Abstract
The development of a sequence-general nucleic acid copying system is an essential step in the assembly of a synthetic protocell, an autonomously replicating spatially localized chemical system capable of spontaneous Darwinian evolution. Previously described nonenzymatic template-copying experiments have validated the concept of nonenzymatic replication, but have not yet achieved robust, sequence-general polynucleotide replication. The 5′-phosphorimidazolides of the 2′-amino-2′,3′-dideoxyribonucleotides are attractive as potential monomers for such a system because they polymerize by forming 2′→5′ linkages, which are favored in nonenzymatic polymerization reactions using similarly activated ribonucleotides on RNA templates. Furthermore, the 5′-activated 2′-amino nucleotides do not cyclize. We recently described the rapid and efficient nonenzymatic copying of a DNA homopolymer template (dC15) encapsulated within fatty acid vesicles using 2′-amino-2′,3′-dideoxyguanosine−5′-phosphorimidazolide as the activated monomer. However, to realize a true Darwinian system, the template-copying chemistry must be able to copy most sequences and their complements to allow for the transmission of information from generation to generation. Here, we describe the copying of a series of nucleic acid templates using 2′-amino-2′,3′-dideoxynucleotide−5′-phosphorimidazolides. Polymerization reactions proceed rapidly to completion on short homopolymer RNA and LNA templates, which favor an A-type duplex geometry. We show that more efficiently copied sequences are generated by replacing the adenine nucleobase with diaminopurine, and uracil with C5-(1-propynyl)uracil. Finally, we explore the copying of longer, mixed-sequence RNA templates to assess the sequence-general copying ability of 2′-amino-2′,3′-dideoxynucleoside−5′-phosphorimidazolides. Our results are a significant step forward in the realization of a self-replicating genetic polymer compatible with protocell template copying and suggest that N2′→P5′-phosphoramidate DNA may have the potential to function as a self-replicating system.
Introduction
A simple synthetic cell, or protocell, must be able to grow, divide, and maintain heritable information. We are attempting to construct such a system through the integration of two complementary components: (1) a spatially localized replicating compartment or vesicle and (2) a spontaneously replicating genetic polymer.1,2 Our group recently demonstrated that fatty acid membranes are permeable to small-molecule solutes including charged mononucleotides, and that activated nucleotides added to the outside of fatty acid vesicles are capable of copying an encapsulated genetic polymer.(3) We have also demonstrated repeated cycles of growth and division for fatty acid vesicles without loss of encapsulated material.(4) However, no general nucleic acid copying mechanism has been demonstrated with the capacity to function as a self-replicating protocellular genome, even though many ribozyme-mediated and nonenzymatic genetic polymer replication schemes have been investigated.5−10 Ribozymes such as the naturally occurring sunY and Tetrahymena self-splicing introns or the in vitro evolved class I and R3C ligases and polymerases are able to assemble short oligonucleotides or mononucleotides in a template-directed manner. All, however, lack the high reaction efficiency and sequence-general copying ability required for the replication of a protocell genome.11−16
The alternative strategy of chemical replication was pioneered by Orgel and colleagues who demonstrated the nonenzymatic template-directed polymerization of 5′-phosphorimidazolide-activated ribonucleotides.17−20 This seminal work validated the principle of enzyme-independent template copying, but also highlighted the difficulties associated with nonenzymatic replication. In general, these chemical-copying reactions are very slow and work best on a limited range of C-rich templates, while A-rich template copying can only be accomplished at subfreezing temperatures.(21) Furthermore, the regiospecificity of the reaction is poor, and the products tend to have a majority of 2′→5′ linkages. Orgel initiated the use of 2′- and 3′-aminonucleotides to increase reaction rates and regiospecificity22−26 due to the enhanced nucleophilicity of the amine relative to the normal sugar hydroxyls. Similar phosphoramidate ligation chemistry was used to demonstrate the amplification of tetramer and hexamer oligonucleotide templates using dimer and trimer substrates, respectively.27,28 Recent work from Richert and colleagues suggests that the rate of the chemical step in the polymerization of 3′-amino nucleotides can be very fast (>1 min−1 per nucleotide) with optimal leaving group chemistry and the use of 3′-oligonucleotides or capping groups to optimize orientation.25,29 However, 3′-aminonucleotides are subject to competing intramolecular cyclization.(30) Moreover, the thermal stability of a N3′→P5′-DNA duplex is even greater than that of its RNA counterpart, suggesting that strand separation for N3′→P5′ phosphoramidate nucleic acids may be difficult or impossible.31,32
More divergent chemistry has been explored in the search for an effective sequence copying system. Recent work by Liu, Lynn and colleagues has demonstrated rapid DNA-template-directed peptide nucleic acid (PNA) synthesis using reductive amination to drive oligonucleotide condensation reactions.33,34 It will be of interest to see if PNA-directed PNA synthesis can be achieved in this way. A radically different approach uses reversible thioester or imine exchange to add nucleobases, in a template-directed manner, to a preformed peptide, which becomes the backbone of the assembled PNA-like polymer.35,36
Despite the promising initial results obtained with activated 2′-amino-ribonucleotides over 30 years ago, Orgel did not further explore the potential of this system because the monomer building blocks did not appear to be prebiotically plausible. Since our immediate goal is the assembly of a self-replicating protocell, we have decided to focus on developing an efficient nucleic acid chemical-copying mechanism unrestrained by prebiotic chemical considerations, and have therefore begun to explore the potential of a series of phosphoramidate-linked nucleic acids and their corresponding amino-nucleotide monomers in chemical-copying reactions.24,37,38 We are particularly interested in the activated 2′-amino, 2′-3′-dideoxyribonucleotides because previously described activated RNA mononucleotide polymerization reactions display a bias for 2′→5′-linkages, and because steric restrictions imposed by the ribose ring prevent the unwanted intramolecular cyclization reaction seen with the activated 3′-amino analogues. In addition, the 2′-amino, 2′-3′-dideoxyribonucleotides have the potential to generate more chemically stable (DNA-like) polymers than the more RNA-like 2′-amino, 3′-hydroxy nucleotides. We recently showed that 2′-amino-2′, 3′-dideoxyguanosine−5′-phosphorimidazolide (2′-NH2−ImpddG) participates in the rapid and efficient template-directed copying of a dC15 template, in a reaction that proceeds to >95% completion in ∼6 h.(3) This reaction is compatible with the copying of templates encapsulated within fatty acid vesicles and could thus be used in a protocell model system. However, to enable a nucleic acid replication system that can proceed for multiple generations, and which is capable of Darwinian evolution, the copying reaction must be efficient and sequence-general, so that most if not all sequences and their complements can be replicated. Here, we present further studies of template-directed nucleic acid copying using 2′-amino-2′, 3′-dideoxynucleoside−5′-phosphorimidazolides (2′-NH2−ImpddNs). We describe the copying of a series of short homopolymer templates composed of DNA, RNA, locked nucleic acid (LNA), and O2′→P5′ DNA and therefore varying in helical geometry, sugar conformation, and base stacking. We describe the use of nucleotides with modified bases to increase the base-pairing strength and copying efficiency of the weak A:T base-pair. Finally we examine the copying of longer mixed-sequence RNA templates by 2′-NH2−ImpddNs.
Results and Discussion
Template-Copying Strategy
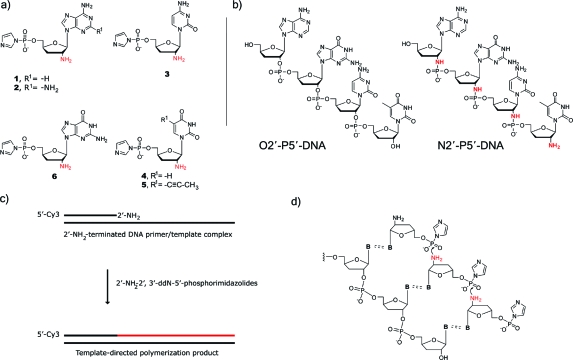
We synthesized the four standard 2′-NH2−ImpddNs (1, 3, 4, and 6) from the corresponding 2′-azido nucleosides of A, U, C, and G, as described in the . We then used a primer-extension assay to test each 2′-NH2−ImpddN in a nonenzymatic template-copying reaction (Figure 1). A 5′-Cy3-labeled 2′-amino-terminated DNA primer was annealed to a complementary oligonucleotide with an overhanging template region. The elongation of the fluorescent primer was used to follow the stepwise chemical addition of activated nucleotide monomers, generating four new N2′→P5′-phosphoramidate DNA linkages. Given the influence of template structure in template-directed chemical-copying reactions, we tested DNA, RNA, LNA, and O2′→P5′ DNA templates, which vary in sugar conformation from C2′-endo in DNA, flexible C3′-endo in RNA, and O2′→P5′ DNA (39) and rigid C3′-endo in LNA (the sugar in LNA is rigidified by a methylene bridge between O2′ and C4′40,41). We also studied the effect of Watson−Crick base-pair strength by comparing copying reactions on templates containing adenine to templates containing diaminopurine which form two vs three hydrogen bonds to the complementary thymidine or uridine nucleotide. We also compared copying reactions on uridine and C5-(1-propynyl)uridine (Up) templates to clarify base-stacking and template preorganization effects.(42) Finally, in preparation for future efforts to copy N2′→P5′-phosphoramidate DNA templates, we examined primer-extension reactions on O2′→P5′-phosphodiester DNA templates.
