minimum of two independently produced protein samples, totaling at least n=6 replicates. Relative activity was determined at 60 minutes by calculating the amount of p-nitrophenol produced (ε=1.78×10−4 M−1·cm−1) (48). Averages of the individual runs are shown.
NMR Experiments
For structural comparisons of PRL-1 variants, samples were prepared in 50 mM sodium phosphate buffer, 100 mM NaCl, pH 6.5, containing 5% D2O. 1H-15N Heteronuclear Single Quantum Coherence spectra were acquired on a Bruker Avance 800 MHz spectrometer using a cryogenic, triple resonance probe equipped with pulse field gradients. Water suppression was accomplished using flip-back pulses. The concentrations of PRL-1-WT and PRL-1-C170S-C171S were 1.0 mM and the spectra were acquired in 8 scans with 2048 points in 1H and 128* increments in 15N. The concentration of PRL-1-C98A was 3 mM and the spectra were acquired in 2 scans with 2048 points in 1H and 256* increments in 15N. Sample reduction was accomplished by the addition of 10 mM DTT at least 72 hours prior to spectral acquisition. All spectra were obtained at 37°C.
For the redox titration, the method previously published by Piotukh et al. (2007) was followed (49). Briefly, PRL-1-C98A was prepared in 50 mM sodium phosphate, 100 mM NaCl, pH 7.5, containing 5% D2O at a final concentration of 0.3 mM. As judged by SDS-PAGE, PRL-1-C98A was fully oxidized by adding 5 mM oxidized DTT (DTT oxidation was verified as described above). The sample was then titrated with increasing amounts of reduced DTT (1–200 mM) and a 1H-15N HSQC spectrum was taken at each point. Spectra were collected on a Bruker Avance 800 spectrometer using a cryogenic, triple resonance probe equipped with pulse field gradients. All spectra were obtained at 37°C and acquired in at least 16 scans with 2048 points in 1H and 256* increments in 15N. For both studies, 1H chemical shifts were referenced with respect to an external DSS standard in D2O. Indirect referencing relative to 1H was determined for 13C and 15N, assuming ratios 13C/1H = 0.251449530 and 15N/1H = 0.101329118 (50). nmrPipe and SPARKY were used for data processing and spectral analysis (51, 52).
Determining the Reduction Potential of PRL-1
Following the method by Piotukh et al. (2007) to determine the reduction potential of PRL-1, the heights of several well-resolved peaks in each NMR spectrum were obtained using SPARKY and plotted versus the corresponding halfcell potential of DTT (49). The half-cell potential of DTT was calculated according to the Nernst equation (42),
| (1) |
where R is the universal gas constant (8.3145 J K−1 mol−1), T is the temperature in Kelvin, n=2 for the two-electron reduction, and F is the Faraday constant (9.6485×104 C mol−1). For the reduced and oxidized DTT pair, the standard midpoint potential, E0’, was previously determined to be −332 mV at pH 7.0 (46). This value was adjusted to pH 7.5 according to the equation (42, 49)
| (2) |
The ΔE/ΔpH value for DTT was taken as −61.5 (42).
PRL-1 undergoes a structural transition upon reduction and sequence assignments for the reduced and oxidized species are published elsewhere (44, 45). This information was used to identify chemical shifts for the same residues in each state and to select reference peaks that have no change in chemical shift upon reduction (G123; 7.9 and 107.6 ppm and Y126; 9.3 and 124.3 ppm). The reference peaks were used to control for dynamics changes between the two spectra. The correction factors were averaged for both reference peaks and then applied to the peaks selected for statistical analysis. 10% of the backbone NH resonances that change intensity significantly upon reduction were selected on the basis that they were well-resolved and assigned in both the reduced and oxidized spectra. These peaks were used to calculate E0’ of PRL-1. The peak heights of individual resonances were normalized, plotted against the half-cell potential and fit to the following function using Graphpad Prism Software for non-linear curve fitting (Version 5) (49)
| (3) |
where A and B are the values for the initial and final plateaus of the sigmoidal decay function and p is the curve increase rate from the plot of each resonance. The resulting individual curves, depicting the relative amounts of the corresponding NH resonance in the reduced and oxidized states, intersect at 50% of the maximal intensity. This point defines the standard potential E’. 15 individual peaks were fit for an E’ value and then averaged to determine the reduction potential.
Results
Purified Full-length PRL-1-WT Is Inactive
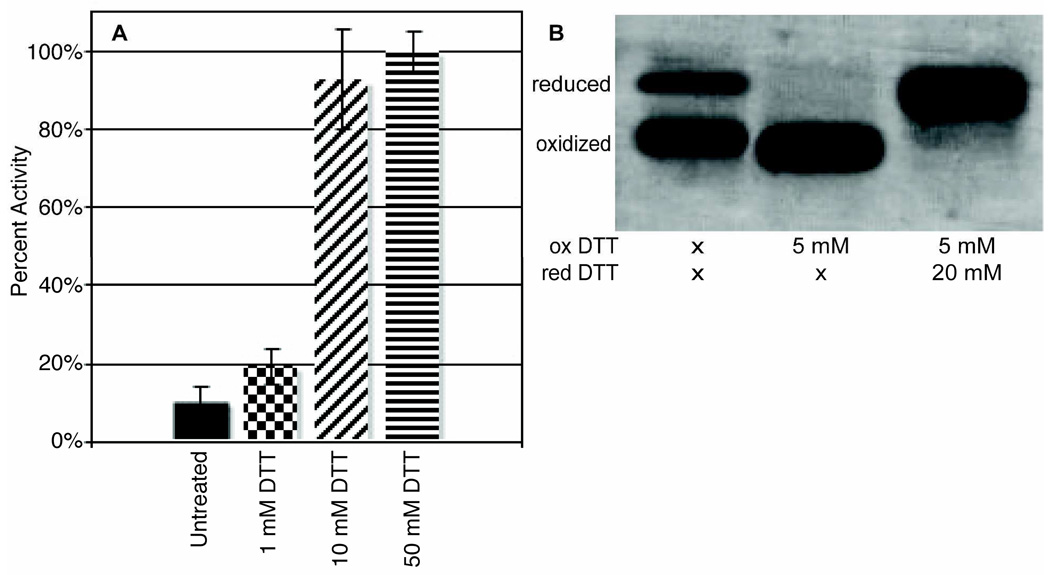
The pNPP activity assay performed on nonreduced, purified wild-type protein revealed that 90% of the protein is inactive under nonreducing conditions. As shown in Figure 1A, PRL-1 requires the addition of a thiol reducing agent for activity. Maximal activity is reached when 10 mM DTT is added to the solution of 1 mg/mL PRL-1. An eight-fold increase in activity is observed when 10 mM DTT is added to the assay mixture. As a negative control, the PRL-1-C104S mutant was constructed. This mutant is completely inactive because the nucleophilic thiol, which is essential for activity, has been removed. Under reducing conditions our activity data is comparable to the previously published kinetic constants, where truncated forms of PRL-1 were found to turn over generic PTPase substrates quite slowly (36).
Figure 1. PRL-1-WT.
A. The activity of PRL-1-WT was measured using a standard pNPP activity assay (see methods section). The amount of product formed at one hour was calculated and used to assess the relative activity of PRL-1-WT and PRL-1-C104S, an inactive form of PRL-1. The activity of PRL-1-C104S was identical to that of background pNPP (data not shown). Data are displayed as percent maximum activity (100%) based on the average of reduced PRL-1-C170S-C171S samples, following subtraction of the blank involving the inactive PRL-1-C104S mutant. PRL-1-WT requires 10 mM reducing agent for optimal activity. The activity of PRL-1-WT with 10 and 50 mM DTT were statistically identical, and no further increase in activity was observed when 100 and 200 mM reduced DTT were added (data not shown). B. 2 mg/mL PRL-1-WT was incubated with various amounts of oxidized or reduced DTT for approximately one hour. PRL-1 runs as two bands on non-reducing SDS-PAGE corresponding to reduced (upper) and oxidized (lower) forms of the protein (lane 1). The higher intensity of the lower band indicates that untreated PRL-1-WT is primarily oxidized. The addition of 5 mM oxidized DTT sufficiently oxidizes PRL-1-WT (lane 2). This was determined by titrating increasing amounts of oxidized DTT (0.01–10 mM) (data not shown). The addition of 20 mM reduced DTT to the reaction mixture recovers reduced PRL-1-WT (lane 3). The final oxidized to reduced DTT concentration ratio was 4:1. All samples contained 50 mM sodium phosphate buffer and 100 mM NaCl at pH 7.5.
