Abstract
Adipocyte lipolysis is controlled by complex interactions of lipases, cofactors, and structural proteins associated with lipid droplets. Perilipin (Plin) A is a major droplet-associated protein that functions as a scaffold, both suppressing basal and facilitating cAMP-dependent protein kinase (PKA)-stimulated lipolysis. Plin is required for the translocation of hormone-sensitive lipase (HSL) from the cytosol to lipid droplets upon stimulation. In these studies, we provide direct evidence for a physical interaction of HSL with Plin. By coexpressing HSL with truncation mutations of Plin, we demonstrate using coimmunoprecipitation that HSL can interact with an N-terminal region located between amino acids 141 and 200 of Plin A as well as with a C-terminal region located between amino acids 406 and 480. The N-terminal construct, Plin 1-200, which does not associate with lipid droplets but interacts with HSL, can function as a dominant negative for PKA-stimulated lipolysis. Using confocal microscopy of Plin truncations, we demonstrate that sequences between amino acids 463 and 517 may be important for or participate in lipid targeting. The results suggest the translocation of HSL to the lipid droplet occurs by virtue of Plin localization to the surface of lipid droplets and a physical interaction of HSL occurring with sequences within the N-terminal region of Plin.
Keywords: translocation, lipid droplet, adipocyte
The major contribution of adipose tissue to whole-body metabolism is the storage of energy in the form of triacylglycerols and the mobilization of this stored energy leading to the release of FFA, the process known as lipolysis. The control of lipolysis represents a complex process involving multiple regulatory events (1, 2). These broadly include lipolytic (ACTH, β-adrenergic agonists, etc.) and anti-lipolytic (insulin, adenosine, etc.) hormones, their cognate receptors and signaling pathways, and lipid droplet-associated proteins, such as perilipins, cytosolic lipid binding proteins, and lipases. The complexity of the regulation of lipolysis has further increased in recent years with the identification of a large number of proteins that are found associated with the lipid droplets stored within adipocytes and with the realization of the dynamic nature of the proteins that populate lipid droplets.
The major lipid droplet-associated protein in differentiated adipocytes is perilipin (Plin) A, which is also the most highly phosphorylated protein in adipose cells following cyclic AMP-dependent protein kinase (PKA) activation (3). Plin A plays an essential role in regulating lipolysis, as evidenced by the 3-fold elevation in basal lipolysis and the absence of PKA-stimulated lipolysis in adipocytes from Plin A null mice (4, 5). Moreover, using model cell systems, it has been shown that Plin A promotes PKA-stimulated lipolysis (6–8). Thus, it appears that Plin A has dual functions of acting as a barrier to suppress basal or constitutive lipolysis and of enhancing PKA-stimulated lipolysis.
Intracellular lipases are instrumental in lipolysis, and there are many proteins that are predicted to have lipase or esterase activity that are expressed in adipose tissue. It had long been considered that hormone-sensitive lipase (HSL) (9, 10) was the rate-limiting enzyme for lipolysis; however, examination of the nonobese HSL null mouse indicated the existence and importance of other lipases (11). Adipose triglyceride lipase (ATGL) (12–14) was identified as an acylhydrolase that catalyzes the initial step of triacylglycerol (TAG) hydrolysis to diacylglycerol (DAG) but lacks DAG hydrolase activity. In combination, HSL and ATGL account for >95% of TAG hydrolase activity in murine adipose cells (15). Generation of ATGL null mice (16) and knockdown of endogenous ATGL in murine adipocytes (6, 17) have provided evidence that ATGL is the rate-limiting enzyme for the initiation of PKA-stimulated lipolysis in murine cells and that ATGL is predominantly responsible for the first step of TAG hydrolysis and HSL primarily responsible for the hydrolysis of DAG. However, some data have suggested that ATGL is less important in human adipose cells and that HSL fulfills the function as the rate-limiting TAG hydrolase in human adipose cells (18, 19). Therefore, both HSL and nonHSL lipase(s), such as ATGL, are involved in the control of lipolysis.
Previous work from our laboratories, using model adipocyte systems, has shown that the C-terminal domain of Plin is responsible for suppressing basal lipolysis mediated by non-HSL lipases, whereas sequences within the N-terminal domain suppress basal lipolysis mediated by HSL (8). Moreover, both the C-terminal and the N-terminal domains enhance PKA-stimulated lipolysis by HSL, whereas only the C-terminal domain enhances PKA-stimulated lipolysis mediated by non-HSL lipases (8). PKA-stimulated lipolysis is paralleled by a translocation of HSL from the cytosol to the surface of the lipid droplet (20, 21), which is dependent on the phosphorylation of HSL (22). Either no or only modest movement of ATGL to the lipid droplet is observed with PKA stimulation (12, 23). Whereas PKA-mediated phosphorylation of Plin is required for stimulated lipolysis, the translocation of HSL to the lipid droplet is dependent on Plin (7) but independent of phosphorylation of Plin (6). This dichotomy is partially explained by the importance of phosphorylation of Plin at Ser-517 to regulate ATGL and the initiation of lipolysis (24). The close cooperation of HSL and Plin in stimulated lipolysis and the involvement of both the N- and C-terminal domains of Plin in HSL-mediated lipolysis imply a potential physical interaction between Plin and HSL that affects lipolysis. Indeed, in support of this, we have shown that HSL could be cross-linked and coimmunoprecipitated with phosphorylated Plin (6). Additionally, recent experiments using fluorescence resonance energy transfer and bimolecular fluorescence complementation suggest that HSL closely, if not directly, interacts with Plin (23).
In this article, we provide additional evidence that HSL and Plin form a physical interacting complex and, using deletion constructs of Plin, we have located two separate regions within Plin, one within the N-terminal portion and one within the C-terminal portion, that are involved in the interaction with HSL. Furthermore, using coexpression of a Plin N-terminal fragment that can bind to HSL, yet does not localize to the lipid droplet (Plin 1-200), we show that the localization of Plin to the lipid droplet and its interaction with HSL are required for hormone-stimulated, HSL-mediated lipolysis.