Figure 1.
N2′→P5′-DNA genetic polymer system. (a) 2′-Amino-2′,3′-dideoxyribonucleoside−5′-phosphorimidazolides. Clockwise from upper-left: 2′-NH2−ImpddA, R1 = -H or 2′-NH2−ImpddD, R1 = -NH2; 2′-NH2−ImpddC; 2′-NH2−ImpddU, R1 = -H or 2′-NH2−Impdd-C5-(1-propynyl)-U, R1 = -C≡C−CH3; 2′-NH2−ImpddG. (b) Structure of O2′→P5′-phosphodiester-linked DNA (right) compared to N2′→P5′-phosphoramidate−DNA (left). The 2′-internucleotide bridging atom substitution is shown in red. (c) Primer-extension reaction scheme. A 5′-Cy3-labeled 2′-amino-terminated DNA primer anneals to a complementary template. 2′-NH2−ImpddN analogues form Watson−Crick base pairs on a complementary template and participate in a chemical chain reaction forming the N2′→P5′-DNA polymer product. (d) Chemistry of the template-directed copying reaction using the activated 2′-NH2−ImpddN monomer. The attacking nucleophile is shown in red.
Template-Directed Activated Mononucleotide Polymerization
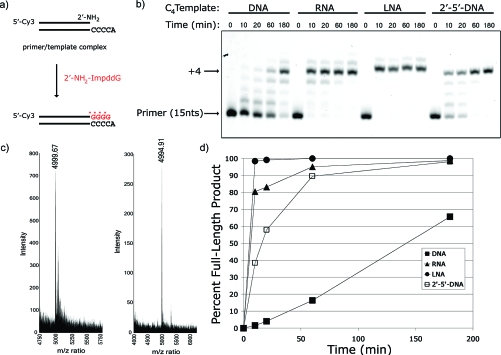
We previously observed efficient template-directed copying of a dC15 template using 2′-NH2−ImpddG (Figure 1a). This result led us to explore structural factors that might affect the reaction efficiency using the set of C4 nucleic acid templates listed above. Primer-extension reaction products were analyzed by high-resolution polyacrylamide gel electrophoresis (Figure 2). Reaction efficiency is summarized as the percent of primer extended to full-length (≥+4 nts) product at a given time point. The LNA template, with a rigid C3′-endo sugar conformation, is the most efficient template, reaching >98% completion within 10 min, while the DNA template is over an order of magnitude less efficient, taking 3 h to achieve 65% completion. Both the RNA and the O2′→P5′-DNA templates have intermediate activity, reaching >98% completion at 3 h. Control reactions on noncomplementary templates showed low levels of +1 product at 3 h, but no full-length product (Figure S3). We confirmed the formation of the proper N2′→P5′-phosphoramidate DNA primer-extension product on the RNA C4 template by MALDI-TOF MS (Figure 2). We further characterized the N2′→P5′-DNA generated on the dC15 template (Figure S1) by showing that it exhibited the expected phosphoramidate bond acid-lability (Figure S1), but is resistant to enzymatic digestion by RQ1 DNase and RNase If (Figure S2).
Figure 2.
Primer-extension reaction using a 2′-NH2−ImpddG as monomer. (a) Primer-extension reaction scheme showing a 5′-Cy3-labeled 2′-amino-terminated DNA primer annealed to a complementary template. 2′-NH2−ImpddG monomer participates in a chemical chain reaction extending the primer by four (4) nucleotides on the complementary template forming a N2′→P5′-DNA polymer product. * indicates new phosphoramidate bond. (b) High-resolution gel electrophoresis analysis of primer-extension products on indicated templates. Primer-extension reactions contained 0.1 μM Cy3-labeled-2′-amino-terminated DNA primer, 0.5 μM template, 100 mM MES-CAPS-HEPES, pH 7.5, 100 mM 1-(2-hydroxyethyl)imidazole, and 5 mM 2′-NH2−ImpddG. The reaction was initiated by addition of 2′-NH2−ImpddG, thermal cycling as described in the , and incubation at 4 °C for the indicated time. Arrows indicate primer and full-length product. (c) MALDI-TOF MS analysis of primer-extension product. 100−200 pmol amino-terminated primer was extended on an RNA template followed by base hydrolysis of the RNA template and preparation for MALDI-TOF MS as detailed in the . DNA control oligonucleotide (left): calcd mass 4999.87 and observed mass 4999.67; N2′→P5′-DNA extension product (right): calcd mass 4994.95 and observed mass 4994.91. (d) Graphical summary of primer-extension reactions. Primer-extension reaction efficiency is expressed as percent full-length product (≥+4) achieved at the given time point.
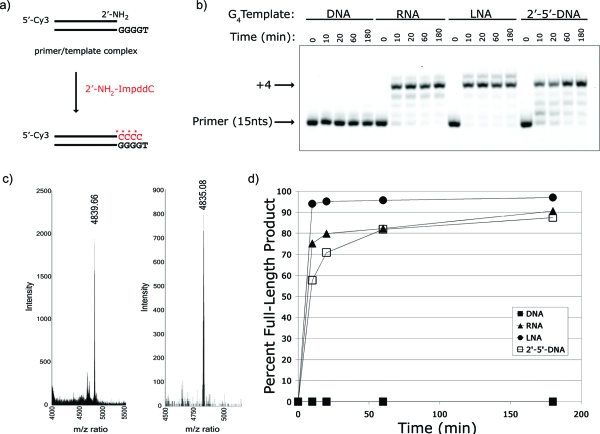
Given the efficient template copying observed with the activated guanosine amino nucleotide, we next assessed the activity of 2′-amino-2′,3′-dideoxycytidine−5′-phosphorimidazolide (2′-NH2−ImpddC) (Figure 1a) on complementary DNA, RNA, LNA, and O2′→P5′-DNA templates with the sequence G4. We observed very efficient copying on the RNA, LNA, and O2′→P5′-DNA templates; in all cases the reactions were essentially complete within 10 min (Figure 3). We observed no extension on the DNA template, possibly due to aggregation of the dG4 region to form G-quartets. Control extension reactions on noncomplementary templates show limited extension to the +1 position with traces of +2 product but do not generate any full-length +4 product (Figure S4, Supporting Information). MALDI-TOF MS analysis confirmed the expected N2′→P5′-DNA primer-extension product (Figure 3).
Figure 3.
Primer-extension reaction using a 2′-NH2−ImpddC as monomer. (a) Primer-extension reaction scheme. * indicates new phosphoramidate bond. (b) High-resolution gel electrophoresis analysis of primer-extension products on indicated templates. Primer-extension reactions were completed as previously described with 5 mM 2′-NH2−ImpddC. Arrows indicate primer and full-length product. (b) MALDI-TOF MS analysis of primer-extension product performed as described in the . DNA control oligonucleotide (left): calcd mass 4839.84 and observed mass 4839.66; N2′→P5′-DNA extension product (right): calcd mass 4834.92 and observed mass 4835.08. (c) Graphical summary of primer-extension reaction efficiency.
