Abstract
Sphingolipids are vital components of eukaryotic membranes involved in the regulation of cell growth, death, intracellular trafficking, and the barrier function of the plasma membrane (PM). While sphingomyelin (SM) is the major sphingolipid in mammals, previous studies indicate that mammalian cells also produce the SM analog ceramide phosphoethanolamine (CPE). Little is known about the biological role of CPE or the enzyme(s) responsible for CPE biosynthesis. SM production is mediated by the SM synthases SMS1 in the Golgi and SMS2 at the PM, while a closely related enzyme, SMSr, has an unknown biochemical function. We now demonstrate that SMS family members display striking differences in substrate specificity, with SMS1 and SMSr being monofunctional enzymes with SM and CPE synthase activity, respectively, and SMS2 acting as a bifunctional enzyme with both SM and CPE synthase activity. In agreement with the PM residency of SMS2, we show that both SM and CPE synthase activities are enhanced at the surface of SMS2-overexpressing HeLa cells. Our findings reveal an unexpected diversity in substrate specificity among SMS family members that should enable the design of specific inhibitors to target the biological role of each enzyme individually.
Keywords: bifunctional enzyme, sphingolipid biosynthesis, ceramide homeostasis, cell growth, lipid asymmetry, cervical carcinoma HeLa cells
Sphingolipids are vital components of eukaryotic membranes that have been implicated in a multitude of cellular processes. Sphingomyelin (SM), the major sphingolipid species in mammalian cells, is concentrated in the outer leaflet of the plasma membrane (PM) where its high packing density and affinity for cholesterol contributes to the barrier function of the organelle (1). SM and cholesterol form concentration gradients along the exocytic pathway that are thought to affect protein sorting through hydrophobic matching of membrane spans (2, 3). Moreover, SM can be hydrolyzed by neutral SMases at the PM to form phosphocholine and the proapoptotic signaling molecule ceramide. This enzymatic reaction is part of an antiproliferative, sphingolipid-mediated signal transduction pathway that controls cell cycle arrest, differentiation, and apoptosis in response to growth factor deprivation, cytokines, ionizing radiation, heat, and chemotherapy (4).
SM synthesis is mediated by a phosphatidylcholine (PC):ceramide cholinephosphotransferase or SM synthase (EC 2.7.8.27). This enzyme catalyzes the transfer of phosphocholine from PC onto ceramide, yielding SM and diacylglycerol (5). Mammalian cells also produce small amounts of the SM analog ceramide phosphoethanolamine (CPE). However, very little is known about the biological role of this analog or about the enzyme(s) responsible for its production. Two CPE synthase activities have been described in mammalian cells, one enriched in a microsomal fraction [presumably endoplasmic reticulum (ER)] and the other one associated with the PM (6–9). As PE serves as the headgroup donor for both activities, the enzyme(s) involved can be classified as PE:ceramide ethanolaminephosphotransferases in analogy to SM synthase. While the identity of the mammalian CPE synthase(s) has not been reported yet, characterization of a trypanosome sphingolipid synthase family that is orthologous to the Leishmania inositol phosphoceramide synthase revealed an enzyme with dual CPE and SM synthase activity (10).
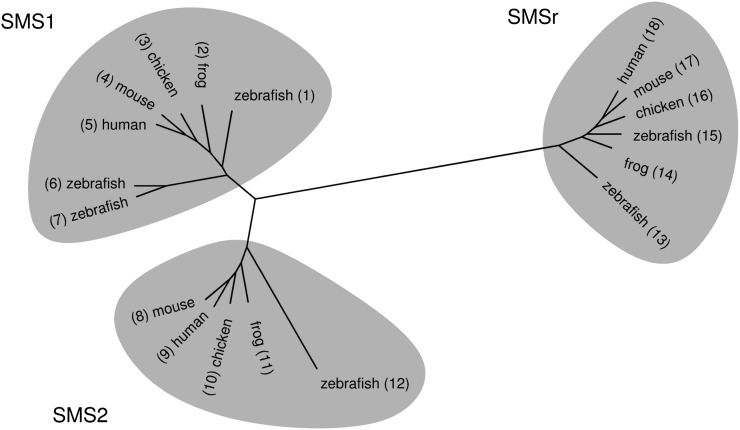
Mammalian cells contain two SM synthase isoforms, namely SMS1, responsible for bulk production of SM in the Golgi, and SMS2 at the PM, serving a role in regenerating SM from ceramides liberated by SMase (11–13). Both SMS1 and SMS2 are required for cell growth, at least in certain types of cancer cells (12, 13). Together with a closely related enzyme, SMSr, they form the SMS protein family (5, 11; Fig. 1). The function of SMSr had so far been unknown, but we recently identified it to be an ER-resident CPE synthase (14). SMSr thus qualifies for the microsomal CPE synthase activity originally described by Malgat et al. (6, 7).
Fig. 1.
SMS1, SMS2, and SMSr are conserved throughout vertebrates. Human, mouse, chicken, zebrafish, and the frog Xenopus laevis have at least one homolog of each SMS1, SMS2, and SMSr. The phylogenetic tree was constructed with Tree Puzzle (24) from protein sequences that were aligned using T-Coffee (25) and displayed using TreeIllustrator (http://www.genohm.com). Some of the sequences were hand-edited before aligning. The UniProt or GenBank accession numbers are: A0JMN0; Q640R5; 54020704; Q8VCQ6; Q86VZ5; Q5U3Z9; A8KBN3; Q9D4B1; Q8NHU3; 50746427; Q5M7L7; Q6DEI3; A4QNV5; Q28CJ3; B0BLX7; 118092693; Q9DA37; and Q96LT4.