Because activity was dramatically affected by the addition of reducing agents, we hypothesized that the lack of signal observed for the untreated PRL-1-WT was due to disulfide bond formation at the active site. To test this, we ran untreated PRL-1-WT on SDS-PAGE under non-reducing conditions. Disulfide bond formation can be visualized using non-reducing SDSPAGE (36). An oxidized protein, containing a disulfide bond, runs at a lower molecular weight than predicted because the sequence is more compact. When left untreated, wild-type PRL-1 runs as two bands on an SDS-PAGE gel, just below the 20-kDa marker (Figure 1B, lane 1). For wild-type PRL-1, the lower band is much more intense, corresponding to a high degree of oxidation. When run under oxidizing conditions, only the lower band is detected (Figure 1B, lane 2). The addition of four-fold excess of reduced DTT to the oxidized sample restores the upper band or reduced species. Disulfide bond formation in PRL-1 has been observed by SDS-PAGE previously, but these studies were performed on a truncated version of PRL-1, and hydrogen peroxide was used to accomplish oxidation (36). Because H2O2 also oxidizes methionine residues rapidly, we chose to use a thiol-specific chemistry for this study. This approach has the advantage of employing an equilibrium reaction and enables quantitative analysis. Because glutathione (GSH) is the primary redox buffer in vivo, these experiments were also performed using the GSH/GSSG redox couple as a thiol modulating agent. Although GSSG efficiently oxidized PRL-1 (data not shown), complete reduction by GSH was only accomplished at concentrations near 50 mM (Supporting Information, Figure 1). Because of this, DTT was used in all studies as both an oxidizing and reducing agent. As a control, the C104S mutant was also examined. At 50 mM oxidized DTT, no band corresponding to the 20 kDa oxidized species was observed (data not shown), indicating the band at 20 kDa reflects formation of a disulfide bond between C104 and C49. This is consistent with the findings of Sun et al., who examined both the C104S and C49S mutant by SDS-PAGE (36).
To confirm the formation and position of disulfide bonds in the untreated full-length protein, PRL-1 was analyzed using mass spectrometry. Several peaks were observed in the mass spectra of intact protein, which indicate wild-type PRL-1 is oxidized. A mass shift of +2 is observed in the spectra of proteins subject to chemical reduction compared to the oxidized samples, suggesting that one disulfide bond has been broken. To identify the specific sites at which oxidation occurs, PRL-1 was digested with trypsin and examined using LC-separated ESIMS under both reducing and oxidizing conditions. Digests of wild-type PRL-1 protein in nonreducing and reducing environments reveal differences in intensity for peaks associated with peptides containing C49 and C104. In the presence of oxygen, PRL-1-WT shows almost exclusively the presence of the C49- and C104-containing fragments joined by a disulfide bond (Supporting Information, Figure 2A). The disulfide-bound fragment is observed as a large m/z +1 peak at 3183.3 in the mass spectrum of oxidized PRL-1. The individual C49-containing fragment expected at m/z +1 1470.6 in oxidized PRL-1-WT is present only as a trace peak. These two fragments are easily distinguished from other peaks because they are located in unpopulated regions of the spectrum. In contrast, the 1714 m/z +1 signal calculated for the C104-containing fragment is buried under other peaks and cannot be easily distinguished among numerous overlapping signals for this sample. Chemical reduction of PRL-1 with reducing agent results in complete loss of signal at 3183.3, indicating the disulfide bond between the two peptides has been broken. Concomitantly, a large peak appears at 1470.6, which corresponds to the individual C49-containing peptide. Additionally, because the nucleophilic cysteine may be oxidized to sulfinic and sulfonic acids, MS data were mined for additional oxidation to the corresponding peptide with two and three oxygen atoms, respectively. Peaks at the predicted masses were not found in any of the samples, indicating that the redox inactivation of PRL-1 occurs predominantly through disulfide bond formation at the catalytic cysteine. A disulfide bond between C104 and C49 has also been reported for PRL-1 previously and is obvious in the crystal structure of the oxidized form (36, 39). Collectively, our data show that purified full-length PRL-1-WT is oxidized and, consequently, requires a reducing agent to restore activity.
Complete Oxidation of Full-length PRL-1-WT Leads to Precipitation
NMR can be used to determine the reduction potential of a protein (49). An accurate calculation of the reduction potential of PRL-1 requires an accurate measurement of the NMR peak heights that correspond to the reduced and oxidized species. This necessitates conversion to either a completely oxidized or reduced species in order to perform a complete titration and to have an accurate reference baseline. We chose to completely oxidize PRL-1 because it is predominately oxidized following purification without treatment of oxidizing agents. Unexpectedly, incubation of wild-type PRL-1 with oxidizing agents (H2O2, GSSG and/or oxidized DTT) resulted in complete precipitation of the protein at one hour. Attempts to re-oxidize PRL-1 following reduction with these components also led to precipitation (data not shown). Because the NMR experiments necessary to complete a redox titration require the protein to remain stable and soluble for a longer period of time, the wild-type protein was not a suitable candidate for this study.
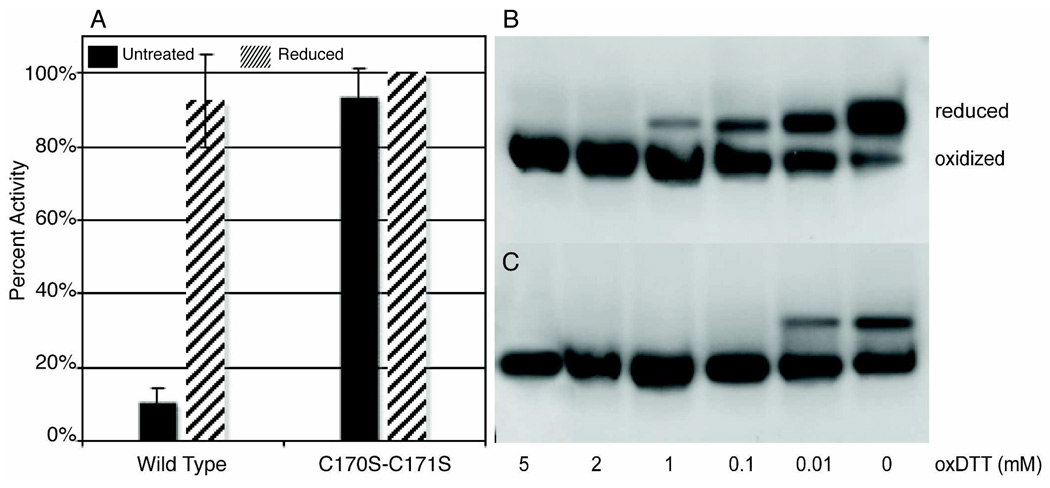
Previous studies of PRL-1 indicated that the C-terminus affects solubility (36, 39). As such, we anticipated that precipitation could be avoided by using a C-terminal mutant, to perform the redox titration. We first investigated the activity of the PRL-1-C170S-C171S mutant and performed a pNPP activity assay. In stark contrast to untreated wild-type PRL-1, the untreated C170S-C171S mutant exhibits an eight-fold increase in activity (Figure 2A). The activity of PRL-1-C170S-C171S is statistically identical to wild-type PRL-1 under reducing conditions, and the activity of this mutant is not modulated by the addition of reducing agents. Taken together, these data indicate that the active-site cysteine in the C170S-C171S mutant remains primarily reduced. To confirm this, we performed a trypsin digest and analyzed the peptides by mass spectrometry. The digest of untreated PRL-1-C170S-C171S shows disulfide-linked peptides only at equivalent intensity to background (Supporting Information, Figure 2B), suggesting no specific disulfide formation between the C49 and C104-containing peptides occurs. Conversely, the individual C49-containing peptide is exclusively detected. The MS spectrum for PRL-1-C170S-C171S is identical under non-reducing and reducing conditions, as no change in the signal for either mass is observed following addition of reducing agent.
Figure 2. PRL-1-C170S-C171S.
A. The activity of PRL-1-C170S-C171S was measured as described in Figure 1. The activity of PRL-1-C170S-C171S is not modulated by the addition of DTT and is statistically identical under both non-reducing and reducing conditions. There is also no statistically significant difference in activity between the reduced wild type and PRL-1-C170S-C171S mutant, indicating that this mutant is primarily in the reduced state even in the absence of thiol modulating agents. B. PRL-1-C170S-C171S was run on SDS-PAGE under various oxidizing conditions (0–5 mM oxidized DTT). Under non-reducing conditions (lane 6), C170S-C171S runs as two bands. In contrast to wild type, however, C170S-C171S is primarily reduced as indicated by the much greater intensity of the upper reduced band. The small amount of oxidized protein is reduced upon addition of 3 mM DTT (data not shown). Complete oxidation of PRL-1-C170-171S occurs near 2 mM oxidized DTT. C. PRL-1-WT was run on SDS-PAGE under the same conditions as in B. Complete oxidation of wild type occurs near 0.1 mM oxidized DTT. In both B and C panels, samples were prepared as in figure 1.
