Abstract
Nuclear receptor (NR) transcriptional activity is controlled by agonist binding and concomitant exchange of receptor-associating corepressor proteins for NR box-containing, receptor AF-2-targeting coactivator proteins. We report here that TNIP1 is an atypical NR coregulator. Requirements for TNIP1-RAR interaction – its NR boxes, ligand, and the receptor's AF-2 domain – are characteristic of coactivators. However, TNIP1 reduces RAR activity. Repression is partially relieved by SRC1, suggesting interference with coactivator recruitment as a mechanism of TNIP1 repression. TNIP1 does not bind RXRα and RARα AF-2 domain, necessary for that receptor's association with TNIP1, is insufficient to confer upon RXRα interaction with TNIP1. Preferential interaction of RARα over RARγ with TNIP1 can be mapped to RARα ligand binding domain helices 5-9 and suggests regions outside the receptor helix 12 modulate interaction of NRs and NR box containing corepressors. TNIP1 repression of RARs in the presence of RA places it in a small category of corepressors of agonist-bound NRs.
Keywords: coregulator, nuclear receptor, coactivator, corepressor, TNIP1
Introduction
Transcriptional activity of nuclear receptors (NRs) is modulated not only by ligands but also by coregulator proteins that act either as coactivators or corepressors of receptor function. Coregulators facilitate or inhibit transcription of NR target genes through chromatin modification or interaction with other components of cellular transcriptional machinery [1]. Typically, in the absence of agonist, the receptor interacts with a corepressor protein such as nuclear receptor corepressor (NCoR) or silencing mediator of retinoid and thyroid receptors (SMRT). Through interaction with histone deacetylase (HDAC) or other repressor proteins, corepressors help maintain the chromatin in a compact state preventing target gene transcription [2]. Agonist binding the receptor causes a conformational change in the activation function 2 (AF-2) domain within helix 12 of the ligand binding domain (LBD) causing release of the corepressor, recruitment of a coactivator, and transcriptional activation. Characteristic of coactivator proteins is the NR box, a leucine rich amino acid motif, LXXLL (L=leucine, X=any amino acid), which is used to bind to the receptor AF-2 domain [3].
The model of the apo-receptor being bound by a corepressor and the holo-receptor by coactivator was challenged with the discovery that receptor interacting protein 140 (RIP140) acts as a corepressor in the presence of bound agonist [4]. Indeed several corepressors of agonist-bound NRs have since been characterized [5]. Their interaction requirements with NRs are those of coactivators: they bind the receptor in the presence of ligand and usually rely on NR boxes to do so. Their method of receptor repression has been shown to involve competition with coactivators for receptor binding, recruitment of HDACs or other transcriptional repressors or a combination of these mechanisms [5, 6].
In search for new coregulators we previously carried out a yeast two-hybrid screen with peroxisome proliferator activated receptor (PPAR) for NR-interacting proteins from a human keratinocyte cDNA library. We identified TNFα-induced protein 3 (TNFAIP3)-interacting-protein 1 (TNIP1), a protein previously identified as Naf, VAN, and ABIN-1 and shown to associate with HIV proteins nef and matrix [7, 8] and to repress the activity of nuclear factor κB (NF-κB) [9], as a PPAR coregulator capable of repressing that receptor's activity in the presence of ligand [Flores et al. submitted]. To improve our understanding of TNIP1's function as NR corepressor we examined its interaction with other group 1 NRs [10] characteristic of keratinocytes [11], retinoic acid receptors α and γ (RARα and RARγ), and a group 2 receptor retinoid×receptor (RXRα), the heterodimer partner [12] for both RARs and PPARs Here we report that TNIP1 does not interact with RXRα but exhibits ligand-dependent association with RARα and γ and acts as RAR corepressor in presence of all-trans-RA (ATRA). The repression is partially relieved by steroid receptor coactivator 1 (SRC1), suggesting interference with coactivator binding as a possible repression mechanism. The AF-2 domain of RARs is necessary but not sufficient for interaction with TNIP1. Among RARs, RARα is preferred over RARγ as TNIP1 interaction partner. In this regard, we identified a region within the LBD of RARα responsible for this subtype difference – region not previously shown to play any role in NR-coregulator association.