EXPERIMENTAL PROCEDURES
Chemicals and reagents
Reagents were obtained from the following sources: BSA (fraction V) from Sigma-Aldrich (St. Louis, MO); FBS from Gemini Bio-Products (Calabasas, CA); Coon's F12/DMEM, Lipofectin reagent from Invitrogen (Carlsbad, CA); ECL Western blotting detection reagents, horseradish peroxidase-linked whole antibody anti-rabbit IgG, cholesteryl [1-14C] oleate, and [35S]methionine from Amersham Life Sciences Products (Arlington Heights, IL); nitrocellulose paper from Schleicher and Schuell (Keene, NH); and organic solvents from J.T. Baker (Phillipsburg, NJ). The glutathione beads and glycerol assay kit were from Stanbio (Boerne, TX).
Cell lines
Chinese hamster ovary (CHO) cells and CHO cells overexpressing HSL (3F9) were grown in Coon's F12/DMEM supplemented with 10% FBS at 37°C under 5% CO2. Fatty acid transport protein 1-acyl-coenzyme A synthase 1 (FATP1-ACS1) cells were infected with retrovirus carrying full-length mouse HSL using polybrene method (25). Expression of HSL was confirmed by Western blotting and assay of HSL activity. A stable clone that overexpresses HSL was selected and maintained.
In vitro protein-protein interaction
Full-length Plin was in vitro translated with [35S]methionine using the TNT® transcription/translation system (26). Glutathione S-transferase (GST)-HSL or GST alone (27) were incubated with glutathione-agarose beads in buffer B (20 mM Tris, pH 8.0, 0.15 M NaCl, 1 mM EDTA, and 0.5% Nonidet P-40). After 1 h of incubation at room temperature, the beads were washed three times in buffer B and then incubated with [35S]methionine-labeled Plin. After 1 h of incubation at room temperature, the beads were washed five times in buffer B, and proteins that bound to the beads were eluted in SDS-PAGE sample buffer, separated on SDS/4–20% PAGE, and visualized on a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Overexpression of full-length and truncated Plin A in cells and lipolysis assays
FATP1-ACS1-HSL cells were transduced with recombinant adenovirus carrying full-length or truncated Plin A constructs (Fig. 1 is a schematic diagram of the structural features of Plin A and the Plin constructs) with LipofectAMINE Plus™ (Invitrogen). The amount of each adenovirus used was selected to assure comparable levels of expression of the different Plin A constructs, which was confirmed by Western blots and densitometry. Twenty-four hours after adenovirus transduction, cells were loaded with lipids using BSA-bound FA for 2 days. Subsequently, cells were washed and changed to DMEM +2% FA-free BSA and treated with/without the PKA activator forskolin (20 µM) to stimulate lipolysis. Glycerol released to the media was quantified using an assay kit from Stanbio.
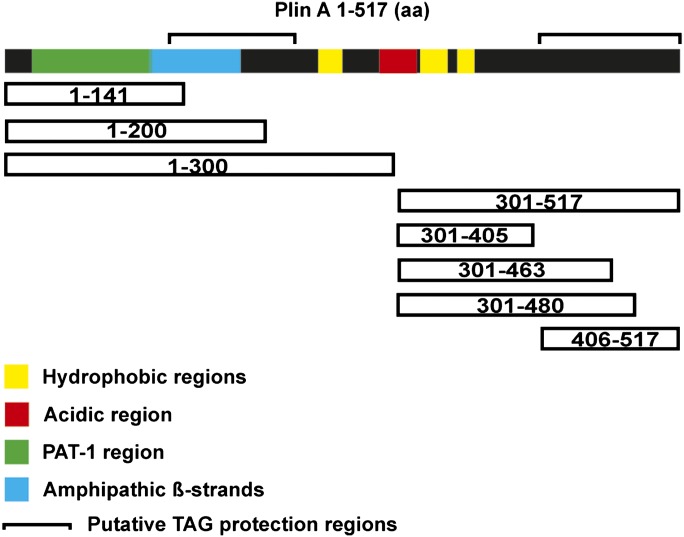
Fig. 1.
Schematic diagrams of structural features of Plin A and Plin constructs. Full-length Plin A is 517 amino acids (aa). The PAT-1 region includes amino acids 17–111, amphipathic β-strands are found between amino acids 111–182, hydrophobic sequences are amino acids 242–260, 320–342, and 349–364, the acidic region is amino acids 291–318, and putative TAG protection regions are amino acids 122–222 and 406–517. Truncations were made from the N and C termini of Pln A. The figure depicting structural features of Plin was adapted from (31).
Immunoprecipitation and immunoblot analyses
Cells that overexpress HSL and Plin were harvested, and immunoblot analyses were performed as previously described (28). Total proteins were homogenized in TES (10 mM Tris, 1 mM EDTA, pH 7.4, with 8% sucrose) with 10 µg/ml leupeptin. Following centrifugation at 10,000 g, 20 µg of protein from tissue homogenates were subjected to SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was incubated with rabbit anti-Plin or anti-Flag antibody (Sigma-Aldrich) and anti-β-actin antibody (Santa Cruz Biotechnology, Santa Cruz, CA) and then with horseradish peroxidase-linked anti-rabbit IgG (Amersham). The membranes were visualized with chemiluminescence reagent ECL (Amersham), exposed to Kodak XAR film, and then analyzed by a Fluor-S multi-image analyzer (Bio-Rad).
Immunofluorescence microscopy
Immunofluorescence confocal microscopy was performed to determine the subcellular localization of Plin truncations as previously described (8). Plin was visualized with MSM antibody (full-length Plin) or PREK antibody (Plin truncations) followed by Alexa Fluor 637-conjugated goat anti-rabbit IgG (Molecular Probes, Eugene, OR). Bodipy 493/503 (Molecular Probes) was used at 10 µg/ml to visualize neutral lipid staining.
Statistical analysis
Results are expressed as the mean ± SD. Statistical significance was tested using ANOVA with Bonferroni as post test with InStat (version 2.03; GraphPad Software, San Diego, CA) software for Macintosh.