We next investigated the ability to oxidize PRL-1-C170S-C171S. When left untreated, PRL-1-C170S-C171S also runs as two bands on an SDS-PAGE gel; however, the upper band, which corresponds to reduced PRL-1, is much more intense than the lower band (Figure 2B-lane 6). We attempted to oxidize PRL-1-C170S-C171S. Based on the intensity of the reduced and oxidized bands on SDS-PAGE, we estimated that this mutant requires at least 20-fold more oxidizing agent than WT (Figure 2, compare panel B to C). This finding indicates that PRL-1-C170S-C171S is much less susceptible to oxidation than WT. We did not attempt to quantify the reduction potential of this protein because, surprisingly, incubation of PRL-1-C170S-C171S with oxidizing agents also results in complete precipitation of the protein at one hour (data not shown). Instead we pursued identification of a PRL-1 variant that would allow us to determine the active-site reduction potential of full-length PRL-1. We investigated the role of the remaining two Cysteines, C98 and C99, in PRL-1 solubility. The crystal structure of PRL-1 reveals that C98 and C99 are largely buried in the hydrophobic core of the protein, but the side chain of C98 is partially surface-accessible. We hypothesized that oxidation of C98 might lead to formation of a second disulfide bond and cause a structural transition, leading to precipitation. To investigate this hypothesis, we mined the NMR and MS data more closely.
The Cβ NMR chemical shift of a cysteine residue can be used to deduce the oxidation state of the thiol moiety. The chemical shift of an oxidized side chain (disulfide bound) should have a downfield shift closer to that of the Cα resonance (53). The Cβ chemical shift of C98 in untreated wild-type PRL-1 indicates that this residue is reduced (26.449 ppm). For comparison, the Cβ chemical shifts of the disulfide bound C49 and C104 residues are 35.21 ppm and 36.36 ppm, respectively. Reduction of PRL-1 leads to a large change in Cβ chemical shifts for the C49 and C104 residues (34.01 ppm and 29.82 ppm, respectively). Cβ chemical shift information is unavailable for residue C99 in the oxidized state and is likely absent due to chemical exchange (45).
To further investigate the propensity of C98 and C99 to be oxidized we examined the MS data of wild type and PRL-1-C170S-C171S. Digested PRL-1-C170S-C171S shows a strong signal for the peptide containing C104 at 1712. The observed value is 2 amu below the predicted mass of 1714. This mass, which is shifted by −2, also is consistently observed in the spectra of the cross-linked species involving the C49- and C104-containing peptides (3181.3) derived from non-reduced wild-type PRL-1. Addition of reducing agent shifts the mass of the C104-containing peptide to 1714 for both wild type and PRL-1-C170S-C171S. Because the C104-containing peptide also includes C98 and C99, it is likely that an intra-peptide disulfide bond between C98 and C99 is also present. MS analysis of intact PRL-1 under non-reducing conditions reveals a peak 4 amu below the calculated mass, and this peak shifts to the expected position in the MS spectrum upon addition of reducing agent when no LC separation is performed. As such, it appears that this additional disulfide bond forms after the reducing agent is removed, likely during the LC step.
PRL-1-C98A Does Not Precipitate and Is Structurally and Functionally Equivalent to PRL-1-WT
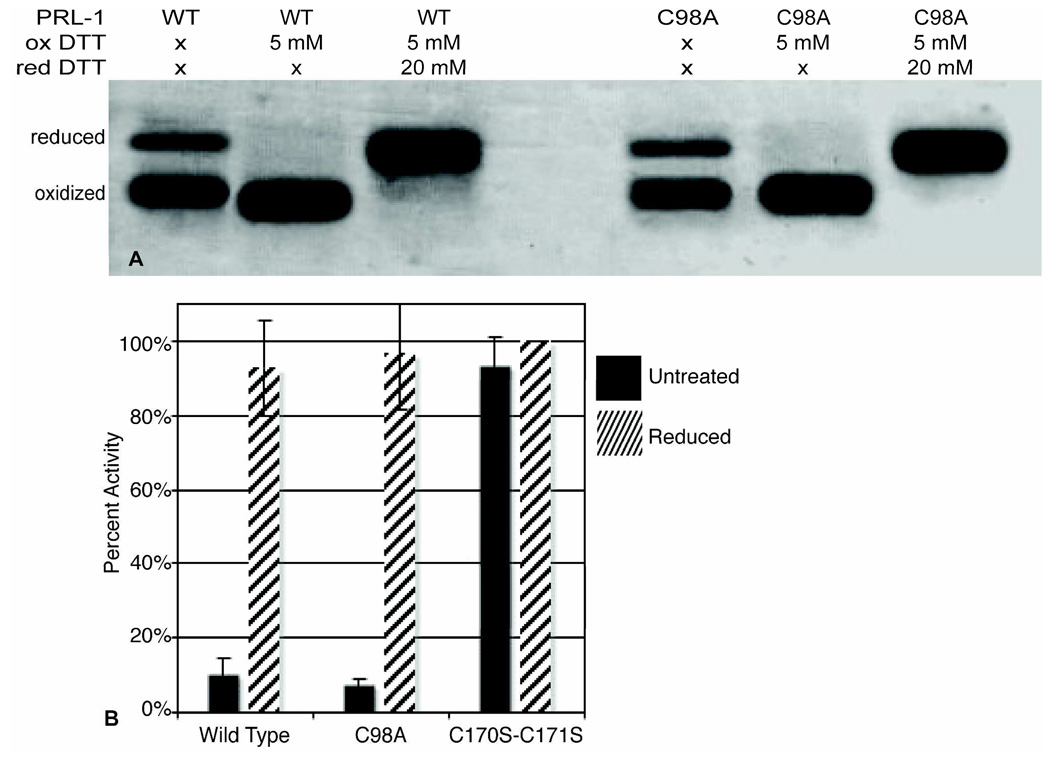
Because the MS data indicate the possibility of C98-C99 disulfide bond formation upon oxidation, we created the PRL-1-C98A mutant. We hypothesized that chemical oxidation by thiol modulating agents leads to oxidation of the C98 and C99 residues in both PRL-1-WT and PRL-1-C170S-C171S, which results in precipitation. Mutation of C98 mitigates this problem. Untreated PRL-1-C98A runs similarly to PRL-1-WT on SDS-PAGE (Figure 3A, compare lane 1 to 4). Like wild-type PRL-1, complete oxidation of PRL-1-C98A occurs with 5 mM oxidized DTT and is re-reduced by addition of a four-fold excess of reduced DTT (Figure 3A, compare lane 2 to 5 and 3 to 6). Unlike wild type, incubation of C98A with oxidizing agents for extended periods of time results in little to no precipitation. Consequently, PRL-1-C98A was found to be an appropriate candidate to use for a reversible, equilibrium redox titration. To ensure that this mutation has not compromised the functional or structural integrity of the protein, we compared the PTPase activity and NMR spectra of PRL-1-WT to PRL-1-C98A.
Figure 3. PRL-1-C98A.
A. Lanes 1–3 are the same samples from the gel shown in Figure 1B and are included here for ease of comparison. Lanes 4–6 were run under the same conditions as described for Figure 1. PRL-1-C98A behaves much like PRL-1-WT. When left untreated, it runs as two bands corresponding to both reduced (upper) and oxidized (lower) forms. Oxidation of PRL-1-C98A also occurs in 5 mM oxidized DTT. This was determined in the same manner as for wild type. Re-reduction of PRL-1-C98A occurs at the same ratio (4:1) as PRL-1-WT. B. The measured activities of PRL-1-WT and PRL-1-C98A are equivalent within error prior to addition of reducing agent, and each is completely activated by the addition of 10 mM reduced DTT. This is in contrast to the PRL-1-C170S-C171S mutant, which is not modulated by the addition of reducing agent. The activities of reduced WT, C98A and C170S-C171S are identical within error. The activity of PRL-1-C98A under oxidizing conditions (5 mM oxidized DTT) was equivalent to that of PRL-1-C104S (data not shown).
Wild type and C98A exhibit almost identical substrate turnover when assayed in equivalent redox conditions. PRL-1-C98A shows an equivalent level of activity when left untreated. Oxidation of the protein with 5 mM oxidized DTT completely abolishes the activity of the enzyme (data not shown), while reduction with 10 mM DTT recovers the full activity of this mutant, as shown by comparison to the reduced wild type and C170S-C171S mutant (Figure 3B). The percent activity of PRL-1-C98A under oxidizing conditions was determined to be 0.05 ± 2% and was not included in the figure. Activity of the wild-type protein under oxidizing conditions was not measured because the formation of the precipitant precludes accurate measurement of the assay product.