The identity of the PM-associated CPE synthase activity (6, 8, 9), however, remains to be established. Because this second CPE synthase shares the same subcellular location as SMS2 and because the enzymatic reactions leading to SM and CPE formation are very similar, we now systematically revisit the substrate specificities of all three SMS family members. Here, we report that while SMS1 and SMSr are monofunctional SM and CPE synthases, respectively, SMS2 is a bifunctional enzyme producing both SM and CPE. Thus, SMS2 very likely accounts for the PM-resident CPE synthase activity described previously. Collectively, our findings reveal a surprising variation in substrate specificity among SMS family members that should facilitate their pharmacological discrimination.
EXPERIMENTAL PROCEDURES
Chemicals
7-nitro-2,1,3-benzoxadiazole (NBD)-C6-ceramide (NBD-Cer) and NBD-SM were from Avanti Polar Lipids, synthetic NBD-CPE was generously provided by Philippe Devaux (Institut de Biologie Physico-Chimique, Paris), and trinitrobenzenesulfonic acid (TNBS) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine were from SIGMA. Organic solvents were distilled before use. Chloroform was stabilized with 2% ethanol.
Yeast strains and culture
Yeast strains IAY11 (MATα ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-52 ade3-Δ853) or 4Δ.CerS5 (MATa ade2-101ochre his3-Δ200 leu2-Δ1 lys2-801amber trp1-Δ63 ura3-52 lag1::TRP1 lac1::LEU2 ydc1::natMX ypc1::kanMX4 p413MET25:CerS5) (originally named 4Δ.Lass5; 15) transformed with human SMS1, SMS2, SMSr, or control insert in pYES2.1/V5-His-TOPO (Invitrogen) (11) were grown in SDUra− medium containing 2% (w/v) galactose and 50 mg/l myo-inositol (in the case of 4Δ.CerS5).
In vitro enzyme assay
A total of 100 OD of IAY11 yeast cells expressing human SMS1, SMS2, SMSr, or control insert was washed with water and lysed by bead bashing in glycerol buffer (10% glycerol, 15 mM KCl, 5 mM NaCl, 1 mM EDTA, 20 mM Hepes-KOH, pH 7.0) containing freshly added protease inhibitors at 4°C. A postnuclear supernatant was prepared by centrifuging at 700 g and 4°C for 10 min. Then 40 nmol 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine was dried under a stream of nitrogen. Glycerol buffer (200 µl) containing 0.002% Triton X-100 was added and sonicated in a water bath for 15 min. A total of 200 µl postnuclear supernatant was added and the mixture was incubated on ice for 1 h. The reaction was started by adding NBD-Cer dissolved in ethanol to a final concentration of 25 µM and incubated at 37°C for 2 h. The reaction was stopped by adding 1 ml of methanol and 0.5 ml of chloroform. Phase separation was induced by further adding 0.5 ml of chloroform and 0.5 ml of 0.45% (w/v) NaCl (16). The lower phase was evaporated under a stream of nitrogen and analyzed by TLC. The TLC was developed first in acetone and dried, and then in chloroform-methanol-25% ammonia (50:25:6, v/v/v). Fluorescent lipids were visualized on a STORM 860 Image Analysis system (Molecular Dynamics). The image was processed with Quantity One software (Bio-Rad) using a γ setting of 0.5.
Derivatization of CPE with TNBS
The bands corresponding to NBD-SM and NBD-CPE in the experiment shown in Fig. 2 were scraped from the TLC and extracted from the silica gel (16). The solvent was evaporated under a stream of nitrogen, and the NBD-labeled lipids were dissolved in a few drops of methanol. Labeling buffer (200 µl; 40 mM NaCl, 120 mM NaHCO3) was added dropwise, followed by 6.5 µl of a 5% (w/v) aqueous solution of TNBS. The reaction was incubated at room temperature for 1 h and then stopped by adding 200 µl of 0.45% (w/v) NaCl, 1 ml of methanol, and 0.5 ml chloroform. Phase separation was induced by further adding 0.5 ml of chloroform and 0.5 ml of 0.45% (w/v) NaCl (16). The lower phase was evaporated under a stream of nitrogen and analyzed by TLC in chloroform-methanol-25% ammonia (50:25:6,v/v/v). Fluorescent lipids were detected as described above.