RESULTS
HSL and Plin form a physical interaction
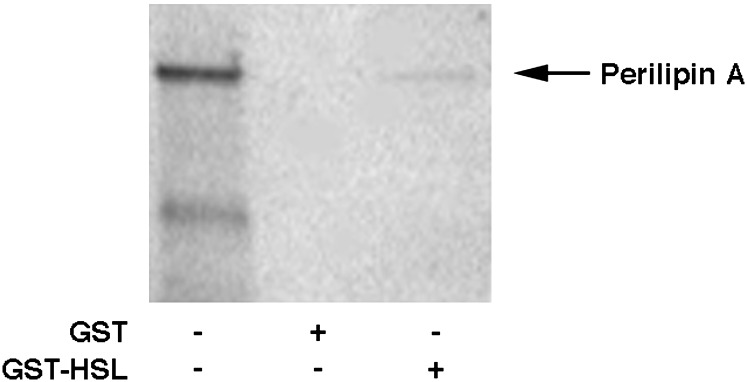
To test the hypothesis that HSL and Plin form a physical interaction, the ability of rat GST-HSL fusion protein, produced in a baculovirus expression system, to interact with [35S]methionine-labeled, in vitro-translated, full-length Plin A was examined in an in vitro GST pull-down assay system. As shown in Fig. 2, GST-HSL fusion protein was able to pull down [35S]methionine-labeled, in vitro-translated Plin specifically, while control GST protein did not. Thus, Plin and HSL are capable of interacting in the absence of other cellular proteins, supporting a direct physical interaction.
Fig. 2.
HSL and Plin form a physical interaction. Full-length Plin was in vitro translated with [35S]methionine using the TNT transcription/translation system (lane 1). Extracts (400 μg) of baculovirus overexpressing GST (lane 2) or GST-HSL fusion protein (GST-HSL, lane 3) were incubated with glutathione-agarose beads and with [35S]methionine-labeled Plin as described in Experimental Procedures. Proteins that bound to the beads were eluted, separated on SDS/4–20% PAGE, and visualized using a PhosphorImager.
HSL interacts with an N-terminal region of Plin
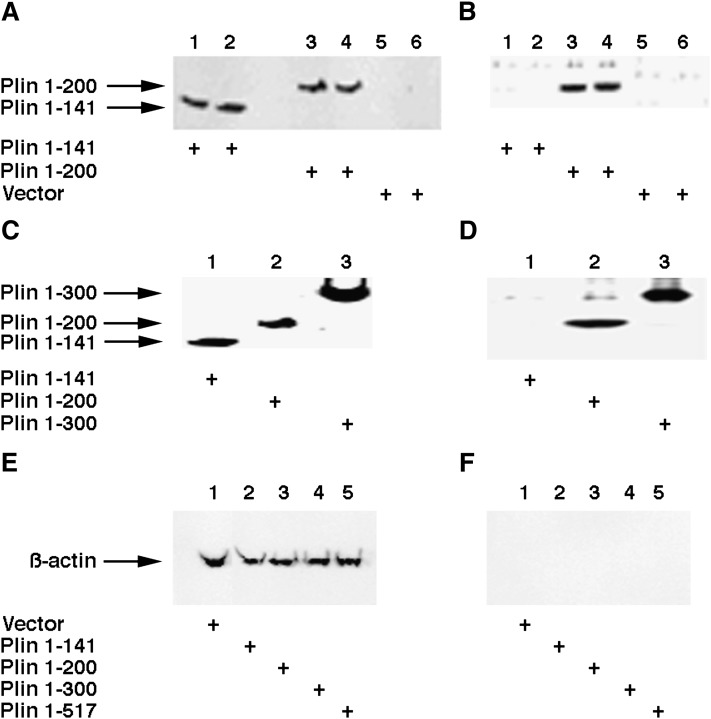
To further characterize the interaction of HSL with Plin and to determine the role of the N terminus of Plin in the interaction with HSL, CHO cells that overexpress HSL (3F9) were transduced with adenovirus containing C-terminal deletions of Plin with a Flag tag. After treatment with forskolin, cells were lysed and HSL was immunoprecipitated with anti-HSL antibodies. The presence of Plin was detected using anti-flag antibody after the immunoprecipitated complex was separated on SDS-PAGE. As shown in Fig. 3, Plin 1-141 and Plin 1-200 were both expressed in the cells at similar levels (Fig. 3A). The anti-Flag antibodies recognize Plin 1-200 in the immune complexes, but Plin 1-141 was not seen in the immune complexes (Fig. 3B). Therefore, HSL appears to interact with the N-terminal portion of Plin through a region involving amino acids between 141 and 200. To further confirm the expression of the Plin constructs, we also used an anti-peptide antibody that can recognize the N-terminal portion of Plin to detect the expression and the presence of the C-terminal deletions in the immunoprecipitated complexes. As shown in Fig. 3C, Plin 1-141, Plin 1-200, and Plin 1-300 were all expressed at similar levels, and the anti-N-Plin antibody recognized Plin 1-200 and Plin1-300 in the immune complexes, but Plin 1-141 was not seen in the immune complexes (Fig. 3D). As evidence that the interactions of Plin and HSL were not due to nonspecific immunoprecipitation, β-actin was used as a control in cell lysates (Fig. 3E) and immunoprecipitated complexes (Fig. 3F). Equivalent amounts of β-actin were seen in the cell lysates, but no β-actin was observed in the anti-HSL precipitated immune complexes.
Fig. 3.