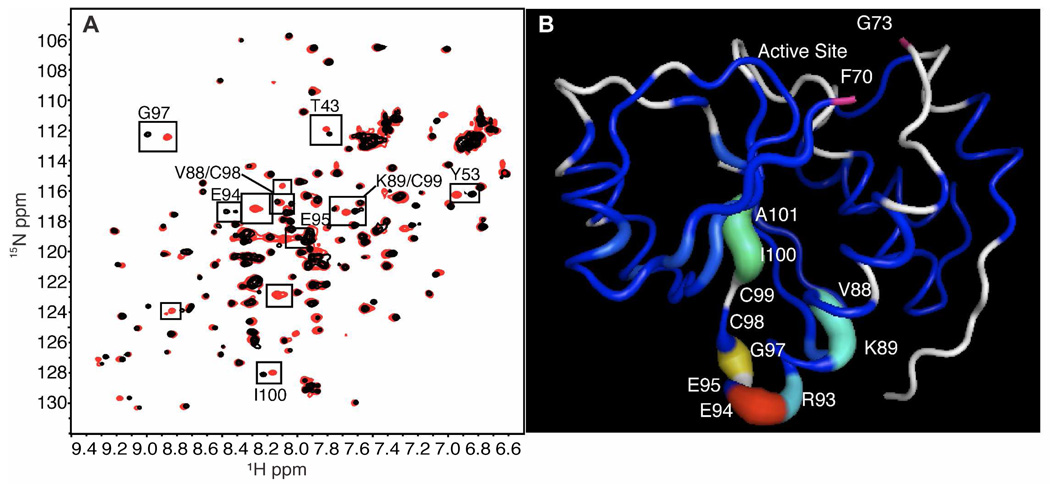
To verify the C98A mutation does not significantly alter the protein structure, we collected 1H-15N HSQC spectra of wild type and PRL-1-C98A. The HSQC experiment is particularly useful for the analysis of protein structure because the experiment selectively detects those protons directly coupled to nitrogen. One cross peak is observed for every amide in the protein at a position characterized by its 1H and 15N chemical shifts. The HSQC spectrum provides an overall map or fingerprint of the protein backbone and serves as the basis for assessing the overall fold and behavior of a protein. Because chemical shift is so sensitive to environment, changes in a protein’s tertiary structure are reflected by changes in the peak positions in the HSQC spectrum (54). Comparison of the HSQC spectra of wild type and PRL-1-C98A reveals relatively small changes between the two variants (Figure 4A). As such, the overall structure of PRL-1-C98A and PRL-1-WT are virtually identical, as indicated by the large number of peaks in common in these two spectra. Based on the previously reported assignments for full-length oxidized PRL-1-WT and the previously published crystal structure of truncated PRL-1-WT (36, 45), significant chemical shift changes correspond to residues located near the site of the mutation (Figure 4A, black boxes and Figure 4B). The average chemical shift deviation (Δδavg) for PRL-1-WT and PRL-1-C98A was calculated by subtracting the chemical shift values of the C98A mutant from those of the wild type and was determined to be 0.07 ppm for 15N and 0.01 ppm for 1H. It should be noted that several new peaks appear in the spectrum of PRL-1-C98A that are not observed for wild type. Peaks for many of the C-terminal residues in PRL-1-WT were absent from the HSQC spectrum, likely because of line broadening due to exchange on the intermediate timescale. We suspect the new peaks in the PRL-1-C98A spectrum are resonances from the C-terminus, and their appearance may reflect a faster rate of exchange on which the motions at these residues occur in the PRL-1-C98A protein. Sequential backbone assignments of the C98A mutant and complete dynamics analysis of both are needed to confirm this, which is beyond the scope of this paper.
Figure 4. Comparisons of PRL-1-WT and C98A.
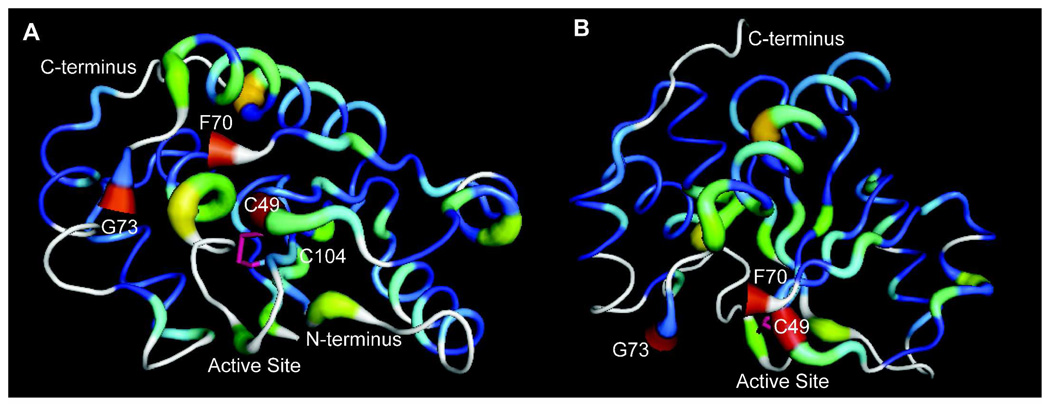
A. The spectrum of untreated PRL-1-WT (red) is overlaid with untreated PRL-1-C98A (black). Small differences in structure exist between these two proteins, and the residues that change significantly with mutation are enclosed in boxes. Unlabeled boxes enclose either new peaks in C98A (black) or unassigned peaks in wild type (red). B. The chemical shift changes observed between these two proteins were mapped to the available crystal structure of the oxidized wild type. A break in the structure exists between residues F70 and G73 because no information is available for D71 and D72 residues. F70 and G73 are labeled and colored in pink to illustrate the break. For chemical shift mapping, the Δδ value for all assigned residues was used as the b-factor in the pdb file (1ZCK). Δδ was determined by subtracting the reduced chemical shift from that of the oxidized. Residues for which an assignment is missing in one of the two states are colored white. The rainbow color scale is applied with the largest changes being highlighted in red and insignificant changes in blue. Additionally, larger changes in chemical shift correspond to an increase in tube diameter. This figure was generated using Pymol.
We previously reported that reduction of PRL-1 results in a conformational change, causing greater than 90% of the peaks in the wild-type spectrum to move. We obtained backbone chemical shift assignments for both the oxidized and reduced forms, which are reported elsewhere (44, 45). The PRL-1-C170S-C171S was used to assign the reduced state because active site residues were absent from or too weak to assign in the spectra of the chemically reduced wild type. The most dramatic change in the spectrum that occurs upon reduction of PRL-1 corresponds to C49. The position of C49 in the HSQC shifts significantly upfield by more than 9 ppm in nitrogen and 1 ppm in proton compared to the oxidized form. Figure 5 maps the observed changes in chemical shifts (Δδ) of assigned resonances to the available crystal structure of oxidized PRL-1. Δδ values were obtained by subtracting the chemical shift of PRL-1-C170S-C171S from those of wild-type PRL-1 (Supporting Information, Table 1). A Δδavg of 0.91 ppm for 15N and 0.24 ppm for 1H was determined for these two forms of the protein. Parallel changes in chemical shifts are observed upon reduction of PRL-1-C98A (Supporting Information, Figure 3). The Δδavg value was determined to be 0.72 for 15N and 0.14 for 1H by subtracting the chemical shifts of the reduced protein from those of the oxidized protein, which is in strong agreement between that observed for wild type and PRL-1-C170S-C171S and confirms that redox modulation at the active site in C98A parallels that of wild type. Collectively, our structural and functional comparisons have shown that PRL-1-C98A is an appropriate analog of the PRL-1 system, from which the reduction potential of the active-site disulfide bond can be determined.
Figure 5. Comparisons of PRL-1-WT and C170S-C171S.
A. Differences in NMR chemical shifts between PRL-1-WT and PRL-1-C170-C171S were mapped to the crystal structure (as in figure 4) and displayed with the active site in front. The disulfide bound (pink bond) C104 and C49 are labeled. F70 and G73 are labeled to identify the break in the crystal structure. B. As in A, but with the active site rotated downward by approximately 90 degress to reveal the large number of chemical shift deviations in the majority of the protein structure, especially in the central β-strand. Both figures were generated using Pymol.