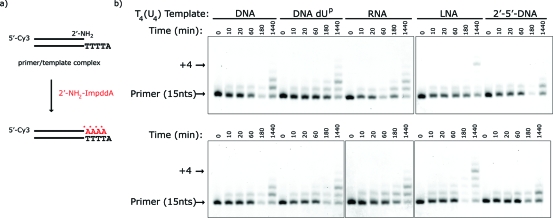
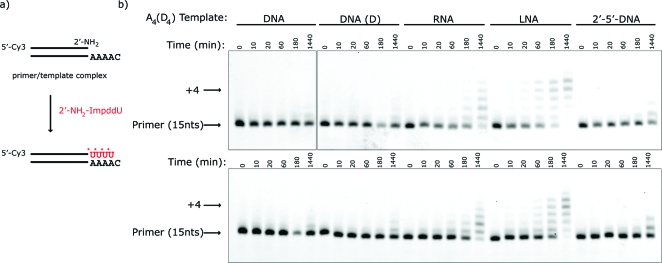
We then asked if 2′-amino-2′,3′-dideoxyadenosine−5′-phosphorimidazolide (2′-NH2−ImpddA) (Figure 1a) would efficiently copy DNA T4, RNA U4, LNA T4, and O2′→P5′-DNA T4 templates. These primer-extension reactions were very slow and inefficient compared to our previous results with G and C. Increasing the concentration of 2′-NH2−ImpddA from 5 mM to 30 mM made very little difference (Figure 4). We observed significant accumulation of the full-length +4 product only on the LNA template at 24 h. Because thymidine and uridine have less effective base-stacking interactions than purine nucleotides, we prepared templates containing four successive residues of the deoxyuridine analogue C5-(1-propynyl)deoxyuridine (dUp) in an attempt to preorganize the template geometry through stabilizing π-stacking interactions between 5-propynyl groups in adjacent nucleotides.(42) However, the dUp4 template only modestly improves reaction efficiency when compared to the dT4 template (Figure 4).
Figure 4.
Primer-extension reaction using a 2′-NH2−ImpddA as monomer. (a) Primer-extension reaction scheme. * indicates new phosphoramidate bond. (b) High-resolution gel electrophoresis analysis of primer-extension products on indicated templates. Template-copying reactions were run as previously described with 5 mM (upper) or 30 mM (lower) 2′-NH2−ImpddA. Arrows indicate primer and full-length extension product.
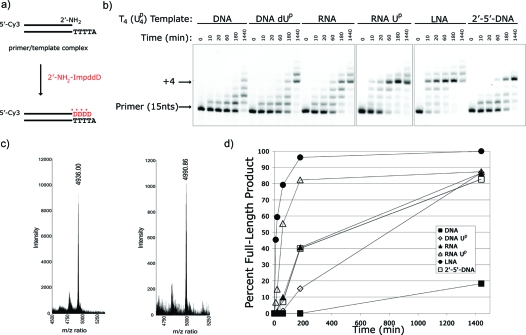
We then tested template-copying reactions based on the reciprocal A:T/U base pair by synthesizing 2′-amino-2′,3′-dideoxyuridine−5′-phosphorimidazolide (2′-NH2−ImpddU) (Figure 1a) and examining primer-extension reactions on DNA A4, RNA A4, LNA A4, and O2′→P5′-DNA A4 templates. As above, all reactions were of low efficiency at both 5 mM and 30 mM 2′-NH2−ImpddU (Figure 5). While slow, both RNA and LNA templates did lead to the accumulation of some full-length +4 product at 24 h (Figure 5). In an effort to improve reaction efficiency, we sought to strengthen the A:T/U base-pair by preparing templates in which adenine was replaced with 2, 6-diaminopurine (D), which contains an exocyclic amine at the C-2 position and has the potential to make three hydrogen bonds with the complementary Watson−Crick base, thymidine or uridine. However, the DNA-D4 template did not enhance reaction efficiency significantly when compared with the dA4 template (Figure 5).
Figure 5.
Primer-extension reaction using the 2′-NH2−ImpddU as monomer. (a) Primer-extension reaction scheme. * indicates new phosphoramidate bond. (b) High-resolution gel electrophoresis analysis of primer-extension products on indicated templates. Primer-extension reactions were run as described with 5 mM (upper) or 30 mM (lower) 2′-NH2−ImpddU. Arrows indicate primer and full-length product.
Improved Template Copying with D:Up Base-Pair
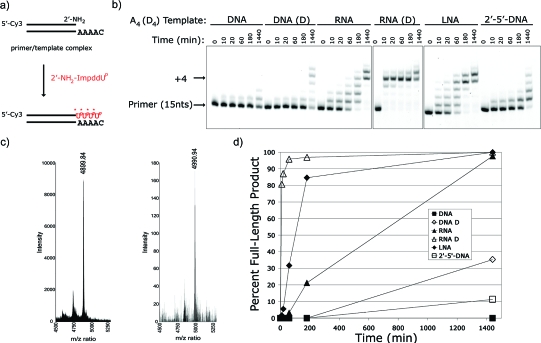
Since the 2′-NH2 A and U nucleotides performed so poorly in template-directed copying reactions, and since the C5-(1-propynyl)-U and diaminopurine nucleobase modifications had such modest effects when present only in the template, we prepared the activated monomers for D (2) and Up (5) and tested these modified nucleotides on templates containing D and Up. The D:Up base-pair has previously been reported to be as effective as a G:C base-pair in the context of the nonenzymatic synthesis of RNA on a DNA template.(43) We observed a wide range of reaction efficiencies with the 2′-NH2−ImpddD monomer (Figure 1a) on DNA T4, DNA dUp4, RNA U4, RNA Up4, LNA T4 and O2′→P5′-DNA T4 templates (Figure 6). The improvement expected from Up containing templates was much more evident when the monomer was 2′-NH2−ImpddD. The dUp4 template achieves 15% full-length product at 3 h compared to 0% full-length product on the standard T4 DNA template. A similar enhancement is seen on the RNA templates, where the RNA Up4 template reaches 82% full-length product compared 40% full-length product on the standard RNA-U4 template in 3 h. Interestingly, the RNA U4 and O2′→P5′-DNA T4 templates display similar reaction efficiencies, with >80% full-length product accumulation over the course of 24 h, again demonstrating that 2′→5′-linked oligonucleotides serve as effective templates. Experiments at 5 and 30 mM 2′-NH2−ImpddD gave similar results (data not shown) suggesting that the template is already saturated with 5 mM monomer. Noncomplementary template copying reactions yield only small amounts of +1 product and no full-length product at 3 h (Figure S5, Supporting Information). We detected the proper N2′→P5′-DNA primer-extension polymerization product by MALDI-TOF MS analysis (Figure 6).
Figure 6.
Primer-extension reaction using the 2′-NH2−ImpddD as monomer. (a) Primer-extension reaction scheme. * indicates new phosphoramidate bond. (b) High-resolution gel electrophoresis analysis of primer-extension products on indicated templates. Primer-extension reactions were completed as previously described with 5 mM 2′-NH2−ImpddD. (c) MALDI-TOF MS analysis of primer-extension product. DNA control oligonucleotide (left): calcd mass 4935.89 and observed mass 4936.00; N2′→P5′-DNA extension product (right): calcd mass 4991.01 and observed mass 4990.86. (d) Graphical summary of primer-extension reactions.
We prepared the second unnatural nucleobase activated analogue, 2′-amino-2′,3′-dideoxy-C5-(1-propynyl)uridine−5′-phosphorimidazole (2′-NH2−ImpddUp) and used it in primer-extension experiments to copy a series of A and D containing templates: DNA A4, DNA D4, RNA A4, RNA D4, LNA A4, and O2′→P5′-DNA A4 templates (Figure 7). Copying of the LNA and RNA A4 templates was largely complete after 3 and 24 h respectively. The effect of enhanced hydrogen bonding on reaction efficiency was clearly visible by comparing the RNA A4 template to the RNA D4 template. The RNA D4 template reaction is largely complete in 10−20 min compared to 24 h for the RNA A4 template. Control copying reactions on noncomplementary templates result in traces of +1 product but no full-length product (Figure S6, Supporting Information). We confirmed synthesis of the proper primer-extension product on the RNA D4 template by MALDI- TOF MS (Figure 7).
Figure 7.
Primer-extension reaction using a 2′-NH2−ImpddUp as monomer. (a) Primer-extension reaction scheme. * indicates new phosphoramidate bond. (b) High-resolution gel electrophoresis analysis of primer-extension products on indicated templates. Primer-extension reactions were completed as previously described with 5 mM 2′-NH2−ImpddUp. (c) MALDI-TOF MS analysis of primer-extension product. DNA control oligonucleotide (left): calcd mass 4899.84 and observed mass 4899.84; N2′→P5′-DNA extension product (right): calcd mass 4990.92 and observed mass 4990.94. (d) Graphical summary of primer-extension reactions.