HSL interacts with an N-terminal region of Plin. CHO cells that overexpress HSL (3F9) were transduced with adenovirus containing C-terminal deletions of Plin that expressed a Flag tag. After treatment with forskolin, cells were lysed and HSL was immunoprecipitated with anti-HSL antibodies. A: Total cell extracts of 3F9 cells that were transduced with Plin 1-141 (lanes 1 and 2), Plin 1-200 (lanes 3 and 4), and empty vector (lanes 5 and 6) were loaded on the gel, and the presence of Plin was detected using anti-Flag antibody. B: Anti-HSL immunoprecipitated complexes of 3F9 cells that were transduced with Plin 1-141 (lanes 1 and 2), Plin 1-200 (lanes 3 and 4), and empty vector (lanes 5 and 6) were loaded on the gel, and the presence of Plin was detected using anti-Flag antibody. C: Total cell extracts of 3F9 cells transduced with Plin 1-141 (lane 1), Plin 1-200 (lane 2), and Plin 1-300 (lane 3) were loaded on the gel, and the presence of Plin was detected using an antipeptide antibody that recognizes the N-terminal region of Plin. D: Anti-HSL immunoprecipitated complexes of 3F9 cells transduced with Plin 1-141 (lane 1), Plin 1-200 (lane 2), and Plin 1-300 (lane 3) were loaded on the gel, and the presence of Plin was detected using an antipeptide antibody that recognizes the N-terminal region of Plin. E: Total cell extracts of 3F9 cells transfected with control vector (lane 1), Plin 1-141 (lane 2), Plin 1-300 (lane 3), Plin 1-405 (lane 4), and full-length Plin (lane 5) were loaded on the gel and immunoblotted with anti-β-actin antibody. F: Complexes immunoprecipitated with anti-HSL antibodies of 3F9 cells transfected with control vector (lane 1), Plin 1-141 (lane 2), Plin 1-300 (lane 3), Plin 1-405 (lane 4), and full-length Plin (lane 5) were loaded on the gel and immunoblotted with anti-β-actin antibody.
HSL interacts with a C-terminal region of Plin
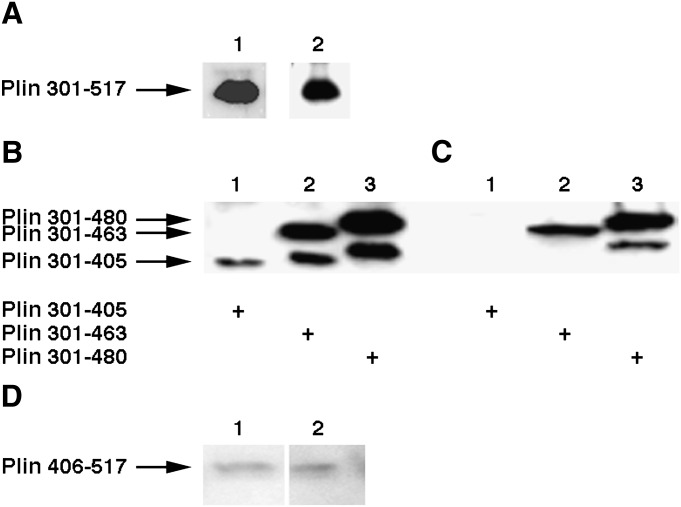
To examine whether the C-terminal region of Plin is involved in the interaction with HSL, a series of N-terminal deletions of Plin was transduced into the 3F9 CHO cells that overexpress HSL, and the transduced cells were lysed and immunoprecipitated with anti-HSL antibodies to detect potential interactions with HSL. The first construct tested contained the complete C terminus (Plin 301-517) and was detected using an antipeptide antibody that can recognize the C terminus of Plin (Fig. 4A, lane 1). To detect potential interactions with HSL, the transduced cells were lysed and immunoprecipitated with anti-HSL antibodies, and the immune complexes were separated on SDS-PAGE and then immunoblotted with anti-Plin antibodies (lane 2). As shown in Fig. 4A, the C-terminal portion of Plin is also present in the immune complex that was precipitated by anti-HSL antibodies. To further characterize the interaction of HSL with the C-terminal region of Plin, the other N-terminal deletions of Plin containing a Flag tag were transduced into the 3F9 CHO cells that overexpress HSL. The transduced cells were lysed and immunoprecipitated with anti-HSL antibodies, and the immune complexes were separated on SDS-PAGE and then immunoblotted with anti-Flag antibodies. Results are shown in Fig. 4B and C. Anti-Flag antibodies recognize Plin 301-463 and Plin 301-480 in the immune complexes and total lysates, whereas Plin 301-405 was not detected in the immune complex. In a separate set of experiments, we tested the ability of HSL to interact with Plin 406-517. As shown in Fig. 4D, anti-Flag antibody was able to detect Plin 406-517 in the immune complex precipitated by anti-HSL antibodies. These results reveal that amino acids between 406 and 480 within the C-terminal portion of Plin can also interact with HSL.
Fig. 4.
HSL interacts with a C-terminal region of Plin. The experimental conditions are the same as described in Fig. 3 except that an adenovirus containing C-terminal regions of Plin was used to transduce 3F9 cells. A: 3F9 cells were transduced with Plin 301-517. Lane 1, total cell extracts were loaded on the gel; lane 2, anti-HSL immunoprecipitated complexes were loaded on the gel. An antibody that recognizes the C-terminal region of Plin was used to detect the presence of Plin 301-517. B: Total cell extracts of 3F9 cells that were transduced with Plin 301-405 (lane 1), Plin 301-463 (lane 2), and Plin 301-485 (lane 3) were loaded on the gel, and the presence of Plin was detected using an anti-flag antibody. C: Anti-HSL immunoprecipitated complexes of 3F9 cells that were transduced with Plin 301-405 (lane 1), Plin 301-463 (lane 2), and Plin 301-485 (lane 3) were loaded on the gel, and the presence of Plin was detected using an anti-Flag antibody. D: 3F9 cells were transduced with Plin 406-517. Lane 1, total cell extracts were loaded on the gel; lane 2, anti-HSL immunoprecipitated complexes were loaded on the gel. An anti-Flag antibody was used to detect the presence of Plin.