Determining the Reduction Potential of the C49-C104 Disulfide Bond in Full-length PRL-1
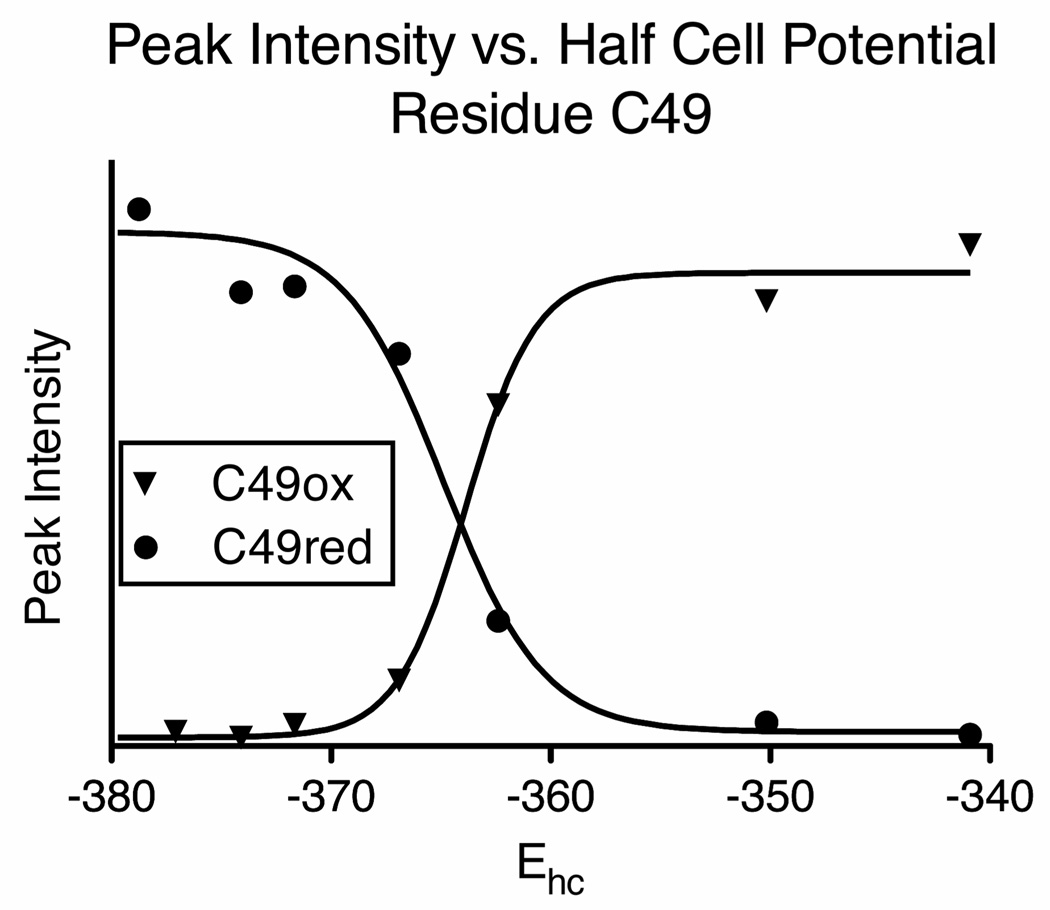
To determine the reduction potential, oxidized PRL-1 (in the presence of 5 mM oxidized DTT) was gradually reduced by the addition of reduced DTT to a concentration of 200 mM. The peak heights of the following residues were monitored and analyzed as described: V12, R18, L30, T43, R47, C49, V65, L66, I80, R93, I100, A101, L114, E127, and Y152. Peak selection was based on spectral resolution, proximity to the disulfide bond, distance from mutation site and chemical exchange behavior (slow exchanging peaks only). Figure 6 displays a representative plot, depicting the results of the titration on residue C49, in which normalized peak height is plotted vs. half-cell potential. Similar results were obtained for all other residues and the chemical shifts of each residue in both states are included in Table 1. An average E’ value of −364.3 ± 1.5 mV was calculated for the C49-C104 disulfide bond using the data from 15 residues.
Figure 6. Representative Plot of Reduction Potential Derived From Residue C49.
The peak heights of residue C49 were normalized and plotted versus the half-cell potential of DTT, which was determined using the Nernst equation. The value determined for this residue was −364.4 ± 0.7 mV. Similar results were obtained for other residues and are listed in Table 1.
Table 1.
Calculated Reduction Potential for PRL-1 Residuesa
| Residue | E' (mV) | ± SD | δ-15N ox | δ-1H ox | δ-15N red | δ-1H red | Δδ (15N) | Δδ (1H) |
|---|---|---|---|---|---|---|---|---|
| V12 | −366.1 | 0.0 | 126.780 | 9.122 | 126.771 | 9.202 | 0.009 | −0.080 |
| R18 | −364.6 | 2.0 | 121.848 | 5.764 | 121.919 | 5.837 | −0.071 | −0.073 |
| L30 | −364.0 | 1.0 | 124.349 | 7.359 | 124.543 | 7.302 | −0.194 | 0.057 |
| T43 | −363.5 | 0.0 | 111.911 | 7.806 | 112.654 | 7.908 | −0.743 | −0.102 |
| R47 | −361.8 | 0.0 | 126.631 | 8.861 | 129.143 | 8.896 | −2.512 | −0.035 |
| C49 | −364.4 | 0.7 | 121.676 | 8.037 | 112.254 | 6.943 | 9.422 | 1.094 |
| V65 | −365.2 | 0.0 | 124.996 | 8.563 | 125.651 | 8.570 | −0.655 | −0.007 |
| L66 | −364.8 | 0.0 | 130.322 | 9.065 | 130.756 | 8.853 | −0.434 | 0.212 |
| I80 | −364.8 | 1.3 | 119.137 | 7.235 | 119.000 | 6.994 | 0.137 | 0.241 |
| R93 | −364.7 | 2.3 | 118.477 | 7.287 | 118.344 | 7.236 | 0.133 | 0.051 |
| I100 | −364.9 | 2.4 | 127.986 | 8.162 | 127.919 | 8.230 | 0.067 | −0.068 |
| A101 | −364.8 | 1.8 | 129.683 | 9.180 | 129.252 | 8.865 | 0.431 | 0.315 |
| L114 | −365.4 | 2.6 | 116.028 | 7.424 | 117.798 | 7.107 | −1.770 | 0.317 |
| E127 | −363.9 | 1.6 | 118.621 | 9.219 | 119.105 | 9.187 | −0.484 | 0.032 |
| Y152 | −362.7 | 1.9 | 122.770 | 7.129 | 123.362 | 7.099 | −0.592 | 0.030 |
| average | −364.3 | 1.5 | ||||||
The reported E’ value for each residue corresponds to the average of both the reduced and oxidized peak heights fit to the equation listed in the methods section. The standard deviation (SD) is also reported in mV. Δδ values were calculated by subtracting the chemical shift of the reduced species from those of the oxidized species.
Discussion
The reduction potential of the active site disulfide bond in full-length PRL-1 was determined to be approximately −365 mV at pH 7.5. The reported reduction potentials determined for various cellular environments range from −320 (most reducing) to −170 mV (most oxidizing), the details of which have been reviewed extensively (42, 55–58). Based on these values, our results indicate that nascent PRL-1 is rapidly oxidized inside the cell to an inactive state. The reduction potential is considerably lower than those reported for proteins that are known to play an important role in maintaining the redox state of the cell and/or specific proteins. The reduction potential of thioredoxin-1 (Trx-1), found in the cytoplasm and nucleus, has been reported to be −230 mV, while that of Trx-2, found in the mitochondria, is near −330 mV (55, 59). Both enzymes would have a substantial fraction of protein in the active state in their respective environments, and Trx-1 would be able to act as a reducing agent more effectively in the nucleus. Reduction potentials for cytoplasmic glutaredoxins (Grx) vary depending on the primary sequence and range from −200 to −240 mV (60). Reduction potentials for proteins that facilitate disulfide bond formation in the endoplasmic reticulum have been determined to be −175 mV (protein disulfide isomerase; PDI) and −275 mV (regulatory disulfide of Ero1α) (61, 62). The role of PDI is to make and break disulfide bonds in target proteins and the reduction potential is appropriate for this enzyme in this compartment. The reduction potential of Ero1α is sufficiently negative to ensure oxidation and facilitate re-oxidation of PDI. To date, analysis of reduction potentials has been focused on a small subset redox modulating enzymes, but measuring the reduction potentials of other proteins that are subject to redox regulation has recently been recognized as important as well. The absence of general and accurate methods for quantifying these potentials has precluded the ability to identify conditions in which proteins are switched on or off by disulfide bond formation. NMR spectroscopy has recently been shown to be a valuable tool for obtaining this information and was used to determine the reduction potential of Trx-1 with great accuracy (49). Other proteins have been studied using this method as well. Zimmerman et al. used NMR to determine the reduction potential of the N-terminal hSH3 domain of the immune cell protein ADAP (adhesion and degranulation promoting adapter protein) (63). Sahu et al. used NMR to quantify the reduction potential of the High Mobility Group B1 (HMGB1) protein, which plays an important role in DNA binding as well as chemotaxis (64). In both examples, disulfide bond formation was shown to lead to a conformational change, which alters the function of the protein. The altered function of the protein also correlates with a change in subcellular location. Likewise, the subcellular location of PRL-1 may affect the activity of the protein, although no cellular compartment is sufficiently negative to ensure activation. Even in the most reducing environment reported (−320 mV), nascent PRL-1 would exist primarily in an inactive, oxidized state (> 96%). This strongly suggests that in vivo, PRL-1 likely requires a posttranslational modification or binding partner to switch on phosphatase activity.
The data presented here demonstrate the importance of the C-terminus to the phosphatase activity of PRL-1 and show that modification of the C-terminus affects the redox state of the active site, making PRL-1 much less susceptible to inactivation (Figure 2). The PRL enzymes each contain a C-terminal prenylation motif or CaaX box (C stands for cysteine, a represents any aliphatic residue and X corresponds to any amino acid). The prenylation motif directs farnesylation at the conserved CaaX box cysteine, followed by proteolytic cleavage of the aaX peptide and methylation of the terminal carboxyl moiety (65). Farnesylation has been shown to occur in vivo for the PRL enzymes (6, 9, 14). In PRL-1, this modification occurs at C170. Based on our data, the shift in the reduction potential of the inactivating disulfide bond in PRL-1 to a less negative value appears to be accomplished through a conformational change that depends on the presence of C-terminal residues in the CaaX box. Because farnesylation occurs on C170 and mutation of this residue makes PRL-1 less prone to inactivation, we expect farnesylation will also alter the protein’s conformation and reduction potential of the active site disulfide bond. Because farnesylation often leads to membrane association and PRL-1 has been shown to localize to several membranes in vivo (6, 8, 14, 16, 36, 39), the protein’s conformation may be altered more substantially in the context of a lipid bilayer than is apparent from solution studies.