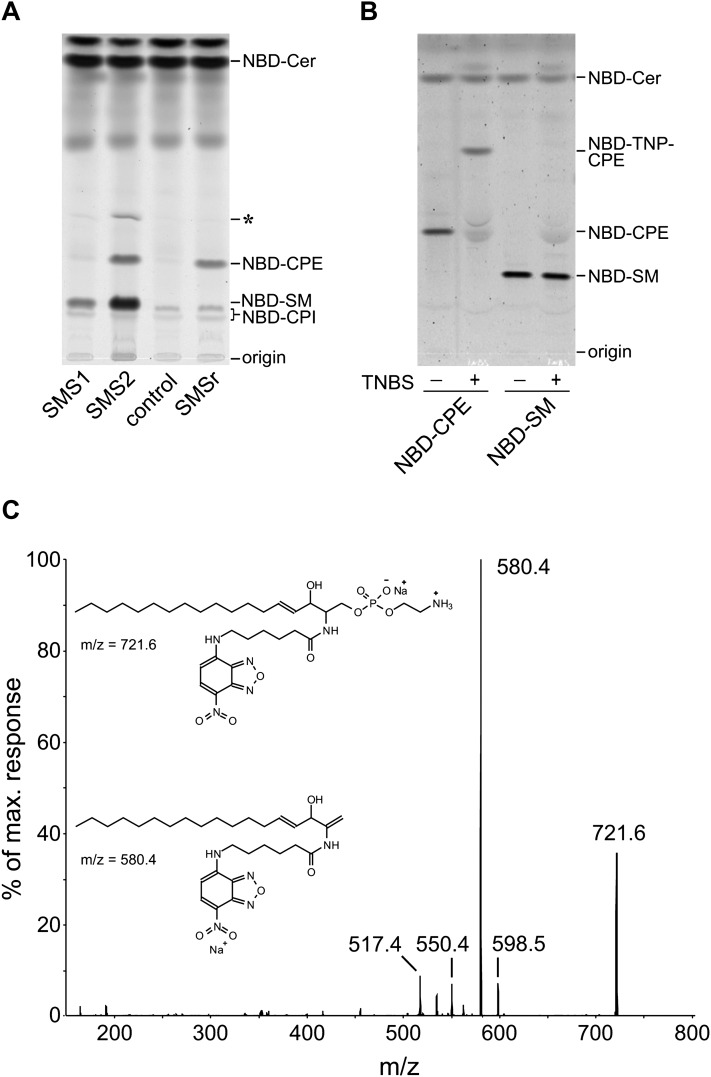
Fig. 2.
SMS2 displays dual activity as SM and CPE synthase in vitro. A: Postnuclear supernatants of yeast strains expressing human SMS1, SMS2, or SMSr or transformed with empty vector (control) were incubated with NBD-Cer. NBD-labeled lipids were separated by TLC and detected by fluorescence scanning. SMS1 and SMSr catalyze the production of only NBD-SM or NBD-CPE, respectively, while SMS2 synthesizes both NBD-SM and NBD-CPE. An unknown by-product that was not consistently observed is marked with an asterisk (*). B: The primary amino group of NBD-CPE can be derivatized with TNBS, a reagent specifically reacting with primary amino groups, which results in a shift of the Rf value (NBD-trinitrophenyl-CPE). NBD-SM, in contrast, does not react with TNBS. This confirms the identity of the products formed in the in vitro enzyme assay shown in A. C: MS/MS fragmentation spectrum of NBD-CPE. NBD-CPE extracted from the TLC shown in Fig. 2A was detected as a sodium adduct (m/z = 721.6). Upon fragmentation, this ion shows a neutral loss of 141 Da corresponding to the loss of the phosphoethanolamine headgroup, resulting in an ion with m/z = 580.4.
MS of lipids produced in yeast
A total of 100 OD of 4Δ.CerS5 yeast cells expressing human SMS1, SMS2, SMSr, or control insert was washed with water, treated with trichloroacetic acid to inactivate lipases, and finally boiled for 10 min (17) before lipid extraction by bead bashing in chloroform-methanol-water (10:10:3, v/v/v) (18). Phase separation was induced by adding chloroform and 0.45% NaCl according to Folch et al. (19). The lower phase was evaporated under a stream of nitrogen. Glycerolipids were removed by mild alkaline hydrolysis in 0.5 M sodium methoxide in methanol (Fluka) for 1 h at room temperature. Lipids were separated by HPLC and analyzed by mass spectrometry on a 4000 Q TRAP system (Applied Biosystems) using electrospray ionization (20). Source temperature was set to 450°C and nitrogen was used as curtain gas. CPE species were identified by scanning for the neutral loss of a fragment of 141 Da corresponding to phosphoethanolamine using an ionspray voltage of −4.3kV, a declustering potential of −110V, and a collision energy of 35V. SM species were detected as precursors of the fragment ion with m/z = +184 corresponding to phosphocholine using an ionspray voltage of +5.5kV, a declustering potential of 80V, and a collision energy of 43V. Product spectra were recorded at a collision energy of 50V and a collision energy spread of 10V.
Mammalian cell culture
HeLa cell lines stably overexpressing human SMS1, SMS2, or SMSr from pcDNA3.1/V5-His TOPO (13, 14) were cultured in DMEM supplemented with 10% Fetal Clone I (HyClone) at 37°C and 5% CO2.
Cell surface enzyme assay
This assay is adopted from van Helvoort et al. (21). HeLa cells stably overexpressing human SMS1, SMS2, or SMSr were grown to 80–90% confluence in a 10 cm dish. Expression of the transgene was enhanced with 10 mM butyrate. The cells were washed with HBSS and preincubated with HBSS containing 1% fatty acid-free BSA at 5°C for 30 min. NBD-Cer dissolved in ethanol was added to a final concentration of 2 µM (0.2% ethanol in the medium) and the assay was incubated at 5°C for 4 h. The supernatant was saved and the cells were washed by incubating with HBSS containing 1% fatty acid-free BSA at 5°C for 30 min. The new supernatant was also saved and the cells were scraped in PBS. Lipids were extracted from the incubation medium, the wash, and the cells (16). The lipid extracts from the medium and the wash were combined, the lipid extract from the cells was kept separate. Lipids were analyzed by TLC in chloroform-acetone-methanol-acetic acid-water (50:20:10:10:5, v/v/v/v/v). Fluorescent lipids were visualized on a STORM 860 Image Analysis system (Molecular Dynamics) and quantified using Quantity One software (Bio-Rad).