Materials and methods
Plasmids
The βRARE-tk-CAT and RXRE-tk-CAT constructs have been previously described [13]. TRE-tk-CAT reporter was made by inserting 3 copies of TRE palindrome [14] into pBLCAT2. The βRARE-tk-Luc was prepared by subcloning the βRARE repeats and the tk promoter from βRARE-tk-CAT into pGL4.10 (Promega, Madison, WI). The pG5-luc reporter was prepared by subcloning the GAL4 binding sites and the E1B promoter from pG5-CAT into pGL4.10. TNIP1 in pOTB7 backbone was obtained from ATCC (Manassas, VA) and subcloned into pVP16 vector for mammalian two-hybrid assays or pCDNA3.1-HA for receptor transactivation and immunoprecipitation assays. The pCDNA3.1-HDAC1-Flag construct was obtained from Dr. Eric Verdin via Addgene (www.addgene.org, Addgene plasmid 13820). RARα/RARγ chimera constructs were generated by subcloning fragments of RARα LBD into existing restriction sites within RARγ using standard techniques. For αγ1 and αγ2 constructs, the restriction sites were engineered into the RARα inserts by PCR using primers shown in Table 1. The αγ3 construct was prepared by cloning a PciI-BamHI fragment of RARα into the corresponding sites in RARγ. RXRα-R construct was prepared by site directed mutagenesis of the RXRα AF-2 domain using primers shown in Table 1 for conversion of the RXRα AF-2 domain (TFLMEMLE) to RARα AF-2 (PLIQEMLE). In all cases, the mammalian two-hybrid constructs were prepared in the pM vector expressing appropriate receptor DEF domains and the constructs for receptor transactivation assays were prepared in the pSG5 vector expressing full length receptors. All site directed mutants were generated using QuikChange XL (Stratagene, LaJolla, CA) kit according to manufacturer instructions. All constructs were verified by sequencing.
Table 1.
Primers used to create the RARαγ and RXRα-R chimeric constructs with the engineered restriction sites or mutations underlined.
| Construct | Primer sequence | |
|---|---|---|
| αγ1 | Forward | 5′-TGCCAGCTGGGCAAGTATACTACGAA-3′ BstZ17I |
| Reverse | 5′-CCGCAGGATCAGGATATCCAGGCA-3′ EcoRV | |
| αγ2 | Forward | 5′-GCCTGCCTGGATATCCTGATCCTG-3′ EcoRV |
| Reverse | 5′-TTGGGGAACATGTGGGGGCGGCT-3′ PciI | |
| RXRα-R | Forward | 5′-CTCTTCTTCTTCAAGCTCATCGGGGACACACCCATTGACCCTCTCATCCAGGAGATGCTGGAGGCGCCGCACCAAATGACTTAG-3′ |
| Reverse | 5′-CTAAGTCATTTGGTGCGGCGCCTCCAGCATCTCCTGGATGAGAGGGTCAATGGGTGTGTCCCCGATGAGCTTG AAGAAGAAGAG-3′ | |
Two-hybrid and receptor transactivation assays
Yeast and mammalian two-hybrid assays were performed as previously described [15]. For receptor transactivation assays Cos-7 or HeLa cells were seeded in 12-well plates and 24h later transfected using calcium phosphate for Cos-7 or FuGENE6 (Roche Applied Science, Indianapolis, IN) for HeLa with receptor constructs in the pSG5 backbone, pSG5-SRC1, pcDNA-HA-TNIP1, and the reporter construct as indicated in figures. RXRα was always cotransfected with RAR expression constructs. Appropriate empty vectors were used to keep the DNA amount constant. Cells were treated 16h later with media containing vehicle (0.1% DMSO) or ligand indicated in the figures at 1μM. After 24h (for luciferase) or 48h (for CAT) of ligand treatment the cells were collected and luciferase assays (Promega) or CAT ELISA (Roche) were performed according to manufacturer instructions.