In all work published to date, the phosphatase activity of PRL-1 has been reported to be necessary for the promotion of cellular proliferation and motility by this protein, regardless of localization (7–9, 26, 30, 33, 34). PRL-1 is typically farnesylated in cells (6, 9, 14, 24) and farnesylated PRL-1 is most often directed to the nucleus (9, 13, 43). The fact that PRL-1 is most often localized to the nucleus seems appropriate with respect to cellular redox conditions (2,5,10), because this compartment is often the most reducing organelle at −320 mV (55). Based on our studies of the C-terminal mutant, it seems that modification at the C-terminus would likely raise the reduction potential sufficiently to turn on phosphatase activity in this environment. Interestingly, when farnesylation of PRL-1 is prevented in cells, the ability of this protein to promote proliferation is abrogated and its localization is often altered. For example, the lack of prenylation inhibits the ability of PRL-1 to promote invasion and motility of SW480 colon carcinoma cells and to promote HEK293 cell growth and migration (8, 24). In these cases, it was not determined whether localization and/or PTPase activity is responsible for the altered phenotype. In mitotic cells, however, PRL-1 localizes to the centrosomes and spindle apparatus independent of whether farnesylation occurs, and despite proper localization, farnesylation deficient mutants result in mitotic defects (9). Interestingly, the inactive mutant produced the same result. During mitosis, the nuclear envelope breaks down and the reduction potential of the cell becomes approximately −240 mV (42, 66, 67). The reduction potential of unmodified PRL-1 (−365 mV) is much too low to be active in this redox environment. As such, farnesylation may enhance PRL-1 activity during mitosis, and this may explain why mitotic defects were observed with both the catalytically inactive C104 mutant and non-farnesylated proteins.
PRL-1 facilitates the regeneration of damaged tissue by promoting proliferation and migration, notably in the liver, which has a highly reducing environment. Increased activity in this case may be due, at least in part, to elevated expression of the protein (2). The expression and phosphatase activity of PRL-1 have also been associated with many types of cancer cell lines (12, 13, 16, 17, 23). It has been well documented that cancer cells are generally more reducing than their normal counterparts (68–70). Because cancer cells have aberrantly regained the ability to proliferate and have lost the ability to maintain redox homeostasis (42), we hypothesize that the redox environment in some cancer cells may be sufficiently negative to confer activity in PRL-1. This is supported by the fact that abolishing the activity of PRL-1 in cancer cell lines by transfecting them with catalytically inactive PRL mutants decreases cell viability, prevents cells from entering into mitosis and also inhibits anchorage independent growth (8, 9). These and other studies collectively suggest that the active form of PRL-1 may be a good target for therapeutic intervention in the treatment of metastatic cancers (12).
It is clear from our in vitro studies that changes in cellular redox conditions will have a profound affect on the activity of PRL-1 in vivo and that modification of residues, especially at the C-terminus, may dramatically alter the propensity of PRL-1 to be oxidized and inactivated inside the cell. Given its diverse localization and multifarious roles in proliferation, differentiation and motility, the data presented here provides the impetus to pursue further studies that investigate the function of PRL-1 under distinct cellular conditions in order to generate a more accurate account of how PRL-1 is regulated in vivo and how its regulation contributes to such diverse cellular outcomes.
Supplementary Material
Supporting information for this work is available free of charge via the Internet at http://pubs.acs.org. This information illustrates the ability of PRL-1-WT to be reduced by GSH (Figure 1), MS data for both PRL-1-WT and PRL-1-C170S-C171S (Figure 2), and 1H-15N HSQCs of PRL-1-C98A under non-reducing and reducing conditions (Figure 3). Additionally, chemical shift information for oxidized (WT) and reduced (C170S-C171S) is provided in Table 1.
Acknowledgements
The authors would like to thank Dr. Dave Vander Velde for his assistance with the NMR spectrometer and Dr. Krista Shipley for helpful discussion of the manuscript.
Abbreviations
- PRL
phosphatase of regenerating liver
- PTPase
protein tyrosine phosphatase
- NMR
nuclear magnetic resonance
- ESI-MS
electrospray ionization mass spectrometry
- RP-HPLC
reverse-phase high performance liquid chromatography
- MeOH
methanol
- BME
β-mercaptoethanol
- DTT
dithiothreitol
- pNPP
para-nitrophenyl phosphate
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HSQC
Heteronuclear Single Quantum Coherence
- DSS
2, 2-dimethylsilapentane-5-sulfonic acid
- WT
wild type
- GSH
glutathione
- GSSG
glutathione disulfide
- ROS
reactive oxygen species
- NLS
nuclear localization signal
Footnotes
This publication was made possible by NIH grant number P20 RR-17708 from the National Center for Research Resources and the Kansas University Center for Research. This work was additionally supported by fellowships for Andria Skinner from Amgen and the Edith and Eleta Ernst Cancer Research Fellowship. The Q-Tof2tm was purchased with support from KSTAR, Kansas administered NSF EPSCoR and the University of Kansas. The CapXL HPLC was obtained with support by KCALSI (www.kclifesciences.org).
References
- 1.Chiarugi P, Buricchi F. Protein tyrosin phosphorylation and reversible oxidation: two cross-talking posttranslation modifications. Antiox and Redox Signal. 2007;9:1–24. doi: 10.1089/ars.2007.9.1. [DOI] [PubMed] [Google Scholar]
- 2.Diamond RH, Cressman DE, Laz TM, Abrams CS, Taub R. PRL-1, a unique nuclear protein tyrosine phosphatase affects cell growth. Mol Cell Biol. 1994;14:3752–3762. doi: 10.1128/mcb.14.6.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takano S, Fukuyama H, Fukumoto M, Kimura J, Xue J-H, Ohashi H, Fujita J. PRL-1, a protein tyrosine phosphatase, is expressed in neurons and oligodendrocytes in the brain and induced in the cerebral cortex following transient forebrain ischemia. Mol Brain Res. 1996;40:105–115. doi: 10.1016/0169-328x(96)00035-6. [DOI] [PubMed] [Google Scholar]
- 4.Rundle CH, Kappen C. Developmental expression of the murine Prl-1 protein tyrosine phosphatase gene. J Exp Zoo. 1999;283:612–617. doi: 10.1002/(sici)1097-010x(19990501)283:6<612::aid-jez14>3.3.co;2-o. [DOI] [PubMed] [Google Scholar]
- 5.Diamond RH, Peters C, Jung SP, Greenbaum LE, Haber BA, Silberg DG, Traber PG, Taub R. Expression of PRL-1 nuclear PTPase is associated with proliferation in liver but with differentiation in intestine. Am J Physiol Gastrointest Liver Physiol. 1996;271:G121–G129. doi: 10.1152/ajpgi.1996.271.1.G121. [DOI] [PubMed] [Google Scholar]
- 6.Zeng Q, Si X, Horstmann H, Xu Y, Hong W, Pallen CJ. Prenylationdependent association of protein-tyrosine phosphatases PRL-1,-2, and -3 with the plasma membrane and the early endosome. J Biol Chem. 2000;275:21444–21452. doi: 10.1074/jbc.M000453200. [DOI] [PubMed] [Google Scholar]
- 7.Werner SR, Lee PA, DeCamp MW, Crowell DN, Randall SK, Crowell PL. Enhanced cell cycle progression and down regulation of p21Cip1/Waf1 by PRL tyrosine phosphatases. Cancer Lett. 2003;202:201–211. doi: 10.1016/s0304-3835(03)00517-2. [DOI] [PubMed] [Google Scholar]