RESULTS
SMS1, SMS2, and SMSr show different substrate specificities in vitro
Human SMS1, SMS2, and SMSr were expressed in budding yeast, and their enzymatic activities were investigated in vitro by incubating postnuclear supernatants with fluorescently labeled ceramide (NBD-Cer) as described in Experimental Procedures. Yeast is naturally lacking SM and CPE and instead produces sphingolipids with a headgroup based on phosphoinositol. However, in postnuclear supernatants of yeast cells expressing SMS1, a fluorescently labeled product with an Rf value corresponding to NBD-SM was formed, confirming previous results that SMS1 is a dedicated SM synthase (11; Fig. 2A). When yeast cells expressing SMSr were investigated, a product with an Rf value higher than that of NBD-SM was detected. This product was extracted from the silica gel and further identified as NBD-CPE by comparing its Rf value with that of synthetic NBD-CPE (data not shown), by derivatization with TNBS (Fig. 2B), and by MS/MS analysis (Fig. 2C), confirming that SMSr is a dedicated CPE synthase (14). In postnuclear supernatants of yeast cells expressing SMS2, both NBD-SM and NBD-CPE were formed (Fig. 2A), indicating that this enzyme possesses dual activity. Together, these results suggest that the SM synthase family comprises both mono- and bifunctional enzymes, with each enzyme displaying a distinct substrate specificity.
SMS1, SMS2, and SMSr show different substrate specificities in vivo
To exclude the possibility that the differences in substrate specificities among SMS family members were an artifact caused by the use of the unnatural ceramide analog NBD-Cer as substrate, we next investigated their ability to convert natural ceramides into SM and/or CPE. To this end, human SMS1, SMS2, and SMSr were expressed in the yeast strain 4Δ.CerS5 and lipid extracts were analyzed by LC/MS/MS. The ceramide composition of the 4Δ.CerS5 strain resembles that of mammalian cells in containing C16 fatty acids due to a replacement of the endogenous ceramide synthases Lag1p and Lac1p by the mouse ceramide synthase CerS5 (15). Normally, yeast ceramides mainly contain very long chain fatty acids (C24–C26).
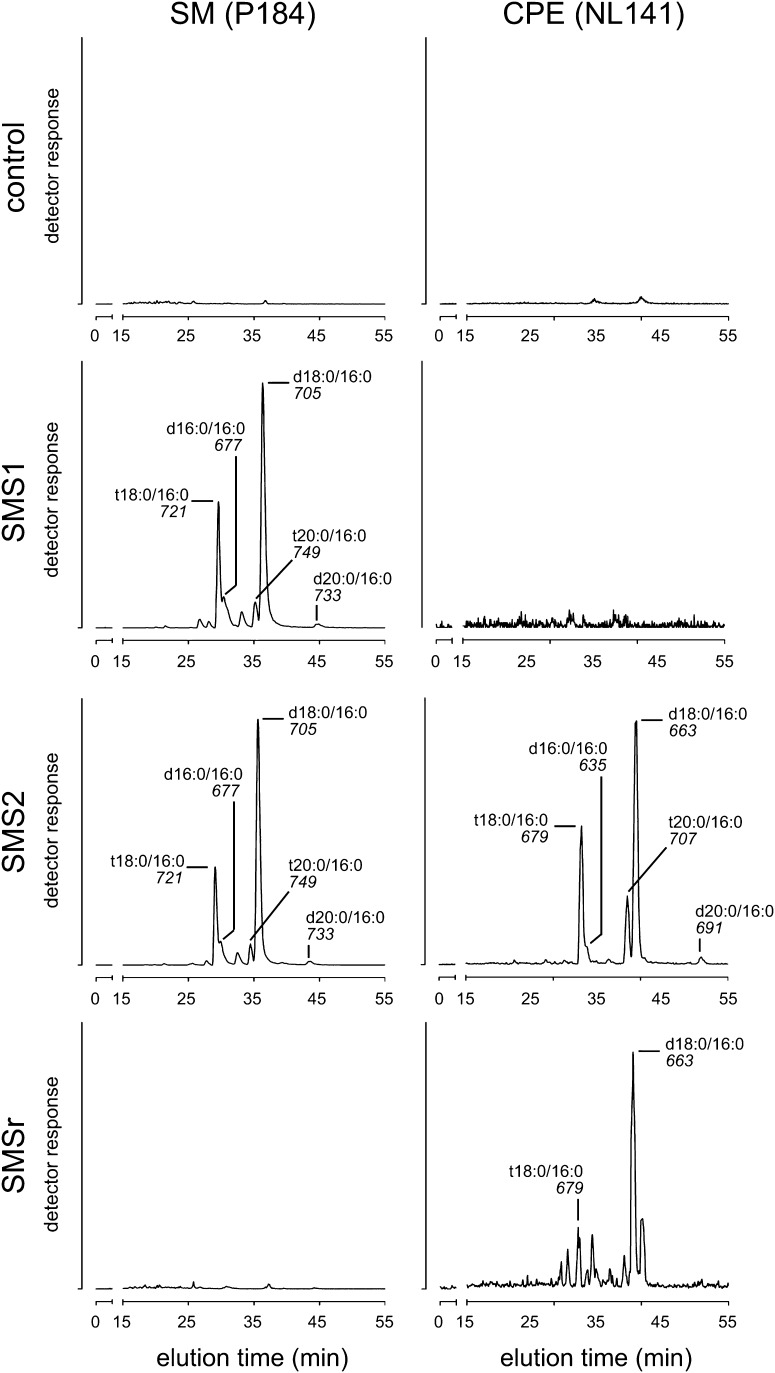
Consistent with the substrate specificities found in the in vitro enzyme assay, cells expressing SMS1 produced only SM while cells expressing SMSr produced only CPE. Importantly, both CPE and SM were detected in cells expressing SMS2, hence confirming that SMS2 is a bifunctional SM/CPE synthase (Fig. 3). The identity of SM and CPE molecular species eluting from the column was determined by on-line acquisition of product spectra (supplementary Fig. I; data not shown). This revealed that the ceramide backbone composition of SM and CPE was very similar in all samples and corresponds to the ceramide composition of the 4Δ.CerS5 strain (15).