Immunoprecipitations and HDAC assays
Cos-7 cells were transfected with pcDNA3.1-HA or pcDNA3.1-HA-TNIP for HDAC activity assays, or cotransfected with pcDNA3.1-HA-TNIP1 and pcDNA3.1-HDAC1-Flag for immunoblot detection of coimmunoprecipitation and 48h later were washed with ice cold PBS and lysed with buffer containing 20mM Tris pH8, 100mM NaCl, 1mM EDTA, 0.5% IGEPAL, and complete protease inhibitor (Roche). Lysates were diluted to 400μg/mL protein concentration with lysis buffer and immunoprecipitated overnight with normal rat IgG (Santa Cruz Biotechnology, Santa Cruz, CA) or HA rat monoclonal antibody (Roche) at 4°C with rotation. The immunoprecipitates were incubated for 4h with Protein A/G agarose beads (Santa Cruz) at 4°C with rotation and washed 4 times with cold lysis buffer. The HDAC activity was determined using an HDAC fluorescent assay kit (Biomol Research Laboratories, Plymouth Meeting, PA) according to manufacturer instructions. For western blots the immunocomplexes were eluted from the beads with Laemmli buffer and blotted with Flag antibody (Santa Cruz) using standard techniques.
Statistical analyses
Data was analyzed using Prism software (GraphPad, San Diego, CA). Student's t-test was used to compare two groups and one-way ANOVA with Newman-Kewls post hoc test was used for comparing more than two groups. P-values of less than 0.05 were considered significant.
Results and Discussion
TNIP1 interaction with RARα and RARγ is dependent on ligand, NR boxes and receptor AF-2 domain
We used the partial TNIP1 cDNA, clone 43a, as well as the full-length TNIP1 cDNA (Fig. 1A) to test for interaction with RARα and RARγ. Clone 43a contains one LXXLL NR box – amino acid sequence well known for its use by coregulators for interaction with liganded NRs [3, 5] – while full length TNIP1 contains two. Thus the partial cDNA provided us an initial tool to examine the role of one TNIP1 NR box versus two in interaction with retinoid receptors. In a yeast-two hybrid assay, clone 43a demonstrated strong interaction with RARα that was wholly dependent on ATRA and an intact NR box. There was no detectable association in the absence of ligand or when assayed in the presence of ligand but with the NR box mutated (m43a) from LKKLL to LKKAA (Fig. 1B). To assess AF-2 contribution to interaction between RARα and 43a, we introduced previously characterized [16] mutations (PLIQEMLE to PAAQEMLE) into this domain of RARα. This completely abolished the receptor's ability receptor to interact with clone 43a (Fig. 1B). Consistent with and extending these results for RARα, interaction between RARγ and clone 43a was also strictly dependent on ATRA, the receptor's AF-2 domain, and the clone's LKKLL NR box (not shown).
Fig. 1.