- 8.Sun J-P, Luo Y, Yu X, Wang W-Q, Zhou B, Liang F, Zhang Z-Y. Phosphatase activity, trimerization, and the C-terminal polybasic region are all required for PRL1-mediated cell growth and migration. J Biol Chem. 2007;282:29043–29051. doi: 10.1074/jbc.M703537200. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Kirby CE, Herbst R. The tyrosine phosphatase PRL-1 localizes to the the endoplasmic reticulum and the mitotic spindle ad is required for normal mitosis. J Biol Chem. 2002;277:46659–46668. doi: 10.1074/jbc.M206407200. [DOI] [PubMed] [Google Scholar]
- 10.Kong W, Swain GP, Li S, Diamond RH. PRL-1 PTPase expression is developmentally regulated with tissue-specific patterns in epithelial tissues. Am J Physiol Gastrointest Liver Physiol. 2000;279:613–621. doi: 10.1152/ajpgi.2000.279.3.G613. [DOI] [PubMed] [Google Scholar]
- 11.Yarovinsky TO, Rickman DW, Diamond RH, Taub R, Hageman GS, Rickman CB. Expression of the protein tyrosine phosphatase, phosphatase of regenerating liver 1, in the outer segments of primate cone photoreceptors. Mol Brain Res. 2000;77:95–103. doi: 10.1016/s0169-328x(00)00045-0. [DOI] [PubMed] [Google Scholar]
- 12.Stephens BJ, Han H, Gokhale V, Von Hoft DD. PRL phosphatases as potential molecular targets in cancer. Mol Cancer Ther. 2005;4:1653–1661. doi: 10.1158/1535-7163.MCT-05-0248. [DOI] [PubMed] [Google Scholar]
- 13.Bessette DC, Qiu D, Pallen CJ. PRL PTPs: mediators and markers of cancer progression. Cancer Metastasis Rev. 2008;27 doi: 10.1007/s10555-008-9121-3. [DOI] [PubMed] [Google Scholar]
- 14.Cates CA, Michael RL, Stayrook KR, Harvey KA, Burke YD, Randall SK, Crowell PL, Crowell DN. Prenylation of oncogenic human PTPcaax protein tyrosine phosphatases. Cancer Lett. 1996;110:49–55. doi: 10.1016/s0304-3835(96)04459-x. [DOI] [PubMed] [Google Scholar]
- 15.Wang Q, Holmes DLR, Powell SM, Lu QL, Waxman Analysis of stromal-epithelial interactions in prostate cancer identifies PTPCAAX2 as a potential oncogene. Cancer Lett. 2002;175:63–69. doi: 10.1016/s0304-3835(01)00703-0. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Li Z-F, He J, Li Y-L, Zhu G-B, Zhang L-H, Li Y-L. Expression of human phosphatases of regnerating liver (PRLs) in colonic adenocarcinoma and its correlation with lymph node metastasis. In J Colorectal Dis. 2007;22:1179–1184. doi: 10.1007/s00384-007-0303-1. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y-Q, Li H-X, Lou X, Lei J-Y. Expression of phosphatase of regnerating liver 1 and 3 mRNA in esophageal squamous cell carcinoma. Arch Pathol Lab Med. 2008;132:1307–1312. doi: 10.5858/2008-132-1307-EOPORL. [DOI] [PubMed] [Google Scholar]
- 18.Miskad UA, Semba S, Kato H, Matsukawa Y, Kodama Y, Mizuuchi E, Maeda N, Yanagihara K, Yokozaki H. High PRL-3 expression in human gastric cancer is a marker of metastasis and grades of malignancies: an in situ hybridization study. Virchows Arch. 2007;450:303–310. doi: 10.1007/s00428-006-0361-8. [DOI] [PubMed] [Google Scholar]
- 19.Polato F, Codegoni A, Fruscio R, Perego P, Mangioni C, Saha S, Bardelli A, Broggini M. PRL phosphatase is implicated in ovarian cancer growth. Clin Cancer Res. 2005;11:6835–6839. doi: 10.1158/1078-0432.CCR-04-2357. [DOI] [PubMed] [Google Scholar]
- 20.Radke I, Gotte M, Kersting C, Mattsson B, Kiesel L, Wulfing P. Expression and prognostic impact of the protein tyrosine phosphatases PRL-1, PRL-2, and PRL-3 in breast cancer. Brit J Cancer. 2006;95:347–354. doi: 10.1038/sj.bjc.6603261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallin AR, Svanvik J, Adell G, Sun X-F. Expression of PRL proteins at invasive margin of rectal cancers in relation to preoperative radiotherapy. Int J Radiat Oncol. 2006;65:452–458. doi: 10.1016/j.ijrobp.2005.12.043. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Peng L, Dong B, Kong L, Meng L, Yan L, Xie Y, Schou C. Overexpression of phospatase of regenerating liver-3 in breast cancer: association with a poor clinical outcome. Ann Oncol. 2006;17:1517–1522. doi: 10.1093/annonc/mdl159. [DOI] [PubMed] [Google Scholar]
- 23.Achiwa H, Lazo JS. PRL-1 tyrosine phosphatase regulates c-Src levels, adherence, and invasion in human lung cancer cells. Cancer Res. 2007;67:643–650. doi: 10.1158/0008-5472.CAN-06-2436. [DOI] [PubMed] [Google Scholar]
- 24.Fiordalisi JJ, Keller PJ, Cox AD. PRL tyrosine phosphatases regulate Rho family GTPases to promote invasion and motility. Cancer Res. 2006;66:3153–3161. doi: 10.1158/0008-5472.CAN-05-3116. [DOI] [PubMed] [Google Scholar]
- 25.Kato H, Semba S, Miskad UA, Seo Y, Kasuga M, Yokozaki H. High expression of PRL-3 promotes cancer cell motility and liver metastasis in human colorectal cancer: a predictive molecular marker of metachronous liver and lung metastases. Clin Cancer Res. 2004;10:7318–7328. doi: 10.1158/1078-0432.CCR-04-0485. [DOI] [PubMed] [Google Scholar]
- 26.Zeng Q, Dong J-M, Guo K, Li J, Tan H-X, Koh V, Pallen CJ, Manser E, Jong W. PRL-3 and PRL-1 promote cell migration, invasion and metastasis. Cancer Res. 2003;63:2716–2722. [PubMed] [Google Scholar]
- 27.Bardelli A, Saha S, Sager JA, Romans KE, Xin B, Markowitz SD, Lengauer C, Velculescu VE, Kinzler KW, Vogelstein B. PRL-3 expression in metastatic cancers. Clin Cancer Res. 2003;9:5607–5615. [PubMed] [Google Scholar]
- 28.Saha S, Bardelli A, Buckhaults P, Belculescu VE, Rago C, St. Croix B, Romans KE, Choti MA, Lengauer C, Kinzler KW, Vogelstein B. A phosphatase associated with metastasis of colorectal cancers. Science. 2001;294:1343–1346. doi: 10.1126/science.1065817. [DOI] [PubMed] [Google Scholar]
- 29.Peng L, Ning J, Meng L, Shou C. The association of the expression level of protein tyrosine phosphatase PRL-3 protein with liver metastasis and prognosis of patients with colorectal cancer. J Cancer Res Clin Oncol. 2004;130:521–526. doi: 10.1007/s00432-004-0563-x. [DOI] [PubMed] [Google Scholar]
- 30.Wu X, Zeng H, Zhang X, Zhao Y, Sha H, Ge X, Zhang M, Gao X, Xu Q. Phosphatase of regenerating liver-3 promotes motility and metastasis of mouse melanoma cells. Am J Pathol. 2004;164:2039–2054. doi: 10.1016/S0002-9440(10)63763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang F, Liang J, Wang W-Q, Sun J-P, Udho E, Zhang Z-Y. PRL3 promotes cell invasion and proliferation by down-regulation of Csk leading to Src activation. J Biol Chem. 2007;282:413–5419. doi: 10.1074/jbc.M608940200. [DOI] [PubMed] [Google Scholar]
- 32.Qian F, Li Y-P, Sheng X, Zhang Z-C, Song R, Dong W, Cao S-X, Hua Z-C, Xu Q. PRL-3 siRNA inhibits metastasis of B16 and Bl6 mouse melanoma cells in vitro and in vivo. Mol Med. 2007;13:151–159. doi: 10.2119/2006-00076.Qian. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daouti S, Li W-H, Qian H, Huang K-S, Holmgren J, Levin W, Reik L, McGady DL, Olivier AR, Sergi JA, Fry D, Danho W, Ritland S, Fotouhi N, Heimbrook D, Niu H. A selective phosphatase of regenerating liver phosphatase inhibitor suppresses tumor cell anchorage-independent growth by a novel mechanism involving p130Cas cleavage. Cancer Res. 2008;68:1162–1169. doi: 10.1158/0008-5472.CAN-07-2349. [DOI] [PubMed] [Google Scholar]
- 34.Guo K, Li J, Tang JP, Koh V, Gan BQ, Zeng Q. Catalytic domain of PRL-3 plays an essential role in tumor metastasis. Cancer Biol Ther. 2004;3:945–951. doi: 10.4161/cbt.3.10.1111. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Z-Y. Chemical and mechanistic approaches to the study of protein tyrosine phosphatases. Acc Chem Res. 2003;36:385–392. doi: 10.1021/ar020122r. [DOI] [PubMed] [Google Scholar]
- 36.Sun J-P, Wang W-Q, Yang H, Liu S, Liang F, Fedorov AA, Almo SC, Zhang Z-Y. Structure and biochemical properties of PRL-1, a phosphatase implicated in cell growth, differentiation, and tumor invasion. Biochem. 2005;44:12009–12021. doi: 10.1021/bi0509191. [DOI] [PubMed] [Google Scholar]