Fig. 3.
SMS2 displays dual activity as SM and CPE synthase in vivo. The substrate specificities of human SMS1, SMS2, and SMSr were investigated by heterologous expression in yeast. The yeast strain 4Δ.CerS5 used in this experiment provides ceramides containing a C16 fatty acid as substrates for the biosynthesis of SM or CPE (15). Several molecular species of SM and CPE were detected in yeast cells expressing SMS2, while only SM or CPE species were detected in cells expressing SMS1 or SMSr, respectively. The ceramide backbone composition of the SM or CPE formed was similar and independent of the enzyme (SMS1, SMS2, or SMSr). SM species were detected by LC/MS as precursors of the fragment ion with m/z = +184 corresponding to phosphocholine (P184) and CPE species by a neutral loss of 141 Da corresponding to phosphoethanolamine (NL141). The nominal mass of the most intense ion and the ceramide backbone composition of the corresponding SM or CPE species are indicated for each peak. The sphingoid base composition (d18:0, sphinganine; t18:0, phytosphinganine, as well as C16 and C20 derivatives thereof) was inferred assuming that the ceramides in the yeast strain 4Δ.CerS5 contain almost exclusively palmitic acid (16:0) (15).
SMS2 displays dual SM and CPE synthase activity on the surface of mammalian cells
To ensure that the apparent dual activity of SMS2 is not a consequence of its heterologous expression in yeast, we next analyzed its substrate specificity in human HeLa cells. The three members of the human SMS family show different subcellular localizations. While SMS1 and SMS2 reside in the Golgi apparatus and at the PM, respectively (11, 13), SMSr is associated with the ER (14). In each case, the active site of the enzyme is situated on the exoplasmic surface of the membrane (11, 13). This implies that CPE formed by SMSr is produced intracellularly, while CPE formed by SMS2 should be synthesized on the cell surface.
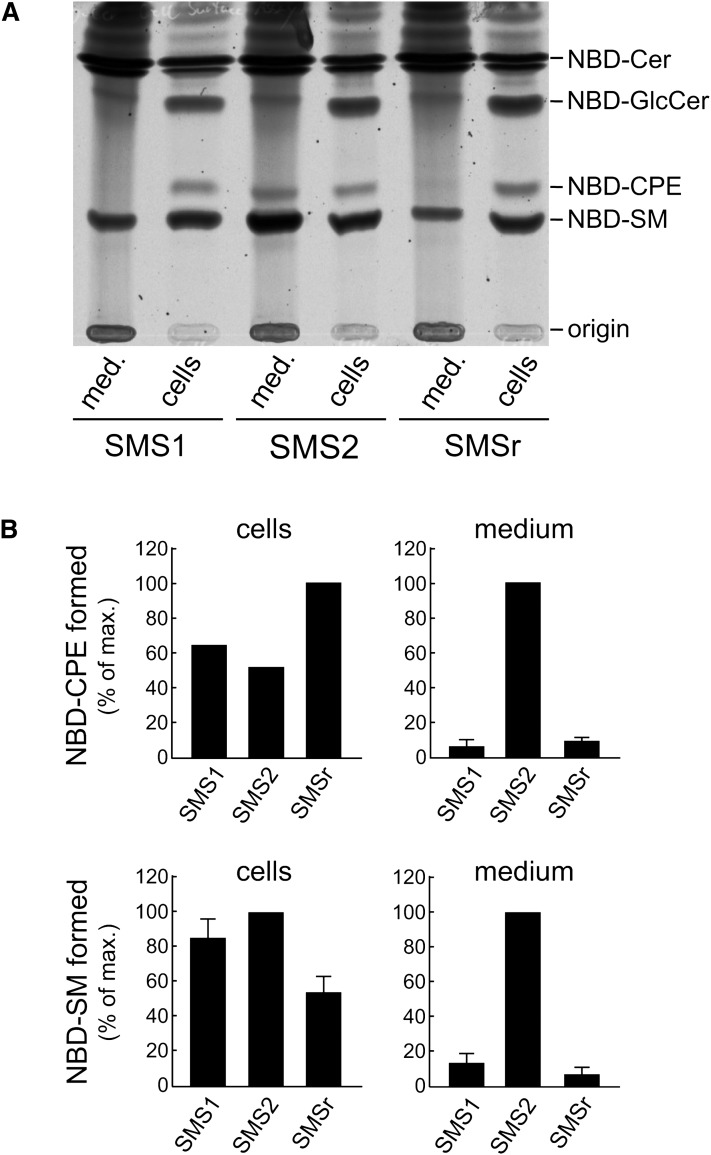