Functional significance of the interaction of HSL with Plin
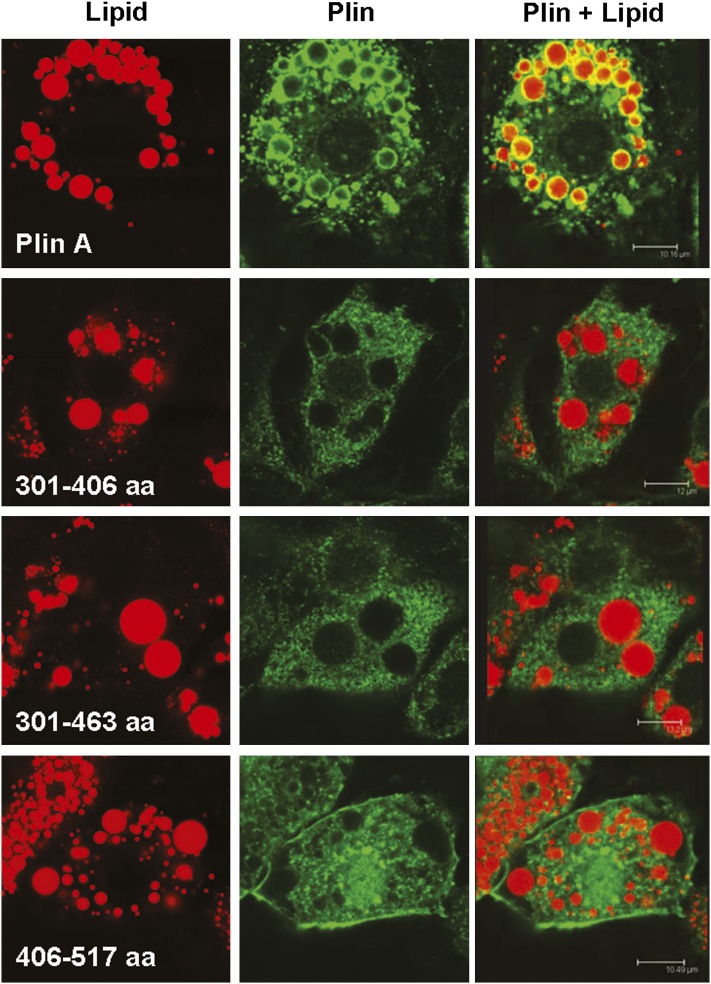
In view of the physical interaction of HSL with Plin, we hypothesized that the translocation of HSL to the lipid droplet occurs by virtue of an interaction with sequences in Plin and that the hydrophobic nature of Plin, with its lipid droplet-targeting domains directing Plin to be localized to the surface of the lipid droplet, determines the final destination of HSL. Therefore, we reasoned that a fragment of Plin, lacking the domains that target it to the lipid droplet and, therefore, localizes in the cytoplasm, but physically interacts with HSL, might function as a dominant negative and prevent the translocation of HSL to the droplet and thus inhibit lipolysis. We have previously shown that, similar to full-length Plin, when Plin 1-300 is expressed in lipid-containing cells, it forms a ring-like pattern localized to the perimeter of lipid droplets, whereas Plin 1-200 shows a diffuse cytoplasmic pattern without association with lipid droplets (8). Thus, these constructs seemed to be appropriate for testing our hypothesis as related to sequences within the N terminus of Plin. For examining sequences within the C terminus of Plin, we have previously shown that Plin 301-517 also localizes to the perimeter of lipid droplets (8). In order to examine the ability of other truncations of Plin to target to and associate with lipid droplets, we transduced ACS1-FATP cells, which stably overexpress ACS-1 and FATP1 (29) and which do not normally express HSL or Plin (25), with the C-terminal constructs. Transduced cells were fixed and prepared for confocal microscopy after incubation with FA for 48 h. Simultaneous fluorescent detection of neutral lipids and Plin allowed us to correlate immunoreactivity with intracellular structures (Fig. 5). Neutral lipids were stained with Bodipy (red fluorescence). Plins were detected with specific antibodies directed against the Flag tag (green fluorescence). In contrast to full-length Plin, which was found in a ring-like pattern, Plin 301-405, Plin 301-463, and Plin 406-517 were all expressed in a diffuse cytoplasmic pattern without association with the lipid droplets. This suggests that sequences between 463 and 517 may be important for lipid targeting or may participate in lipid targeting since Plin 301-517 was previously shown to associate with lipid droplets (8) but are not by themselves sufficient for lipid targeting since Plin 406-517 does not associate with lipid droplets.
Fig. 5.
Localization of C-terminal Plin mutants within cells. ACS1/FATP1 cells were transduced with adenoviruses expressing Plin or Plin truncations. After incubation with FA for 48 h, the cells were fixed and neutral lipids (red) stained with Bodipy and Plin (green) detected with specific antibodies and Alexa Fluor.
To examine the functional role of the interaction of HSL with Plin on lipolysis, we studied forskolin-stimulated lipolysis in a reconstituted lipid-loaded cell system, which we have previously used to examine lipolysis (8), except we now stably overexpress HSL, named ACS1-FATP1-HSL cells. These cells were derived from ACS1-FATP1 cells by stably overexpressing rat HSL. For these sets of experiments, ACS1-FATP1-HSL cells were transfected with either native Plin alone or with full-length Plin along with the different truncated Plin mutants and loaded with TAG using BSA-bound FA. Subsequently, cells were treated with forskolin (20 µM) to stimulate lipolysis, followed by measurement of released glycerol. As shown in Table 1, cells transfected with a full-length construct of Plin, showed a 2-fold stimulation of lipolysis in response to forskolin treatment. When full-length Plin was cointroduced in the cells with Plin 1-141, which does not associate with lipid droplets and does not interact with HSL, there was a trend for a reduction in forskolin-stimulated lipolysis, but this did not reach statistical significance. However, when full-length Plin was cointroduced in the cells with Plin 1-200, which does not associate with lipid droplets but does interact with HSL, there was a loss of essentially all forskolin-stimulated lipolysis (P < 0.05). In contrast, cells expressing Plin 1-300 and Plin 1-405, which both associate with lipid droplets and interact with HSL, displayed normal forskolin-stimulated lipolysis. The results of coexpression of C-terminal and full-length Plin A constructs were less dramatic. Cells expressing full-length Plin along with Plin 301-517, which contains the entire C terminus of Plin and which associates with lipid droplets and interacts with HSL, displayed normal forskolin-stimulated lipolysis, whereas cells coexpressing full-length Plin along with Plin 301-405, Plin 301-463, or Plin 406-517, which do not associate with lipid droplets, had variable, but no statistically significant, effects on forskolin-stimulated lipolysis whether or not there was evidence for interaction with HSL in vitro. Thus, these results are consistent with the hypothesis that the translocation of HSL to the lipid droplet, in our model system, occurs by virtue of an interaction with sequences within the N-terminal region of Plin and Plin's localization to the surface of lipid droplets.
TABLE 1.