Mixed-Sequence RNA Template-Copying Reactions
Because our goal is to develop a sequence-general self-replicating nucleic acid system, we examined the ability of 2′-NH2−ImpddN monomers to copy longer mixed-sequence templates. We used RNA templates for these experiments because of their synthetic accessibility; in future work we plan to examine N2′→P5′-DNA templates. We synthesized RNA templates containing the natural guanosine and cytidine nucleobases and the unnatural diaminopurine and C5-(1-propynyl)uridine nucleobases. We first examined RNA templates containing two noncomplementary nucleotides: 5′-GGGUpUpUp- and 5′-CCCDDD- downstream of the primer binding site (Figures S7 and S8, Supporting Information). On both templates, primer-extension reactions led to the appearance of full-length product (≥+6 nts) within 60 min and were largely complete by 3 h. Because N2′→P5′-DNA cannot be sequenced using standard sequencing techniques that require transcription into DNA or RNA, we used single-nucleotide dropout primer-extension reaction experiments to begin to assess the fidelity of the template copying reaction. On both of these templates, the primer-extension reaction stops at the expected location in the absence of one of the 2′-NH2−ImpddN monomers, with only small amounts of primer extended by a single mismatched base present at 3 h.
We then proceeded to examine two more complex RNA templates containing three different nucleotides in the templating region. Both sequences (5′-GGDDCCDDCCCDDGG- and 5′-AGCCGCCGCC-3′- adjacent to the primer binding region) allowed full-length product accumulation, but the primer-extension reaction was significantly slower than the previously examined templates. Only a trace amount of full-length product is present at 1 h, while the reactions required over 20 h to accumulate significant full-length product (Figures S9 and S10, Supporting Information). Individual 2′-NH2−ImpddN dropout reactions indicate the potential for significant problems with the fidelity of template copying. In the absence of the G monomer, primer-extension stops cleanly before the first C residue on both templates. However, in the absence of the C monomer, there is extensive read-through of G residues on the 5′-GGDDCCDDCCCDDGG-template (Figure S9, Supporting Information), presumably due to wobble-pairing of template Gs with Up monomers but also possibly due to the formation of G:G mismatches. In the absence of the Up monomer, ∼50% of the primer stalled before the first template D residue, as expected, but a similar amount is stalled after the incorporation of a mismatched base (either G or C, or both) opposite the first template D (Figure S9, Supporting Information). On the template sequence 5′-CCGCCGCCGC-, which was copied with just the ImpddG and ImpddC monomers, we see extensive primer extension in the absence of the C monomer, which must be due to the incorporation of G opposite G (Figure S10, Supporting Information). These G:G mismatches appear to be more slowly extended than G:C base-pairs, resulting in the accumulation of multiple stalled primer-extension products with altered gel mobility (Figure S10, Supporting Information), presumably because the G:G pair distorts the helix and prevents the proper alignment of the 2′-amino group of the terminal G residue for attack on the 5′-phosphorimidazolide of the incoming activated nucleotide.
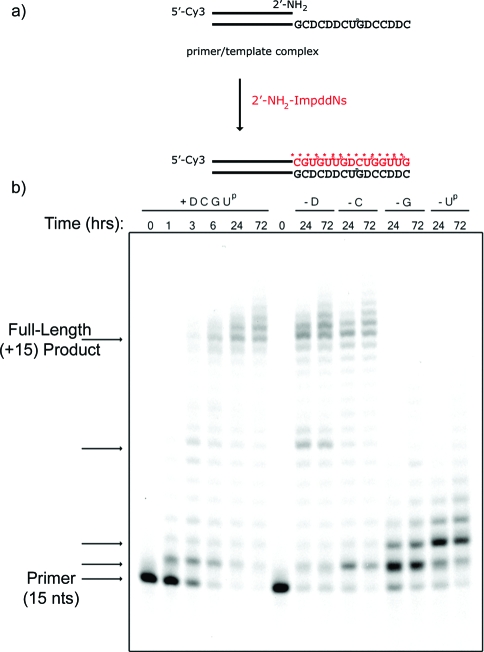
Finally, we asked if a mixed-sequence RNA template containing all four nucleotides could be copied. We used a template with the sequence 5′-CDDCCDGUpCDDCDCG-3′ adjacent to the primer binding site to model a sequence-general-copying reaction (Figure 8). We used a mixture of the 2′-NH2−ImpddG, 2′-NH2−ImpddC, 2′-NH2−ImpddD, and 2′-NH2−ImpddUp monomers at optimized concentration ratios. These ratios were derived from a series of primer-extension experiments carried out at various concentration ratios, in which we attempted to maximize full-length product accumulation and minimize stalling. Using these optimized monomer ratios, we were able to observe conversion of 61% of primer to full-length primer-extension product through nonenzymatic template-directed synthesis of N2′→P5′-DNA. Full-length (≥+15 nts) product is visible at the 3 h time point and continues to accumulate up to 24 h (Figure 8).
Figure 8.
High-resolution gel electrophoresis analysis of mixed-sequence RNA template-copying reaction. (a) Primer-extension reaction scheme. A 5′-Cy3-labeled 2′-amino-terminated DNA primer annealed to a complementary RNA template. 2′-NH2−ImpddN monomers participate in a chemical chain reaction extending the primer by 15 nucleotides on the complementary template, forming a N2′→P5′-DNA polymer product. * indicates new phosphoramidate bond. (b) Primer-extension reactions were completed as previously described with 0.5 μM template (5′-CDDCCDGUpCDDCDCG-3′-primer binding site), 5 mM 2′-NH2−ImpddG, 5 mM 2′-NH2−ImpddC, 10 mM 2′-NH2−ImpddD, and 10 mM 2′-NH2−ImpddUp. The reaction was initiated with addition of the monomers, thermal cycling as described in the , and incubation at 4 °C for the indicated time. Arrows indicate primer, full-length product, and stalled products.
We used single-nucleotide dropout experiments as above to assess potential problems with fidelity (Figure 8). In the absence of the D or C monomers, full-length product is still formed, presumably due to the G:Up wobble base-pairing. In the absence of D, a +7 nucleotide primer-extension product accumulates, suggesting that copying of the single Up in the template by G monomer is slow compared to copying by the correct D monomer. In the absence of the C monomer, full-length primer-extension product accumulates to similar levels as in the complete reaction mix. In this case, a stalled +1 product is visible at 24 h but is decreased in intensity by 72 h. This suggests that the Up monomer may incorporate opposite the first G residue in the template, but that this wobble base-pair is further extended relatively slowly. Very little stalled product is visible either before or at the position of the second G residue in the template. Thus, the incorporation of U opposite template G residues may be a significant source of errors. In contrast, in the absence of G or Up, very strong stalling is observed prior to the first template C and D residue, respectively, suggesting that copying of C and D is likely to proceed with relatively high fidelity.
Conclusions
Our exploration of the spontaneous chemical copying of a range of nucleic acid templates by activated 2′-amino 2′,3′-dideoxyribonucleotide monomers highlights both the advantages and potential problems associated with this approach to template copying. Here we discuss the implications of copying experiments using templates of varying length and sequences, composed of varying nucleic acids. On the basis of these results we suggest directions for future work that may lead to improved sequence-general copying and ultimately an effective self-replication system.