To test whether this indeed is the case, HeLa cells stably overexpressing SMS1, SMS2, or SMSr were incubated with NBD-Cer for 4 h at 5°C in medium containing BSA. Under these conditions, vesicular traffic is blocked, so that fluorescent lipids produced in the ER or Golgi lumen are unable to reach the cell surface. At the same time, any fluorescent lipid formed at the cell surface will be extracted by BSA present in the medium. TLC analysis of cell- and medium-derived lipid extracts showed that NBD-CPE was produced intracellularly in all three cell lines, while NBD-CPE formed at the cell surface was detectable only in SMS2-overexpressing cells (Fig. 4, A and B). Similarly, while intracellular production of NBD-SM was roughly equal in all three cell lines, the level of NBD-SM synthesized at the cell surface was greatly increased in SMS2-overexpressing cells. These results are not due to leakage from damaged cells, because NBD-glucosylceramide, a product synthesized from NBD-Cer on the cytosolic surface of the Golgi, could not be detected in the medium of SMS-overexpressing cells. Collectively, these data demonstrate that SMS2 is indeed capable of catalyzing both CPE and SM production on the surface of mammalian cells.
Fig. 4.
SMS2-overexpressing HeLa cells display elevated SM and CPE synthase activities at the cell surface. A: HeLa cells overexpressing human SMS1, SMS2, or SMSr were incubated with NBD-Cer for 4 h at 5°C in medium containing BSA to extract any fluorescent lipid formed at the cell surface. Total lipid extracts from cells and medium (med) were separated by TLC and detected by fluorescence scanning. While both NBD-SM and NBD-CPE are detected inside the cells in all three cell lines, NBD-CPE can be detected only in medium derived from SMS2-overexpressing cells. NBD-SM is strongly enhanced in the medium of the SMS2-overexpressing cell line as compared with the other cell lines. B: Quantification of the results shown in A, revealing a 10-fold increase for both cell surface-associated SM and CPE synthase activities in SMS2-overexpressing cells.
DISCUSSION
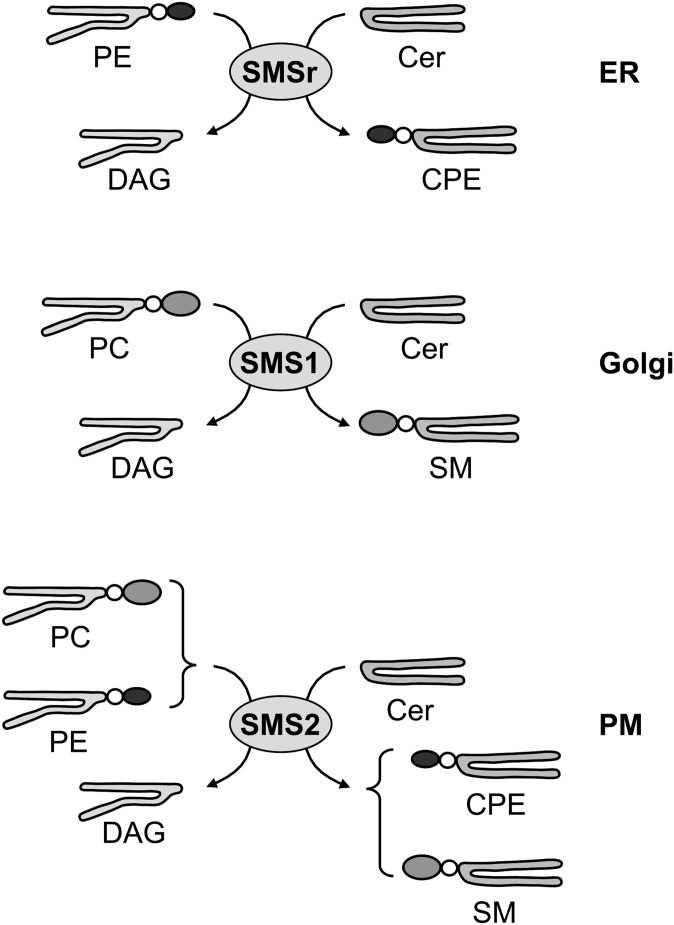
In this study, we show that the mammalian SMS family contains both single and dual activity enzymes, with SMS1 being a monofunctional SM synthase, SMSr a monofunctional CPE synthase, and SMS2 a bifunctional enzyme with both SM and CPE synthase activity. These results, combined with our previous observation that each enzyme resides in a different organelle along the secretory pathway, establish an unexpected level of complexity in the organization of sphingolipid biosynthesis in mammalian cells (Fig. 5).