Effects of Plin mutants on lipolysis
| Fold Stimulation | |
|---|---|
| Plin-full + empty vector | 1.99 ± 0.12 |
| Plin-full + 1-141 | 1.42 ± 0.01 |
| Plin-full + 1-200 | 1.07 ± 0.05* |
| Plin-full + 1-300 | 1.73 ± 0.39 |
| Plin-full + 1-405 | 1.75 ± 0.25 |
| Plin-full + 301-517 | 1.91 ± 0.25 |
| Plin-full + 301-405 | 1.34 ± 0.43 |
| Plin-full + 406-517 | 1.63 ± 0.34 |
| Plin-full + 301-463 | 1.38 ± 0.34 |
ACS1-FATP1-HSL cells were transfected with full-length Plin along with either empty vector or the different truncated Plin mutants (as listed with the starting and the ending amino acids of the fragment) and loaded with TAG using BSA-bound FA. Subsequently, cells were treated with forskolin (20 µM) to stimulate lipolysis. Lipolysis is presented as fold stimulation of glycerol release after forskolin treatment compared with no forskolin treatment. Results are the summary of at least three sets of independent assays and are given as the mean ± SD. *, P < 0.05.
DISCUSSION
Plin is a lipid droplet-associated protein that is involved in the control of both basal and PKA-stimulated lipolysis. Plin expression, but not its phosphorylation, is necessary for maximal translocation of PKA-phosphorylated HSL (6, 7). It has been reported that the N-terminal region (amino acids 1–300) of Plin A inhibits basal lipolysis and promotes PKA-stimulated lipolysis by HSL, whereas the C-terminal (amino acids 301–517) of Plin A inhibits basal lipolysis via non-HSL lipase and promotes PKA-stimulated lipolysis by both HSL and non-HSL lipases (6, 8). These data have suggested a potential physical interaction between Plin and HSL that has been supported by the demonstration that HSL could be cross-linked and coimmunoprecipitated with Plin (6) and by fluorescence resonance energy transfer and bimolecular fluorescence complementation studies that suggest that HSL closely, if not directly, interacts with Plin (23). In our studies, we provide direct evidence for a physical interaction of HSL with Plin A. Our studies demonstrate that HSL can form a physical interaction with an N-terminal region located between amino acids 141 and 200 of Plin A, as well as with a C-terminal region located between amino acids 406 and 480. These sequences lie outside of the PAT-1 region, which displays sequence similarity with adipophilin and TIP47 (30) and do not overlap with the previously identified hydrophobic and acidic regions, but the N-terminal interacting region does overlap with a region of amphipathic β-strands (31) (see Fig. 1 for a schematic representation of structural features of Plin A). However, sequences that overlap with these two HSL-interacting regions of Plin A, namely amino acids 122–222 and 406–517, have been shown to play a role in shielding stored TAG from cytosolic lipases, thereby facilitating TAG storage (32). The demonstration of an interaction of HSL with a similar region of Plin A suggests that the interaction of HSL with Plin A at these sites might have a dual function to facilitate the movement of HSL in close proximity to the lipid droplet as well as to prevent HSL from hydrolyzing TAG. This is in agreement with recent findings that PKA-stimulated HSL translocation was independent of Plin phosphorylation, while the lipolytic action of HSL at the lipid droplet requires the phosphorylation of Plin (6).
It is noteworthy that the sequences within Plin that we have identified as interacting with HSL not only overlap with sequences that have been reported to contribute to the barrier function of Plin, but also overlap with N-terminal (amino acids 1–250) (33) and C-terminal sequences (amino acids 382–429) (34) that have been reported to interact with comparative gene identification 58 (CGI-58). CGI-58 is a member of the a/β hydrolase family that lacks enzymatic activity but appears to function as a coactivator of ATGL (35). CGI-58 is localized to the surface of lipid droplets under basal conditions through its interaction with Plin and rapidly moves off the lipid droplet following PKA stimulation (34). It is not known whether the interaction of Plin with HSL and CGI-58 is mutually exclusive, but one possibility is that these regions of Plin could interact with CGI-58 and HSL with different affinities. Under PKA-stimulated conditions, the release of CGI-58 from the lipid droplet might make sites available for HSL to interact with Plin and to function at the surface of the lipid droplet. Further studies to identify the amino acids that are involved in the Plin interaction with HSL and CGI-58 will be needed to explore this in more detail.
Evidence that the interaction observed between HSL and Plin has functional significance within the cell is provided by our experiments of cotransfection of lipid-loaded, HSL-expressing cells with full-length and mutant constructs of Plin. As predicted, a Plin construct that interacts with HSL, but does not associate with the lipid droplet, should inhibit lipolysis by binding to and preventing the movement, which normally occurs following PKA stimulation, of HSL to the droplet. Indeed, Plin 1-200, which does not associate with the lipid droplet, but does interact with HSL, was able to abrogate all forskolin-stimulated lipolysis in the presence of full-length Plin, thereby functioning as a dominant negative. In contrast, Plin 1-141, which neither associates with the lipid droplet nor interacts with HSL, did not significantly affect PKA-stimulated lipolysis supported by full-length Plin. In addition, neither Plin 1-300, Plin 1-405, nor Plin 301-517, each of which associates with the lipid droplet and interacts with HSL, significantly affected PKA-stimulated lipolysis supported by full-length Plin. Thus, it appears that for a Plin fragment to function as a dominant negative that it is necessary for it to interact with HSL, but not to associate with the lipid droplet. Although our coimmunoprecipitations and studies examining lipolysis with small C-terminal fragments of Plin were not entirely consistent with this conclusion, it is possible that the affinity of HSL for interacting with N- and C-terminal regions of Plin might be different or that the small C-terminal fragments of Plin might have had nonspecific effects in the cells that we used as our model system. The latter would be in agreement with previous studies showing that the N-terminal region of Plin A is involved in HSL-mediated lipolysis, whereas the C-terminal region of Plin works with non-HSL lipases for stimulated lipolysis (8). It should be noted that neither CGI-58 nor ATGL is present in our reconstituted cell system and, thus, the contributions and any additional interactions with CGI-58 and ATGL would not be seen (data not shown). Nonetheless, when taken together, these results lend credence to the notion that the localization of Plin to the lipid droplet through its hydrophobic sequences is an important component of its ability to function as a scaffold for orchestrating lipolysis.