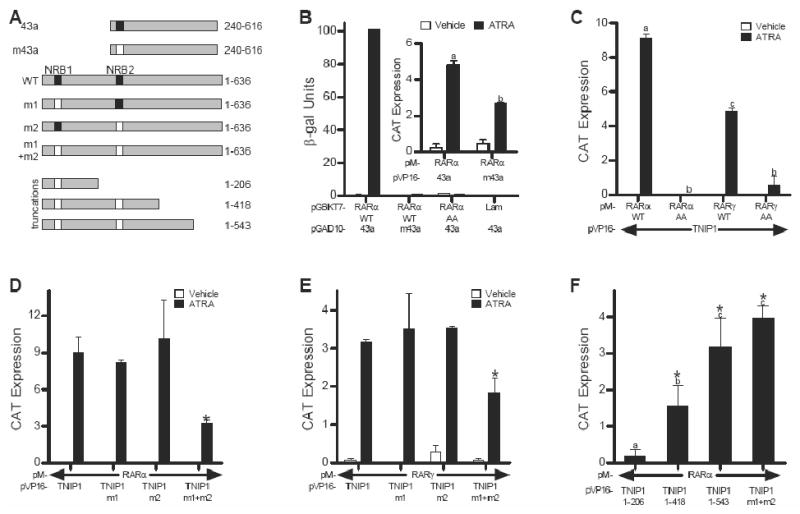
TNIP1-RAR interaction is entirely dependent on ligand and receptor AF-2 domain and partially dependent on TNIP1 NR Boxes. (A) Schematic of TNIP1 constructs showing partial clone 43a, NR Box (NRB) mutant 43a (m43a), and the full-length (amino acids 1-636) wild type (WT) and NR box mutants m1, m2, and m1+m2. Wild type NRB: solid box, mutant NRB: open box. Lower portion of schematic: TNIP1 truncations in the context of the NRB mutation(s). (B) Two-hybrid assays of Y187 yeast transformed with pGAD10-43a or m43a mated to AH109 yeast transformed with pGBKT7-RARα wild-type (WT), AF-2 mutant (AA), or lamin C (Lam) as negative control. Inset: Mammalian two-hybrid (M2H) assay in Cos-7 cells transfected with pM-RARα and pVP-16-43a or m43a. (C) M2H assays in Cos-7 cells transfected with pM-RARα or γ WT or AA constructs and pVP16-TNIP1. (D) and (E) M2H assays in Cos-7 cells showing effects of TNIP1 NR Box mutations on interaction with RARα (D) and RARγ (E); *: different from WT. (F) M2H assays showing interaction of RARα with TNIP1 truncations in Fig. 1A treated with ATRA; *: different from empty vector control. All M2H data are presented as difference in reporter activation between TNIP1 constructs and empty vector control. In (B, inset), (C) and (F) bars with different letter indicate significantly different values.
In mammalian two-hybrid assays, clone 43a demonstrated the same interaction requirements of ligand presence and receptor AF-2 integrity seen in yeast, but retained some association with RARα (Fig. 1B, inset) and RARγ (not shown) when expressed as an NR box mutant. This suggests that other non-canonical motifs contribute to RAR-TNIP1 interaction. Further, the difference between the absolute and partial necessity of an intact NR box in yeast versus mammalian cells may be attributable to the presence of the auxiliary factors in mammalian cells that cooperate with non-LXXLL motifs in TNIP1 to contribute to association with RARs, factors of which the yeast system is devoid [17]. Therefore, full-length TNIP1-RARs interaction was examined in mammalian cells only.
In perfect agreement with our yeast two-hybrid results with the partial TNIP1 clone 43a, both RARs exhibited strict ligand- and receptor AF-2-dependent interaction with full-length TNIP1 in mammalian two-hybrid assays (Fig. 1C). Full-length TNIP1 contains two LXXLL NR boxes and their contribution to interaction with RARs was examined by mutating the last two leucines of the first, second, and both NR boxes to alanines to generate TNIP1 m1, TNIP1 m2 and TNIP1 m1+m2, respectively (Fig. 1A). Mutating individual NR boxes had no detectable effect on RAR interaction but mutation of both motifs significantly reduced the ability of TNIP1 to interact with RARα and RARγ (Fig. 1D and 1E, respectively). To address the possibility that regions outside the canonical NR boxes contribute to interaction with receptor, we generated a series of TNIP1 truncations in the context of NR box mutant TNIP1 m1+m2 (Fig. 1A) and examined their ability to interact with holo-RARα in mammalian two-hybrid assays. There was no significant reporter activation over empty vector control for liganded RARα with the amino third of the protein, amino acids 1-206. Extending the TNIP1 m1+m2 protein through amino acids 1-418 and 1-543 increased reporter