Fig. 5.
SMS family members possess different substrate specificities. While SMSr and SMS1 represent dedicated CPE and SM synthases in the lumen of the ER and Golgi complex, respectively, SMS2 is a bifunctional enzyme that uses both PC and PE as headgroup donors to synthesize SM and CPE at the cell surface.
In vitro enzyme assays previously demonstrated the presence of CPE synthase activities in microsomes and the PM from rat liver and brain (6–9). While SMSr qualifies for the microsomal CPE synthase activity (14), our present findings indicate that SMS2 most likely accounts for the PM-localized CPE synthase activity. Common features include the use of PE as headgroup donor, the PM residency of the enzyme as well as its membrane topology, with the active site facing the cell surface. While the SM synthase activity of SMS2 was investigated by our group before (11, 13), its CPE synthase activity had been missed. This is presumably due to the high PC-PE ratio in the exoplasmic leaflet of the PM (see below), the shorter incubation times used in previous in vitro enzyme assays, and the presence of Triton X-100 in the reaction buffer. It appears that the CPE synthase activity of SMS2 is more sensitive to detergent than its SM synthase activity (P. Ternes, unpublished observations).
What determines the striking variation in substrate specificity among SMS family members? Because these enzymes use the same reaction chemistry (5, 11, 14, 22), the specificity for SM and/or CPE synthesis is most likely defined by differences in the geometry of the substrate-binding site. These differences would favor binding of either PC (for SM synthesis), phosphatidylethanolamine (for CPE synthesis), or both. The high degree in sequence conservation between SMS1, SMS2, and SMSr (11) combined with their single and dual enzyme activities described here offer unique opportunities to map residues involved in substrate recognition and to further elucidate the reaction mechanism of this important class of enzymes. Moreover, the variation in substrate specificity should enable the development of specific inhibitors to target the biological role of each enzyme individually.
Even though mammalian cells contain two distinct CPE synthases, SMSr and SMS2, their CPE content is strikingly low (i.e., <0.05 mol% of total phospholipid or 300-fold lower than SM) (14). In the case of SMSr, this enigma cannot be ascribed to a limited supply of substrates (ceramide, PE) to the enzyme’s active site. Instead, it appears that SMSr is unable to synthesize bulk amounts of CPE, because its catalytic activity is inhibited by CPE (14). For SMS2, the situation may be different. In healthy mammalian cells, PE is largely confined to the cytosolic leaflet of the PM, whereas PC in the exoplasmic leaflet should be plentiful (23). As the active site of SMS2 faces the exoplasmic surface of the PM (11), this would favor production of SM over CPE.
Previous work showed that both SMS1 and SMS2 are required for cell growth, at least in certain types of cancer cells (12, 13). This indicates that their functions are nonredundant. While Golgi-associated SMS1 consumes newly synthesized ceramide to generate the bulk of SM needed for cell growth, SMS2 at the PM keeps ceramide levels low by consuming ceramide produced by SMase. Our present finding that SMS2 is a bifunctional SM/CPE synthase raises the question of how its dual activity integrates into this functional context.
An intriguing possibility is that, in analogy to SMSr, the catalytic activity of SMS2 is negatively controlled by its product CPE. Owing to the high PC-PE ratio in the exoplasmic leaflet of the PM, SMS2 would retain full activity. However, when PM lipid asymmetry is dissipated, e.g., during apoptosis, the higher proportion of PE in the exoplasmic leaflet would stimulate SMS2-mediated CPE production and potentially trap the enzyme in a CPE-bound, inactive form. This, in turn, would block the enzyme’s ability to neutralize the action of SMase and to attenuate proapoptotic ceramide signaling at the PM. The dual role of SMS2 as CPE synthase might therefore serve to accelerate the onset of apoptosis in cells that have lost PM lipid asymmetry. The potential impact of transbilayer lipid organization on the activity of SMS2 as a negative regulator of ceramide signaling at the PM is currently under investigation.
Supplementary Material
Acknowledgments
The authors thank Ana Vacaru and Fikadu Geta Tafesse for stimulating discussions, and Martin Hermansson for helpful comments on the manuscript. We also thank Philippe Devaux for providing synthetic NBD-CPE and Andreas Conzelmann for the yeast strain 4Δ.CerS5.
Footnotes
Abbreviations:
- CPE
- ceramide phosphoethanolamine
- ER
- endoplasmic reticulum
- NBD
- 7-nitro-2,1,3-benzoxadiazole
- NBD-Cer
- 7-nitro-2,1,3-benzoxadiazole ceramide
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PM
- plasma membrane
- SM
- sphingomyelin
- TNBS
- trinitrobenzenesulfonic acid
- TNP
- trinitrophenyl
This work was supported by a Marie Curie Intra-European Fellowship and an EMBO Long-Term Fellowship (to P.T.) and by grants from the Dutch Organization of Sciences (NWO-CW) and the Utrecht University High Potential Program (to J.C.M.H.).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure.