It has previously been reported that three hydrophobic sequences (amino acids 242–260, 320–342, and 349–364), along with an acidic region (amino acids 291–318) and N-terminal sequences predicted to form amphipathic β-strands (see Fig. 1), within Plin are involved in targeting and anchoring Plin A to the lipid droplet (31). Based on deletional experiments, it has been suggested that the first two hydrophobic sequences are important for lipid droplet targeting, while the third has a weak contribution. In our work using Plin truncations, we observed that Plin 301-517 targets to and associates with lipid droplets (8); however, in this report, Plin 301-406 and Plin 301-462 displayed diffuse cytoplasmic localization, failing to associate with lipid droplets. Therefore, at least in our cell model system, sequences within the C-terminal region of Plin can be important in targeting Plin to lipid droplets, yet these sequences alone are insufficient to associate with lipid droplets since Plin 406-517 also displayed diffuse cytoplasmic localization.
In summary, we have provided direct evidence for a physical interaction of HSL with Plin at two different regions, one within the N-terminal region and one within the C-terminal region. An N-terminal construct, Plin 1-200, which does not associate with lipid droplets, but does interact with HSL, can function as a dominant negative for PKA-stimulated lipolysis. The results are consistent with the notion that the translocation of HSL to the lipid droplet occurs by virtue of an interaction with sequences within the N-terminal region of Plin and Plin's localization to the surface of lipid droplets.
Footnotes
Abbreviations:
- ACS-1
- acyl-coenzyme A synthase 1
- ATGL
- adipose triglyceride lipase
- CGI-58
- comparative gene identification 58
- CHO
- Chinese hamster ovary
- DAG
- diacylglycerol
- FATP1
- fatty acid transport protein 1
- GST
- glutathione S-transferase
- HSL
- hormone-sensitive lipase
- PKA
- cAMP-dependent protein kinase
- Plin
- perilipin
- TAG
- triacylglycerol
This work was supported in part by the Research Service of the Department of Veterans Affairs and National Institutes of Health Grant AG-028908 (to F.B.K.), National Institutes of Health Grant DK-50647, and USDA Research Service Co-Operative Agreement 58 1950-4-401 (to A.S.G.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health, the VA, or the USDA.
REFERENCES
- 1.Londos C., Brasaemle D. L., Schultz C. J., Adler-Wailes D. C., Levin D. M., Kimmel A. R., Rondinone C. M. 1999. On the control of lipolysis in adipocytes. Ann. N. Y. Acad. Sci. 892: 155–168. [DOI] [PubMed] [Google Scholar]
- 2.Holm C., Osterlund T., Laurell H., Contreras J. A. 2000. Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Annu. Rev. Nutr. 20: 365–393. [DOI] [PubMed] [Google Scholar]
- 3.Greenberg A. S., Egan J. J., Wek S. A., Garty N. B., Blanchette-Mackie E. J., Londos C. 1991. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J. Biol. Chem. 266: 11341–11346. [PubMed] [Google Scholar]
- 4.Martinez-Botas J., Anderson J., Tessier D., Lapillonne A., Chang B., Quast M., Gorenstein D., Chen K., Chan L. 2000. Absence of perilipin results in leanness and reverses obesity in Lepr(db/db) mice. Nat. Genet. 26: 474–479. [DOI] [PubMed] [Google Scholar]
- 5.Tansey J. T., Sztalryd C., Gruia-Gray J., Roush D. L., Zee J. V., Gavrilova O., Reitman M. L., Deng C-X., Li C., Kimmel A. R., et al. 2001. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc. Natl. Acad. Sci. USA. 98: 6494–6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyoshi H., Souza S. C., Zhang H. H., Strissel K. J., Christoffolete M. A., Kovsan J., Rudich A., Kraemer F. B., Bianco A. C., Obin M. S., et al. 2006. Perilipin promotes hormone-sensitive lipase-mediated adipocyte lipolysis via phosphorylation-dependent and -independent mechanisms. J. Biol. Chem. 281: 15837–15844. [DOI] [PubMed] [Google Scholar]
- 7.Sztalryd C., Xu G., Dorward H., Tansey J. T., Contreras J. A., Kimmel A. R., Londos C. 2003. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J. Cell Biol. 161: 1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H. H., Souza S. C., Muliro K. V., Kraemer F. B., Obin M. S., Greenberg A. S. 2003. Lipase-selective functional domains of perilipin A differentially regulate constitutive and protein kinase A-stimulated lipolysis. J. Biol. Chem. 278: 51535–51542. [DOI] [PubMed] [Google Scholar]
- 9.Kraemer F. B., Shen W-J. 2002. Hormone-sensitive lipase: control of intracellular tri-(di-) acylglycerol and cholesteryl ester hydrolysis. J. Lipid Res. 43: 1585–1594. [DOI] [PubMed] [Google Scholar]
- 10.Yeaman S. J. 1990. Hormone-sensitive lipase - a multipurpose enzyme in lipid metabolism. Biochim. Biophys. Acta. 1052: 128–132. [DOI] [PubMed] [Google Scholar]
- 11.Osuga J-i., Ishibashi S., Oka T., Yagyu H., Tozawa R., Fujimoto A., Shionoira F., Yahagi N., Kraemer F. B., Tsutsumi O., et al. 2000. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc. Natl. Acad. Sci. USA. 97: 787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zimmermann R., Strauss J. G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., et al. 2004. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 306: 1383–1386. [DOI] [PubMed] [Google Scholar]
- 13.Villena J. A., Roy S., Sarkadi-Nagy E., Kim K. H., Sul H. S. 2004. Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: ectopic expression of desnutrin increases triglyceride hydrolysis. J. Biol. Chem. 279: 47066–47075. [DOI] [PubMed] [Google Scholar]