In our initial set of experiments, we synthesized the 2′-amino-2′,3′-dideoxy analogues of the four standard ribonucleotides, A, G, C, and U, and activated each of these by forming the corresponding 5′-phosphorimidazolides. We used each of these compounds to extend a primer across a complementary four-nucleotide template region using DNA, RNA, LNA, and O2′→P5′-DNA templates. We observed that optimal template-copying efficiency is a strong function of template sugar conformation and preorganization. In all cases, the order of template activity was LNA > RNA ∼ O2′→P5′-DNA > DNA, strongly suggesting that a preorganized A-type helical structure provides the optimal context for template-directed polymerization. These initial experiments also showed that G4 and C4 templates were very rapidly copied by activated C and G monomers, respectively. On LNA and RNA templates, these primer-extension reactions were complete in less than 10 min, which is at least 100-fold faster than comparable primer extension reactions on RNA templates using identically activated ribonucleotides.(44) Thus, we see the expected rate enhancement due to the replacement of the normal hydroxyl nucleophile of ribonucleotides with a 2′-amino nucleophile. This rate enhancement is similar to that reported earlier for 3′-amino nucleotides.(25)
When we examined A4 and U4 (or T4) templates, we found that these were copied very inefficiently by activated 2′-amino U and A monomers; only LNA and to a lesser extent RNA templates supported full-length primer-extension, and even in these cases the full-length product accumulated very slowly. The A:T(U) base-pair is well-known to be less stabilizing to duplex formation for two reasons: the presence of only two hydrogen bonds between A and T(U) and the weaker stacking interactions of U (and T) compared to the other nucleotides. We attempted to correct these deficiencies by replacing A with diaminopurine (D), which allows for the formation of a base-pair with three hydrogen bonds, and by replacing U(T) with C5-(1-propynyl)-U, where the propynyl substituent enhances stacking interactions. Either modification alone had little effect, but together the results were more dramatic; the copying of the RNA D4 template by the 2′-amino Up monomer was complete in 10−20 min. However, the copying of the RNA Up template was much slower and was not complete until 3 h. Although the Up monomer is clearly superior to U or T, a further improved derivative would clearly be useful. The electron-withdrawing propynyl substituent has previously been noted to reduce the pKa of N3 by nearly one unit relative to the N3 of uridine, thus reducing the strength of an A:Up base-pair relative to A:T in another system.(45) A less electron-withdrawing substituent at the 5 position, that could still enhance the stacking properties of the nucleobase, might yield improved template-copying performance.
We noticed a significant reduction in the rate of primer-extension on our mixed-sequence RNA templates. For example, the G, C, and D 4-base homopolymer templates were all copied in ∼10 min, but the 6-base mixed templates took 1−3 h and the 15- and 16-base templates were not completely copied after 24 h. There are several possible explanations for this effect. Template secondary structures or template−template interactions may prevent the activated 2′-NH2−ImpddNs from aligning on the template, although the sequences we used should support only weak interactions. If this explanation is correct, decreased template concentration and/or elevated temperature during the primer-extension reaction may facilitate the copying reaction. Alternatively, binding of 2′-NH2−ImpddNs to incorrect bases on the template may decrease the polymerization rate, either by preventing primer-extension when a mismatched base is bound, or because the incorporation of mismatched bases into the growing chain slows subsequent chain elongation. It may be possible to test this hypothesis using specially designed templates and varying monomer concentrations and ratios. Another possibility is that the slow copying of longer templates may be due to the weak pairing of the newly synthesized N2′→P5′-DNA region with the RNA template, so that the terminus of the growing chain becomes increasingly likely to dissociate from the template as the chain grows longer. This hypothesis could be tested with homopolymer templates of varying lengths. Finally, it is possible that the regular helical geometry of homopolymer templates is disrupted with mixed sequences, perhaps due to irregular stacking interactions of template bases or bound monomers. Modified nucleobases with improved stacking interactions might be necessary to overcome such an effect.
Successful genetic inheritance requires high fidelity replication. Early work by Eigen and more recent work by Nowak and Ohtsuki has explored the fidelity required for successful template replication.46,47 In essence, successful transmission of genetic information can only occur if the sequence mutation rate, u, is less than the inverse of the length of the sequence (1/n), no matter how great the selective advantage conferred by that sequence. Copying a functional + strand into a complementary − strand, then back to a new + strand, requires two rounds of strand copying, and thus the per nucleotide, per strand error rate should be less than 1/2n. This requirement may be alleviated by a factor of approximately 2 when considering realistic functional sequences, such as ribozymes, where not every base need be strictly defined to retain function.(48) Thus, replication of functional sequences of 30 to 35 nucleotides would seem to require an error rate of less than about 3%. As noted above, the most likely source of errors during template copying with 2′-amino nucleotides is G:Up wobble pairing. Primer-extension past template G residues in the absence of C monomer suggests that G:Up wobble pairing may compete effectively with G:C Watson−Crick pairing. However, it is not possible from our current results to estimate the error frequency resulting from wobble-pair-mediated misincorporation, since this depends on the competition between correct and incorrect monomer in reactions where both are present. Accurate measurement of copying fidelity will require the ability to examine the products of primer-extension by techniques that are more informative then gel electrophoresis. It is unfortunate that the standard methods of molecular biology such as cloning and sequencing are not applicable in this case, since N2′→P5′-DNA cannot at present be accurately copied into DNA by any known DNA polymerase. However, high-resolution MS/MS methods should allow the composition of N2′→P5′-DNA oligonucleotides to be accurately determined. We note that there are at least two possibilities for a reduction in errors due to G:Up wobble pairing. First, an improved version of C5-(1-propynyl)-U with stronger base-pairing to D would allow the use of lower concentrations of the U monomer, thus decreasing G:Up wobble pairing. A second, more speculative possibility, is that fidelity may be higher on N2′→P5′-DNA templates. The geometry of the N2′→P5′-DNA duplex, which is still unknown, may be such that wobble pairing greatly disfavors the chemical step in chain elongation. We hope to be able to investigate this possibility in the near future.
On the basis of the results discussed above, we believe that N2′→P5′-DNA is an attractive candidate for development into a self-replicating genetic system. Most significantly, the activated 2′-NH2−ImpddN monomers copy a variety of nucleic acid templates on the scale of minutes in efficient primer-extension reactions. Furthermore, since the monomers do not cyclize, the major competing reaction is hydrolysis, which is quite slow (kobsd<10−2 h−1 at 37 °C, pH 7.5).(49) This allows ample time for the completion of template-copying chemistry before the substrates are depleted by hydrolysis. Finally, in the context of multiple replication cycles, where duplex strand separation must occur before the next round of template copying can begin, the N2′→P5′-DNA duplex is likely to melt at a lower temperature than the corresponding N3′→P5′-DNA duplex.(50) Oligonucleotides containing 12 to 16 O2′→P5′ linkages have Tm values between 10 and 30 °C lower than that of O3′→P5′ linked sequences, depending on the G-C content. Facile strand separation, and possible slower strand reannealing, could greatly facilitate the emergence of cycles of chemical replication. Our current work is focused on the synthesis of N2′→P5′-DNA oligonucleotides so that we can explore the possibility of true self-replication in this system.
Regardless of whether or not the N2′→P5′-DNA system ultimately proves capable of robust self-replication, the work presented here provides guidance for the design of alternative and potentially superior systems. Most relevant is the striking superiority in template-copying reactions of the conformationally constrained, and therefore preorganized, LNA templates. This suggests that other conformationally constrained templates may show enhanced chemical replication properties. N2′→P3′-TNA has been synthesized by the Eschenmoser laboratory and is known to be a Watson−Crick base-pairing system.(38) It is conformationally constrained relative to N2′→P5′-DNA because there is one less rotatable bond per backbone repeat unit. A second alternative is the morpholino nucleic acid system, which is conformationally constrained in a very different way, due to the presence of a six-membered ring in the backbone repeat unit. We suggest that these and other conformationally constrained nucleic acids are excellent candidates for chemically self-replicating genetic systems.
Experimental Section
General Methods
Analytical thin layer chromatography (TLC) was performed on Merck silica gel 60 matrix plates containing a fluorescent indicator. Compounds were characterized by proton (1H), carbon (13C), and phosphorus (31P) nuclear magnetic resonance (NMR) spectroscopy on a Varian NMR spectrometer (Oxford AS-400) (1H: 400 MHz, 13C: 100 MHz, and 31P: 80 MHz). In each case the chemical shift of the corresponding solvent was used as a reference. Chemical shift values are reported in ppm. Electrospray mass spectrometry was recorded on a Bruker Daltonics Esquire 6000 ESI-MS. Chemicals and solvents were purchased from Sigma-Aldrich unless otherwise indicated.
Crude 2′-amino-2′,3′-dideoxynucleoside−5′-phosphorimidazolides were purified by preparative HPLC (Varian ProStar Preparative LC) on a Prep-C18 column 21.2 mm × 150 mm, 5 μ particle size (Agilent Technologies) equilibrated with 25 mM triethylammonium bicarbonate/2.5% acetonitrile, pH 8.0 and eluted with an acetonitrile gradient followed by further purification by reverse-phase HPLC (Agilent 1100 series LC) on a Zorbax Extend-C18 column 9.4 mm × 250 mm, 5 μ particle size (Agilent Technologies) equilibrated with 30 mM triethylammonium bicarbonate/2% acetonitrile, pH 8.0, and eluted with an acetonitrile gradient. Purified imidazolides were lyophilized to dryness and stored at −80 °C.