REFERENCES
- 1.van Meer G., Voelker D. R., Feigenson G. W. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bretscher M. S., Munro S. 1993. Cholesterol and the Golgi apparatus. Science. 261: 1280–1281. [DOI] [PubMed] [Google Scholar]
- 3.Patterson G. H., Hirschberg K., Polishchuk R. S., Gerlich D., Phair R. D., Lippincott-Schwartz J. 2008. Transport through the Golgi apparatus by rapid partitioning within a two-phase membrane system. Cell. 133: 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hannun Y. A., Obeid L. M. 2008. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9: 139–150. [DOI] [PubMed] [Google Scholar]
- 5.Tafesse F. G., Ternes P., Holthuis J. C. M. 2006. The multigenic sphingomyelin synthase family. J. Biol. Chem. 281: 29421–29425. [DOI] [PubMed] [Google Scholar]
- 6.Malgat M., Maurice A., Baraud J. 1986. Sphingomyelin and ceramide-phosphoethanolamine synthesis by microsomes and plasma membranes from rat liver and brain. J. Lipid Res. 27: 251–260. [PubMed] [Google Scholar]
- 7.Malgat M., Maurice A., Baraud J. 1987. Sidedness of ceramide-phosphoethanolamine synthesis on rat liver and brain microsomal membranes. J. Lipid Res. 28: 138–143. [PubMed] [Google Scholar]
- 8.Maurice A., Malgat M., Baraud J. 1989. Sidedness of ceramide-phosphoethanolamine synthesis on rat liver plasma membrane. Biochimie. 71: 373–378. [DOI] [PubMed] [Google Scholar]
- 9.Hinkovska-Galcheva V. T., Petkova D. H., Nikolova M. N. 1989. Sphingomyelin and ceramide: phosphoethanolamine synthesis in ram spermatozoa plasma membrane. Int. J. Biochem. 21: 1153–1156. [DOI] [PubMed] [Google Scholar]
- 10.Sutterwala S. S., Hsu F-F., Sevova E. S., Schwartz K. J., Zhang K., Key P., Turk J., Beverley S. M., Bangs J. D. 2008. Developmentally regulated sphingolipid synthesis in African trypanosomes. Mol. Microbiol. 70: 281–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huitema K., van den Dikkenberg J., Brouwers J. F. H. M., Holthuis J. C. M. 2004. Identification of a family of animal sphingomyelin synthases. EMBO J. 23: 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaoka S., Miyaji M., Kitano T., Umehara H., Okazaki T. 2004. Expression cloning of a human cDNA restoring sphingomyelin synthesis and cell growth in sphingomyelin synthase-defective lymphoid cells. J. Biol. Chem. 279: 18688–18693. [DOI] [PubMed] [Google Scholar]
- 13.Tafesse F. G., Huitema K., Hermansson M., van der Poel S., van den Dikkenberg J., Uphoff A., Somerharju P., Holthuis J. C. M. 2007. Both sphingomyelin synthases SMS1 and SMS2 are required for sphingomyelin homeostasis and growth in human HeLa cells. J. Biol. Chem. 282: 17537–17547. [DOI] [PubMed] [Google Scholar]
- 14.Vacaru A. M., Tafesse F. G., Ternes P., Kondylis V., Hermansson M., Brouwers J. F. H. M., Somerharju P., Rabouille C., Holthuis J. C. M. 2009. Sphingomyelin synthase-related protein SMSr controls ceramide homeostasis in the ER. J. Cell Biol. 185: 1013–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerantola V., Vionnet C., Aebischer O. F., Jenny T., Knudsen J., Conzelmann A. 2007. Yeast sphingolipids do not need to contain very long chain fatty acids. Biochem. J. 401: 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 17.Smith S. W., Lester R. L. 1974. Inositol phosphorylceramide, a novel substance and the chief member of a major group of yeast sphingolipids containing a single inositol phosphate. J. Biol. Chem. 249: 3395–3405. [PubMed] [Google Scholar]
- 18.Zhao C., Beeler T., Dunn T. 1994. Suppressors of the Ca(2+)-sensitive yeast mutant (csg2) identify genes involved in sphingolipid biosynthesis. Cloning and characterization of SCS1, a gene required for serine palmitoyltransferase activity. J. Biol. Chem. 269: 21480–21488. [PubMed] [Google Scholar]
- 19.Folch J., Lees M., Stanley G. H. S. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 20.Retra K., Bleijerveld O. B., van Gestel R. A., Tielens A. G., van Hellemond J. J., Brouwers J. F. 2008. A simple and universal method for the separation and identification of phospholipid molecular species. Rapid Commun. Mass Spectrom. 22: 1853–1862. [DOI] [PubMed] [Google Scholar]
- 21.van Helvoort A., van’t Hof W., Ritsema T., Sandra A., van Meer G. 1994. Conversion of diacylglycerol to phosphatidylcholine on the basolateral surface of epithelial (Madin-Darby canine kidney) cells. Evidence for the reverse action of a sphingomyelin synthase. J. Biol. Chem. 269: 1763–1769. [PubMed] [Google Scholar]
- 22.Yeang C., Varshney S., Wang R., Zhang Y., Ye D., Jiang X. C. 2008. The domain responsible for sphingomyelin synthase (SMS) activity. Biochim. Biophys. Acta. 1781: 610–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holthuis J. C. M., Levine T. 2005. Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 6: 209–220. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt H. A., Strimmer K., Vingron M., von Haeseler A. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 18: 502–504. [DOI] [PubMed] [Google Scholar]
- 25.Notredame C., Higgins D. G., Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302: 205–217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.