- 14.Jenkins C. M., Mancuso D. J., Yan W., Sims H. F., Gibson B., Gross R. W. 2004. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J. Biol. Chem. 279: 48968–48975. [DOI] [PubMed] [Google Scholar]
- 15.Schweiger M., Schreiber R., Haemmerle G., Lass A., Fledelius C., Jacobsen P., Tornqvist H., Zechner R., Zimmermann R. 2006. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J. Biol. Chem. 281: 40236–40241. [DOI] [PubMed] [Google Scholar]
- 16.Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., et al. 2006. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 312: 734–737. [DOI] [PubMed] [Google Scholar]
- 17.Kershaw E. E., Hamm J. K., Verhagen L. A., Peroni O., Katic M., Flier J. S. 2006. Adipose triglyceride lipase: function, regulation by insulin, and comparison with adiponutrin. Diabetes. 55: 148–157. [PMC free article] [PubMed] [Google Scholar]
- 18.Langin D., Dicker A., Tavernier G., Hoffstedt J., Mairal A., Ryden M., Arner E., Sicard A., Jenkins C. M., Viguerie N., et al. 2005. Adipocyte lipases and defect of lipolysis in human obesity. Diabetes. 54: 3190–3197. [DOI] [PubMed] [Google Scholar]
- 19.Ryden M., Jocken J., van Harmelen V., Dicker A., Hoffstedt J., Wiren M., Blomqvist L., Mairal A., Langin D., Blaak E. E., et al. 2007. Comparative studies of the role of hormone sensitive lipase and adipose triglyceride lipase in human fat cell lipolysis. Am. J. Physiol. Endocrinol. Metab. 292: E1847–E1855. [DOI] [PubMed] [Google Scholar]
- 20.Egan J. J., Greenberg A. S., Chang M-K., Wek S. A., Moos M. C., Jr., Londos C. 1992. Mechanism of hormone-stimulated lipolysis in adipocytes: translocation of hormone-sensitive lipase to the lipid storage droplet. Proc. Natl. Acad. Sci. USA. 89: 8537–8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clifford G. M., Londos C., Kraemer F. B., Vernon R. G., Yeaman S. J. 2000. Translocation of hormone-sensitive lipase and perilipin upon lipolytic stimulation of rat adipocytes. J. Biol. Chem. 275: 5011–5015. [DOI] [PubMed] [Google Scholar]
- 22.Su C-L., Sztalryd C., Contreras J. A., Holm C., Kimmel A. R., Londos C. 2003. Mutational analysis of the hormone-sensitive lipase translocation reaction in adipocytes. J. Biol. Chem. 278: 43615–43619. [DOI] [PubMed] [Google Scholar]
- 23.Granneman J. G., Moore H. P., Granneman R. L., Greenberg A. S., Obin M. S., Zhu Z. 2007. Analysis of lipolytic protein trafficking and interactions in adipocytes. J. Biol. Chem. 282: 5726–5735. [DOI] [PubMed] [Google Scholar]
- 24.Miyoshi H., Perfield J. W., 2nd, Souza S. C., Shen W. J., Zhang H. H., Stancheva Z. S., Kraemer F. B., Obin M. S., Greenberg A. S. 2007. Control of adipose triglyceride lipase action by serine 517 of perilipin A globally regulates protein kinase A-stimulated lipolysis in adipocytes. J. Biol. Chem. 282: 996–1002. [DOI] [PubMed] [Google Scholar]
- 25.Souza S. C., Muliro K. V., Liscum L., Lien P., Yamamoto M. T., Schaffer J. E., Dallal G. E., Wang X., Kraemer F. B., Obin M., et al. 2002. Modulation of hormone-sensitive lipase and protein kinase A-mediated lipolysis by perilipin A in an adenoviral reconstituted system. J. Biol. Chem. 277: 8267–8272. [DOI] [PubMed] [Google Scholar]
- 26.Shen W-J., Sridhar K., Bernlohr D. A., Kraemer F. B. 1999. Interaction of rat hormone-sensitive lipase with adipocyte lipid-binding protein. Proc. Natl. Acad. Sci. USA. 96: 5528–5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen W-J., Liang Y., Hong R., Patel S., Natu V., Sridhar K., Jenkins A., Bernlohr D. A., Kraemer F. B. 2001. Characterization of the functional interaction of adipocyte lipid-binding protein with hormone-sensitive lipase. J. Biol. Chem. 276: 49443–49448. [DOI] [PubMed] [Google Scholar]
- 28.Shen W-J., Patel S., Natu V., Kraemer F. B. 1998. Mutational analysis of structural features of rat hormone-sensitive lipase. Biochemistry. 37: 8973–8979. [DOI] [PubMed] [Google Scholar]
- 29.Gargiulo C. E., Stuhlsatz-Krouper S. M., Schaffer J. E. 1999. Localization of adipocyte long-chain fatty acyl-CoA synthetase at the plasma membrane. J. Lipid Res. 40: 881–892. [PubMed] [Google Scholar]
- 30.Lu X., Gruia-Gray J., Copeland N. G., Gilbert D. J., Jenkins N. A., Londos C., Kimmel A. R. 2001. The murine perilipin gene: the lipid droplet-associated perilipins derive from tissue-specific, mRNA splice variants and define a gene family of ancient origin. Mamm. Genome. 12: 741–749. [DOI] [PubMed] [Google Scholar]
- 31.Subramanian V., Garcia A., Sekowski A., Brasaemle D. L. 2004. Hydrophobic sequences target and anchor perilipin A to lipid droplets. J. Lipid Res. 45: 1983–1991. [DOI] [PubMed] [Google Scholar]
- 32.Garcia A., Subramanian V., Sekowski A., Bhattacharyya S., Love M. W., Brasaemle D. L. 2004. The amino and carboxyl termini of perilipin A facilitate the storage of triacylglycerols. J. Biol. Chem. 279: 8409–8416. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi T., Omatsu N., Matsushita S., Osumi T. 2004. CGI-58 interacts with perilipin and is localized to lipid droplets. Possible involvement of CGI-58 mislocalization in Chanarin-Dorfman syndrome. J. Biol. Chem. 279: 30490–30497. [DOI] [PubMed] [Google Scholar]
- 34.Subramanian V., Rothenberg A., Gomez C., Cohen A. W., Garcia A., Bhattacharyya S., Shapiro L., Dolios G., Wang R., Lisanti M. P., et al. 2004. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3–L1 adipocytes. J. Biol. Chem. 279: 42062–42071. [DOI] [PubMed] [Google Scholar]
- 35.Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J. G., Gorkiewicz G., Zechner R. 2006. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 3: 309–319. [DOI] [PubMed] [Google Scholar]