Oligonucleotides were synthesized using standard β-cyanoethyl phosphoramidite chemistry on an Expedite 8909 DNA synthesizer (Applied Biosystems) unless otherwise indicated. β-cyanoethyl phosphoramidites used in oligonucleotide synthesis were purchased from Glen Research unless otherwise noted. DNA synthesizer reagents were purchased from Glen Research and American Bioanalytical. Oligonucleotide quantification was done on a Nanodrop ND1000 Spectrophotometer.
Abbreviations
Pd/C, palladium on carbon; DMF, dimethylformamide; TOM, [(triisopropylsilyl)oxy]methyl; lcaa-CPG, long-chain alkylamine-Controlled Pore Glass bead; MES, 2-(N-morpholino)ethanesulfonic acid; CAPS, N-cyclohexyl-3-aminopropanesulfonic acid; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid.
2′-Amino-2′,3′-dideoxy-adenosine−5′-Phosphorimidazole (2′-NH2−ImpddA) (1)
2′-Azido-2′,3′-dideoxyadenosine was synthesized according to the reported protocol.(51) 2′-Azido-2′,3′-dideoxyadenosine (276.1 mg, 1 mmol) was dissolved in freshly distilled triethylphosphate (10 mL) and cooled to 0 °C in an ice bath under an argon atmosphere. Phosphorus oxychloride (1.2 equiv) was added dropwise and the solution stirred at 0 °C until complete consumption of the nucleoside, as determined by TLC. Imidazole (4 equiv) was then added, the solution was stirred at 0 °C for 30 min and then allowed to warm to room temperature and stirred for an additional 3 h. The volatiles were evaporated, and DMF (1 mL) was added to dissolve the residue. The 2′-azido adenosine phosphorimidazolide was precipitated by dropwise addition into a solution of diethyl ether/acetone/triethylamine (17 mL, 2:1.2:0.15) containing NaClO4 (25 mg) and collected by centrifugation. The crude 2′-azido adenosine phosphorimidazolide was dissolved in methanol (10 mL) containing triethylamine (1 mL), and the azido function was reduced by catalytic hydrogenation on Pd/C (10% Pd loading, 0.1 mmol) over a period of 2 h. The suspension was filtered over Celite, the volatiles were evaporated, and the product was purified by preparative RP-HPLC, yielding 193.7−220.9 mg (70−80%) of 1 as a white solid. 1H NMR δ (D2O): 8.01 (1H, s), 7.52 (1H, s), 6.82 (1H, s), 6.65 (1H, s), 5.70 (1H, s), 4.44 (1H, m), 3.91 (1H, m), 3.84 (1H, m), 3.68 (1H, m), 2.15 (1H, m), 1.91 (1H, m). 31P δ (D2O): −6.80. ESI-MS: mass calcd: (−) 379.10; found: (−) 379.1.
2′-Amino-2′,3′-dideoxy-2,6-diaminopurine−5′-Phosphorimidazole (2′-NH2−ImpddD) (2)
2′-Azido-2′,3′-dideoxy-2,6-diaminopurine was prepared according to the reported protocol.(51)2 was prepared according to the protocol for compound 1. 1H NMR δ (D 2O): 7.52 (1H, s), 7.35 (1H, s), 6.61 (1H, s), 6.47 (1H, s), 4.27 (1H, m), 3.76 (1H, m), 3.65 (1H, m), 3.52 (1H, m), 2.03 (1H, m), 1.75 (1H, m). 31P δ (D2O): −6.89. ESI-MS: mass calcd: (−) 394.11; found: (−) 394.1.
2′-Amino-2′,3′-dideoxycytidine−5′-Phosphorimidazole (2′-NH2−ImpddC) (3)
2′-Azido-2′,3′-dideoxycytidine was prepared according to the reported protocol.(51)3 was prepared according to the protocol for compound 1 with the following exceptions. The crude 2′-azido-2′,3′-dideoxycytidine−5′-phosphorimidazolide was dissolved in a solution of pyridine/triethylamine/water (10 mL, 85:13:2) and cooled to 0−5 °C in an ice bath. The solution was then saturated with hydrogen sulfide gas, and the resulting brown mixture was stirred for 30 min. The volatiles were removed by evaporation under reduced pressure (CAUTION: vacuum pump must be inside fume hood. Hydrogen sulfide is extremely toxic. Low concentrations have a strong odor, but higher levels ablate the sense of smell and lead to loss of consciousness followed by death) and the residue coevaporated with anhydrous toluene (2×, 10 mL). The crude product was then purified by RP-HPLC. 1H NMR δ (D2O): 7.72(1H, s), 7.49 (1H, d, J = 7.6 Hz), 6.91 (1H, s), 5.78 (1H, d, J = 8 Hz), 5.52 (1H, br s), 4.35 (1H, m), 3.96 (1H, m), 3.72 (1H, m), 3.35 (1H, m), 1.85 (1H, m), 1.69 (1H, m). 31P δ (D2O): −6.83. ESI-MS: mass calcd: (−) 355.09; found: (−) 355.1.
2′-Amino-2′,3′-dideoxyuridine−5′-Phosphorimidazole (2′-NH2−ImpddU) (4)
2′- Azido-2′,3′-dideoxyuridine was prepared according to the reported protocol.(51)4 was prepared according to the protocol for compound 1. 1H NMR δ (D2O): 7.72 (1H, s), 7.38 (1H, d, J = 7.6 Hz), 7.09 (1H, s), 6.91 (1H, s), 5.58 (1H, d, J=7.8 Hz), 5.54 (1H, d, 4 Hz), 4.30 (1H, m), 3.91 (1H, m), 3.71 (1H, m), 3.34 (m, 1H), 1.87 (m, 1H), 1.71 (m, 1H). 31P δ (D2O): −6.85. ESI-MS: mass calcd: (−) 356.08; found: (−) 356.1.
2′-Amino-2′,3′-dideoxy-5-C-propynyluridine−5′-Phosphorimidazole (2′-NH2−ImpddUp) (5)
2′-Trifluoroacetamide-2′,3′-dideoxy-C5-propynyluridine synthesis details will be described elsewhere. 2′-Trifluoroacetamide-2′,3′-dideoxy-C5-propynyluridine (361.1 mg, 1 mmol) and Proton-Sponge (2 equiv) were dissolved in freshly distilled triethylphosphate (10 mL) and cooled to 0 °C in an ice bath under an argon atmosphere. Phosphorus oxychloride (1.2 equiv) was then added dropwise and the solution stirred at 0 °C until complete consumption of the nucleoside, as determined by TLC. Imidazole (4 equiv) was then added, the solution was stirred at 0 °C for 30 min and then allowed to warm up to room temperature and stirred for additional 3 h. The volatiles were then evaporated, and DMF (1 mL) was added to the residue. The phosphorimidazolide was precipitated by dropwise addition to a solution of diethyl ether/acetone/triethylamine (17 mL, 2:1.2:0.15) containing NaClO4 (25 mg) and collected by centrifugation. The crude product was dissolved in methanol (10 mL) containing K2CO3 (200 mM) and stirred overnight at room temperature. After deprotection, the desired product was purified by RP-HPLC as previously described.1H NMR δ (D2O): 7.57 (1H, s), 7.54 (1H, s), 6.96 (1H, s), 6.72 (1H, s), 5.35 (1H, d, J = 2.8 Hz), 4.19 (1H, m), 3.75 (1H, m), 3.61 (1H, m), 3.26 (1H, m), 1.76 (1H, m), 1.67 (3H, s), 1.59 (1H, m). 31P δ (D2O): −7.0. ESI-MS: mass calcd: (−) 394.09; found: (−) 394.1.
2′-Amino-2′,3′-dideoxyguanosine−5′-Phosphorimidazole (2′-NH2−ImpddG) (6)
2′-Azido-2′,3′-dideoxyguanosine was synthesized according to the reported protocol.(51)6 was prepared according to the protocol for compound 1. 1H NMR δ (D2O): 7.47 (1H, s), 7.37 (1H, s), 6.66 (1H, s), 6.49 (1H, s), 5.32 (1H, d, J = 3.2 Hz), 4.21 (1H, m), 3.71−3.62 (2H, m), 3.50 (1H, m), 1.98 (1H, m), 1.70 (1H, m). 31P (D2O) −6.87. ESI-MS: mass calcd: (−) 395.1; found: (−) 395.1.