- 37.den Hertog J, Groen A, van der Wijk T. Redox regulation of protein-tyrosine phosphatases. Arch Biochem Biophys. 2005;434:11–15. doi: 10.1016/j.abb.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 38.van Montfort RLM, Congreve M, Tisi D, Carr R, Jhoti H. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature. 2003;423:773–777. doi: 10.1038/nature01681. [DOI] [PubMed] [Google Scholar]
- 39.Jeong DG, Kim SJ, Kim JH, Son JH, Park MR, Lim SM, Yoon T-S, Ryu SE. Trimeric structure of PRL-1 phosphatase reveals an active enzyme conformation and regulation mechanisms. J Mol Biol. 2005;345:401–413. doi: 10.1016/j.jmb.2004.10.061. [DOI] [PubMed] [Google Scholar]
- 40.Kozlov G, Cheng J, Ziomek E, Banville D, Gehring K, Ekiel I. Structural insights into molecular function of the metastasis-associted phosphatase PRL-3. J Biol Chem. 2004;279:1182–1189. doi: 10.1074/jbc.M312905200. [DOI] [PubMed] [Google Scholar]
- 41.Kim K-A, Song J-S, Jee JG, Sheen MR, Lee C, Lee TG, Ro S, Cho JM, Lee W, Yamazaki T, Jeon YH, Cheong C. Structure of human PRL-3, the phosphatase associated with cancer metastasis. FEBS Lett. 2004;565:181–187. doi: 10.1016/j.febslet.2004.03.062. [DOI] [PubMed] [Google Scholar]
- 42.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Rad Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 43.Gnainsky Y, Spira G, Paizi M, Bruck R, Nagler A, Genina O, Taub R, Halevy O, Pines M. Involvement of the tyrosine phosphatase early gene of liver regeneration (PRL-1) in cell cycle and in liver regeneration and fibrosis effect of halofuginone. Cells Tissue Res. 2006;324:385–394. doi: 10.1007/s00441-005-0092-1. [DOI] [PubMed] [Google Scholar]
- 44.Skinner A, Laurence JS. 1H, 15N, 13C resonance assignments of the reduced and active form of human protein tyrosine phosphatase, PRL-1. Biomol NMR Assign online first. 2009 doi: 10.1007/s12104-008-9142-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laurence JS, Hallenga K, Stauffacher CV. 1H, 15N, 13C resonance assignments of the human protein tyrosine phosphatase PRL-1. J Biomol NMR. 2004;29:417–418. doi: 10.1023/B:JNMR.0000032506.16792.c6. [DOI] [PubMed] [Google Scholar]
- 46.Cleland WW. Dithiothreitol, a new protective reagent for SH groups. Biochem. 1964;3:480–482. doi: 10.1021/bi00892a002. [DOI] [PubMed] [Google Scholar]
- 47.Iyer KS, Klee WA. Direct spectrophotometric measurement of the rate of reduction of disulfide bonds: the reactivity of the disulfide bonds in bovine alpha-lactalbumin. J Biol Chem. 1973;248:707–710. [PubMed] [Google Scholar]
- 48.Landrieu I, da Costa M, Veylder LD, Dewitte F, Vandepoele K, Hassan S, Wieruszeski J-M, Corellou F, Faure J-D, Montagu MV, Inze D, Lippens G. A small CDC25 dual specificity tyrosine-phosphatase isoform in Arabidopsis thaliana. Proc Nat Acad Sci USA. 2004;101:13380–13385. doi: 10.1073/pnas.0405248101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piotukh K, Kosslick D, Zimmermann J, Krause E, Freund C. Reversible disulfide bond formation of intracellular proteins probed by NMR spectroscopy. Free Rad Biol Med. 2007;43:1263–1270. doi: 10.1016/j.freeradbiomed.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 50.Wishart DS, Bigam CG, Yoa J, Abildgaard F, Dyson HH, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 51.Goddard TD, Kneller DG. SPARKY. San Francisco: University of California; 2004. [Google Scholar]
- 52.Delaglio F, Grezesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensonal spectra processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 53.Sharma D, Rajarathnam K. 13C NMR chemical shits can predict disulfide bonds. J Biomol NMR. 2000;18:165–171. doi: 10.1023/a:1008398416292. [DOI] [PubMed] [Google Scholar]
- 54.Wishart DS. NMR spectroscopy and protein structure determination: applications to drug discovery and development. Curr Pharm Biotechnol. 2005;6:105–120. doi: 10.2174/1389201053642367. [DOI] [PubMed] [Google Scholar]
- 55.Hansen JM, Go Y-M, Jones DP. Nuclear and mitochondrial compartmentaion of oxidative stress and redox signaling. Annu Rev Pharmacol. 2006;46:215–234. doi: 10.1146/annurev.pharmtox.46.120604.141122. [DOI] [PubMed] [Google Scholar]
- 56.Hwang C, Sinskey AJ, Lodish HF. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1497. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 57.Ellgaard L. Catalysis of disulphide bond formation in the endoplasmic reticulum. Biochem Soc Trans. 2004;32:663–667. doi: 10.1042/BST0320663. [DOI] [PubMed] [Google Scholar]
- 58.Kosower NS, Kosower EM. The gluathione status of cells. Int Rev Cytol. 1978;54:109–160. doi: 10.1016/s0074-7696(08)60166-7. [DOI] [PubMed] [Google Scholar]
- 59.Watson WH, Pohl J, Montfort WR, Stuchlik O, Reed MS, Powis G, Jones DP. Redox potential of human thioredoxin 1 and identification of second dithiol/disulfide motif. J Biol Chem. 2003;278:33408–33415. doi: 10.1074/jbc.M211107200. [DOI] [PubMed] [Google Scholar]
- 60.Aslund F, Berndt KD, Holmgren A. Redox potential of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J Biol Chem. 1997;272:30780–30786. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]
- 61.Lundstrom J, Homgren A. Determination of the reduction-oxidation potential of the thioredoxin-like domains of protein disulfide-isomerase from the equilibrium with glutathione and thioredoxin. Biochem. 1993;32:6649–6655. doi: 10.1021/bi00077a018. [DOI] [PubMed] [Google Scholar]
- 62.Baker KM, Chakravarthi S, Langton KP, Sheppard Am, Lu H, Bulleid NJ. Low reduction potential of Ero1a regulatory disulphides ensures tight control of substrate oxidation. EMBO J. 2008;27:2988–2997. doi: 10.1038/emboj.2008.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zimmerman J, Kuhne R, Sylvester M, Freund C. Redox-regulated conformational changes in an SH3 domain. Biochem. 2007;46:6971–6977. doi: 10.1021/bi700437r. [DOI] [PubMed] [Google Scholar]
- 64.Sahu D, Debnath P, Takayama Y, Iwahara J. Redox properties of the Adomain of the HMGB1 protein. FEBS Lett. 2008;582:3973–3978. doi: 10.1016/j.febslet.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moores SL, Schaber MD, Mosser SD, Rands E, O'Hara MB, Garsky VM, Marshall MS, Pompliano DL, Gibbs JB. Sequence dependence of protein isoprenylation. J Biol Chem. 1991;266:14603–14610. [PubMed] [Google Scholar]
- 66.Menon SG, Sarsour EH, Spitz DR, Higashikubo R, Sturm M, Zhang H, Goswami PC. Redox regulation of the G1 to S phase transition in the mouse embryo fibroblast cell cycle. Cancer Res. 2003;63:2109–2117. [PubMed] [Google Scholar]
- 67.Harvey AJ, Kind KL, Thompson JG. Redox regulation of early embryo development. Reproduction. 2002;123:479–486. doi: 10.1530/rep.0.1230479. [DOI] [PubMed] [Google Scholar]
- 68.Batist G, Behrens BC, Makuch R, Hamilton TC, Katki AG, Louie KG, Myers CE, Ozols R. Serial determinations of glutathione levels and glutathione-related enzyme activities in human tumor cells in vitro. Biochem Pharmacol. 1986;35:2257–2259. doi: 10.1016/0006-2952(86)90601-5. [DOI] [PubMed] [Google Scholar]
- 69.McEligot AJ, Yang S, Meyskens FLJ. Redox regulation by intrinsic species and extrinsic nutrients in normal and cancer cells. Annu Rev Nutr. 2005;25:261–295. doi: 10.1146/annurev.nutr.25.050304.092633. [DOI] [PubMed] [Google Scholar]
- 70.Wolf CR, Lewis AD, Carmichael J, Adams DJ, Allan SG, Ansell DJ. The role of glutathione in determining the response of normal and tumour cells to anticancer drugs. biochem Soc Trans. 1987;15:728–730. doi: 10.1042/bst0150728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information for this work is available free of charge via the Internet at http://pubs.acs.org. This information illustrates the ability of PRL-1-WT to be reduced by GSH (Figure 1), MS data for both PRL-1-WT and PRL-1-C170S-C171S (Figure 2), and 1H-15N HSQCs of PRL-1-C98A under non-reducing and reducing conditions (Figure 3). Additionally, chemical shift information for oxidized (WT) and reduced (C170S-C171S) is provided in Table 1.