5′-O-Dimethoxytrityl-5-propyn-1-yl-uridine−2′-O-Triisopropylsilyloxymethyl-3′-[(2-cyanoethyl)-(N,N-diisopropyl)]phosphoramidite (7)
5′-O-DMT-2′-O-TOM-5-C-propynyluridine nucleoside was synthesized in four steps as previously described.(52) 5′-O-DMT-2′-O-TOM-5-C-propynyluridine nucleoside (953 mg, 1.23 mmol) was dissolved in anhydrous dichloromethane (20 mL) and cooled to 0−5 °C in an ice bath under an argon atmosphere. Diisopropylethylamine (0.64 mL, 3.7 mmol) and cyanoethyl diisopropylphosphoramidochloridite (0.43 g, 1.85 mmol) were then added and the resulting solution stirred at room temperature. After 2 h, TLC analysis showed complete conversion of the starting material into a more polar compound, and the reaction mixture was diluted with additional dichloromethane (60 mL) and extracted with aqueous NaHCO3 (30 mL) and washed with water (30 mL). The organic phase was dried with MgSO4, and after filtration the volatiles were removed under reduced pressure. The residue was purified by column chromatography using a gradient of 10−60% ethyl acetate in hexane. The product was obtained as a pair of diastereomers. 1H NMR δ (CDCl3): 7.98 and 7.92 (2 s, 1H), 7.15−7.48 (m, 10 H), 6.77−6.87 (m, 4H), 6.16 and 6.11 (2 d, 1H, J = 6.8 Hz and J = 6.0 Hz), 4.92−5.02 (2 m, 2H), 4.51 and 4.57 (2 m, 2H), 4.44 and 4.38 (2 m, 1H), 4.27 and 4.17 (2 m, 1H), 3.80−3.96 (2 m, 1H), 3.77 and 3.76 (2 s, 6H), 3.32−3.7 (m, 6H), 2.60−2.66 (m, 1H), 2.3−2.4 (m, 1H), 1.65−1.68 (m, 3H), 1.11−1.18 (m, 12H), 0.96−1.06 (m, 18 H).13C NMR δ (CDCl3): 162.16, 162.07, 149.69, 149.59, 144.73, 144.53, 142.06, 141.79, 135.87, 135.81, 135.56, 130.35, 130.25, 130.21, 130.17, 128.27, 128.24, 128.15, 128.11, 127.15, 127.13, 113.52, 113.49, 101.77, 101.58, 91.26, 91.09, 87.60, 87.53, 87.33, 84.34, 83.23, 70.56, 69.98, 63.19, 55.47, 55.45, 24.87, 24.85, 24.81, 24.77, 18.05, 17.97, 12.15, 12.10, 12.07, 4.68, 4.66. 31P NMR δ (CDCl3): 151.77, 151.24. ESI-MS (−): 969.5; (+): 971.3.
Oligonucleotide Synthesis
DNA and LNA template oligonucleotides (5′-NxGCAGTCAGTCTACGC-3′) were synthesized using standard β-cyanoethyl phosphoramidite chemistry. RNA oligonucleotides were either purchased from Dharmacon, Inc. or synthesized in-house using TOM-protected RNA phosphoramidites. DNA and LNA oligonucleotides were cleaved from the synthesis column and base-deprotected in 40% aqueous ammonium hydroxide at 55 °C for 17 h. dUp DNA templates were synthesized with dmf-dG phosphoramidite to reduce ammonium hydroxide deprotection time, thereby avoiding cyclization of the dUp residues. Integrity of the dUp templates was confirmed by MALDI-TOF MS and lack of fluorescence. RNA oligonucleotides were cleaved from the synthesis column and base-deprotected in 1:1 aqueous methylamine:ethanolic methylamine (40% methylamine) solution for 10 min at 65 °C. RNA oligonucleotide 2′-TOM protecting groups were removed using 1 M tetrabutylammonium fluoride in tetrahydrofuran for at least 6 h at 35 °C. DNA and LNA oligonucleotides were purified by anion-exchange HPLC on a DNAPac PA-200 column (Dionex) in 10 mM Tris-Cl, pH 8.0 in a gradient up to 2 M KCl. RNA oligonucleotides were purified by denaturing polyacrylamide gel electrophoresis. The amino-terminated DNA primer (5′-Cy3-GCGTAGACTGACTGC-2′-amino) was synthesized as previously described.(3) 5′-H-phosphonate-2′-deoxythymidine-3′-lcaa-CPG (total loading, 20 μmol) was charged with 5′-dimethoxytrityl-2′-amino-2′,3′-dideoxycytidine51 (100 μmol, 5 equiv) via oxidative amination in 1:1 carbon tetrachloride/acetonitrile. The primer synthesis proceeded by standard β-cyanoethyl phosphoramidite chemistry with a 5′-terminal Cy3-phosphoramidite dye added last. Primer was cleaved from the column and base-deprotected in 40% aqueous ammonium hydroxide at room temperature for at least 36 h. The primer was then treated with 80% acetic acid for 16 h at room temperature to hydrolyze the 2′-phosphoramidate linkage and generate the 2′-amino-5′-Cy3-labeled DNA primer. The primer was purified by anion-exchange HPLC on the DNAPac PA-200 column in 10 mM NaOH, pH 12, against a 1.5 M NaCl gradient. All oligonucleotides were subsequently purified on an Oligonucleotide Purification Cartridge (Applied Biosystems) to remove excess salt from DNA and LNA oligonucleotides and traces of acrylamide, urea, and salt from the RNA oligonucleotides.
Primer-Extension Reactions
Template-copying reactions contained 0.1 μM Cy3-labeled 2′-amino-terminated primer, 0.5 μM template oligonucleotide, 150 mM NaCl, 100 mM 1-(2-hydroxyethyl)-imidazole, 100 mM Na+-MES-CAPS-HEPES, pH 7.5 and 2′-amino-2′,3′-dideoxynucleoside−5′-phosphorimidazolide at indicated concentrations. Reactions were initiated by addition of monomer followed by thermal cycling at 55 °C for 30 s→4 °C for 2 min repeated twice followed by incubation at 4 °C for the duration of the reaction time. Primer extension reactions incubated at 4 °C without thermal cycling left a significant amount of nonutilized 2′-amino-terminated DNA primer. Thermal cycling yielded full primer utilization presumably due to rearrangement of primer/template complexes trapped in nonproductive conformations. Nonreactive DNA primer/template complexes may require a shift of the 2′-amino terminus into a C3′-endo conformation, a possibility confirmed in pilot primer extension reactions using a 2′-amino-terminated RNA primer where all the sugars are in the C3′-endo conformation, which does not require thermal cycling for full utilization (data not shown). Aliquots were removed and stopped at indicated time points by adding three volumes formamide and heating to 95 °C for 2 min followed immediately by ethanol precipitation on dry ice. Stopped reactions were resuspended in formamide gel loading buffer and heated to 95 °C for 5 min. Samples were analyzed by electrophoresis on 7 M urea, 17% polyacrylamide sequencing gels. Reaction products were visualized by fluorescence imaging on a Typhoon 9410 PhosphorImager using the Cy3 fluorophore filter set. Product quantification and analysis was performed using ImageQuant TL software (GE Healthcare Life Sciences).
Primer Extension Product Mass Spectrometry
Primer-extension product mass was determined by extending 100−200 pmol of the amino-terminated DNA primer on a complementary RNA template in 150 mM NaCl, 100 mM 1-(2-hydroxyethyl)-imidazole, 100 mM Na+-MES-CAPS-HEPES, pH 7.5 and 5 mM 2′-amino-2′,3′-dideoxynucleoside−5′-phosphorimidazolide at 4 °C. The completed primer-extension reaction was subjected to base hydrolysis in 600 mM KOH at 50 °C for 2 h to degrade the RNA template leaving the DNA primer with four 2′-terminal 2′-5′-phosphoramidate linkages and a new 2′-amino terminus. The KOH hydrolysis reaction mixture was then ethanol precipitated and resuspended in water in preparation for MALDI-MS. The sample was desalted and washed with 0.1 M triethylamine acetate on a C18 ZipTip (Millipore). MALDI matrices were 50 mg/mL 3-hydroxypicolinic acid or 10 mg/mL 2,4,6-hydroxyacetophenone with 0.1 M diammonium citrate in 50% acetonitrile. Samples were spotted directly on the MALDI-TOF stainless steel plate and the mass acquisition was completed in the positive mode on a MALDI-TOF mass spectrometer (PerSeptive Biosystems, Model Voyager DE).
Acknowledgments
We are grateful to Jesse J. Chen, Yollete V. Guillen-Schlippe, Ben Heuberger, Sylvia Tobé, Doug Treco, Tina Yuan, and Shenglong Zhang for helpful discussions. A.R. is a Research Associate and J.W.S. is an Investigator of the Howard Hughes Medical Institute. This work was supported in part by NSF Grant CHE 0434507 to J.W.S.
Supporting Information Available
Phosphoramidate acid and enzymatic sensitivity analysis and experimental section (Figures S1 and S2); primer extension reactions on noncomplementary templates (Figures S3−S6); additional mixed-sequence RNA template-copying reactions (Figures S7−S10). This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- Chen I. A.; Roberts R. W.; Szostak J. W. Science 2004, 305, 1474–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W.; Bartel D. P.; Luisi P. L. Nature 2001, 409, 387–390. [DOI] [PubMed] [Google Scholar]
- Mansy S. S.; Schrum J. P.; Krishnamurthy M.; Tobe S.; Treco D. A.; Szostak J. W. Nature 2008, 454, 122–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T. F.; Szostak J. W. J. Am. Chem. Soc. 2009, 131, 5705–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.; Kool E. T. Nucleic Acids Res. 1999, 27, 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuberger B. D.; Switzer C. Org. Lett. 2006, 8, 5809–5811. [DOI] [PubMed] [Google Scholar]
- Benner S. A.; Ricardo A.; Carrigan M. A. Curr. Opin. Chem. Biol. 2004, 8, 672–689. [DOI] [PubMed] [Google Scholar]
- Grossmann T. N.; Strohbach A.; Seitz O. Chembiochem 2008, 9, 2185–2192. [DOI] [PubMed] [Google Scholar]
- Lee D. H.; Severin K.; Yokobayashi Y.; Ghadiri M. R. Nature 1997, 390, 591–594. [DOI] [PubMed] [Google Scholar]
- Sievers D.; von Kiedrowski G. Nature 1994, 369, 221–224. [DOI] [PubMed] [Google Scholar]
- Doudna J. A.; Usman N.; Szostak J. W. Biochemistry 1993, 32, 2111–2115. [DOI] [PubMed] [Google Scholar]
- Muller U. F.; Bartel D. P. RNA 2008, 14, 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul N.; Joyce G. F. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 12733–12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston W. K.; Unrau P. J.; Lawrence M. S.; Glasner M. E.; Bartel D. P. Science 2001, 292, 1319–1325. [DOI] [PubMed] [Google Scholar]
- Lincoln T. A.; Joyce G. F. Science 2009, 323, 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.; Szostak J. W. Science 1992, 258, 1910–1915. [DOI] [PubMed] [Google Scholar]
- Lohrmann R.; Orgel L. E. J. Mol. Evol. 1979, 12, 237–257. [DOI] [PubMed] [Google Scholar]
- Fakhrai H.; van Roode J. H.; Orgel L. E. J. Mol. Evol. 1981, 17, 295–302. [DOI] [PubMed] [Google Scholar]
- Sleeper H. L.; Lohrmann R.; Orgel L. E. J. Mol. Evol. 1979, 13, 203–214. [DOI] [PubMed] [Google Scholar]
- Sulston J.; Lohrmann R.; Orgel L. E.; Miles H. T. Proc. Natl. Acad. Sci. U.S.A. 1968, 59, 726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel S. R.; Richert C. Chem. Commun. 2007, 1896–1898. [DOI] [PubMed] [Google Scholar]
- Lohrmann R.; Orgel L. E. J. Mol. Evol. 1976, 7, 253–267. [DOI] [PubMed] [Google Scholar]
- Zielinski W. S.; Orgel L. E. Nucleic Acids Res. 1985, 13, 2469–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. J.; Cai X.; Szostak J. W. J. Am. Chem. Soc. 2009, 131, 2119–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothlingshofer M.; Kervio E.; Lommel T.; Plutowski U.; Hochgesand A.; Richert C. Angew. Chem., Int. Ed. 2008, 47, 6065–6068. [DOI] [PubMed] [Google Scholar]
- Lohrmann R.; Orgel L. E. Nature 1976, 261, 342–344. [DOI] [PubMed] [Google Scholar]
- Zielinski W. S.; Orgel L. E. Nature 1987, 327, 346–347. [DOI] [PubMed] [Google Scholar]
- von Kiedrowski G. Angew. Chem., Int. Ed. Engl. 1986, 25, 932–935. [Google Scholar]
- Rojas Stutz J. A.; Richert C. Chemistry 2006, 12, 2472–2481. [DOI] [PubMed] [Google Scholar]
- Hill A. R. Jr.; Nord L. D.; Orgel L. E.; Robins R. K. J. Mol. Evol. 1988, 28, 170–171. [DOI] [PubMed] [Google Scholar]
- Gryaznov S. M.; Winter H. Nucleic Acids Res. 1998, 26, 4160–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansy S. S.; Szostak J. W. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 13351–13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner R. E.; Brudno Y.; Birnbaum M. E.; Liu D. R. J. Am. Chem. Soc. 2008, 130, 4646–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Zhan Z. Y.; Knipe R.; Lynn D. G. J. Am. Chem. Soc. 2002, 124, 746–747. [DOI] [PubMed] [Google Scholar]
- Ura Y.; Beierle J. M.; Leman L. J.; Orgel L. E.; Ghadiri M. R. Science 2009, 325, 73–77. [DOI] [PubMed] [Google Scholar]
- Heemstra J. M.; Liu D. R. J. Am. Chem. Soc. 2009, 131, 11347–11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbuch P.; Kervio E.; Hochgesand A.; Plutowski U.; Richert C. Angew. Chem., Int. Ed. 2005, 44, 6588–6592. [DOI] [PubMed] [Google Scholar]
- Ferencic M.; Reddy G.; Wu X.; Guntha S.; Nandy J.; Krishnamurthy R.; Eschenmoser A. Chem. Biodiversity 2004, 1, 939–979. [DOI] [PubMed] [Google Scholar]
- Sheppard T. L.; Breslow R. J. Am. Chem. Soc. 1996, 118, 9810–9811. [Google Scholar]
- Obika S.; Nanbu D.; Hari Y.; Morio K.-I.; In Y.; Ishida T.; Imanishi T. Tetrahedron Lett. 1997, 38, 8735–8738. [Google Scholar]
- Koshkin A.; Singh S. K.; Nielsen P.; Rajwanshi V. K.; Kumar R.; Meldgaard M.; Olsen C. E.; Wengel J. Tetrahedron 1998, 54, 3607–3630. [Google Scholar]
- He J.; Seela F. Nucleic Acids Res. 2002, 30, 5485–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaput J. C.; Sinha S.; Switzer C. Chem. Commun. 2002, 1568–1569. [DOI] [PubMed] [Google Scholar]
- Ninio J.; Orgel L. E. J. Mol. Evol. 1978, 12, 91–99. [DOI] [PubMed] [Google Scholar]
- Chen J. J.; Tsai C. H.; Cai X.; Horhota A. T.; McLaughlin L. W.; Szostak J. W. PLoS ONE 2009, 4, e4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M.; Schuster P. Naturwissenschaften 1977, 64, 541–565. [DOI] [PubMed] [Google Scholar]
- Nowak M. A.; Ohtsuki H. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 14924–14927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kun A.; Santos M.; Szathmary E. Nat. Genet. 2005, 37, 1008–1011. [DOI] [PubMed] [Google Scholar]
- Kanavarioti A.; Bernasconi C. F.; Doodokyan D. L.; Alberas D. J. J. Am. Chem. Soc. 1989, 111, 7247–7257. [DOI] [PubMed] [Google Scholar]
- Hashimoto H.; Switzer C. J. Am. Chem. Soc. 1992, 114, 6255–6256. [Google Scholar]
- Kawana M.; Kuzuhara H. J. Chem. Soc. Perkin Trans. 1 1992, 4, 469–478. [Google Scholar]
- Berry D. A.; Jung K.-Y.; Wise D. S.; Sercel A. D.; Pearson W. H.; Mackie H.; Randolph J. B.; Somers R. L. Tetrahedron Lett. 2004, 45, 2457–2461. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.