activation. Notably, for the 1-543 construct activation was similar to the full length TNIP1 m1+m2 (Fig. 1F) suggesting that some amino acid sequence from 418-543 of TNIP1 enhanced association with liganded RARα. However, this region of TNIP1 itself provided for no interaction despite its production level similar to the longer constructs (Ramirez and Aneskievich, unpublished) suggesting that RAR-TNIP1 interaction is more complex than might be predicted from simple linear amino acid sequence consideration of the coregulator. The requirement to experimentally negate multiple NR boxes to reduce the liganded NR-coregulator interaction level is not unprecedented. Interaction between the glucocorticoid receptor (GR) and dosage-sensitive sex reversal-adrenal hypoplasia congentia critical region on the X-chromosome, gene 1 (DAX-1) involves three DAX-1 NR boxes [18]. Mutagenesis of the first or second NR boxes had no significant effect on DAX-1 association with GR and it required inactivation of all three NR boxes to return reporter activity to the control level seen in the absence of DAX-1 cotransfection. The apparent partial reliance on non-canonical sequences for TNIP1 interaction with liganded receptors (Fig. 1F) makes it unlike other LXXLL-dependent corepressors of agonist-bound NRs like LCoR and PRAME, where these motifs are wholly responsible for interaction with target NRs [5] Complete definition of TNIP1 amino acid motifs required for interaction with NRs calls for a series of detailed studies, like those carried out for RIP140 [6].
TNIP1 represses RAR activity in the presence of ligand; the repression is partially relieved by coactivator SRC1 but appears HDAC-independent
TNIP1's interaction with RARα and RARγ in a ligand-dependent and receptor AF-2-dependent manner and at least partially relying on NR boxes are all characteristics of NR coactivators. Despite these traits, recombinant expression of TNIP1 in HeLa cells together with RARα/RXRα heterodimer resulted in a decrease of RAR-driven reporter activation in the presence of ATRA (Fig. 2A). Coexpressing increasing amounts of the coactivator SRC1 relieved some but not all TNIP1 repression of RAR activity (Fig. 2B), suggesting TNIP1's effects may be partially mediated by interference with coactivator binding to holo-receptor, similar to that observed for other corepressors of agonist-bound NRs REA and RIP140 [4, 19]
Fig. 2.
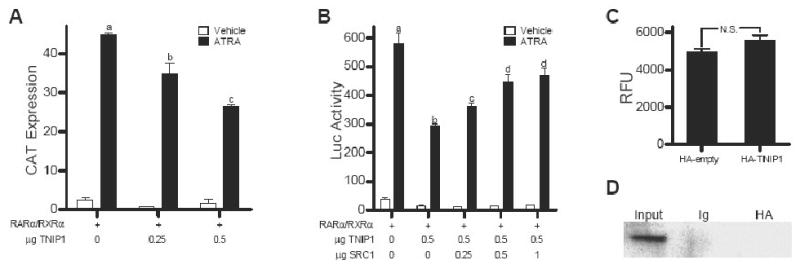
TNIP1 is a repressor of holo-RAR activity whose repression is partially relieved by coactivator SRC1 but appears HDAC-independent. (A and B) Receptor transactivation assays in HeLa cells cotransfected with indicated amounts of pcDNA3.1-HA-TNIP1, pSG5-RARα and RXRα, SRC1 (B), and βRARE-tk-CAT (A) or βRARE-tk-Luc (B) reporter construct. DNA amount kept constant with empty pcDNA3.1-HA or pSG5. Bars with different letter indicate significantly different values. (C) Fluorimetric HDAC activity assay performed with HA immunuprecipitates from Cos-7 cells transfected as indicated. RFU: relative fluorescence units, N.S.: not significant. (D) Coimmunoprecipitation assay of TNIP1 and HDAC1 in Cos-7 cells cotransfected with pcDNA3.1-HA-TNIP1 and pCDNA3.1-HDAC1-Flag, immunoprecipitated as indicated and immunoblotted with Flag antibody.
Corepressors frequently exert their effect through recruitment of HDACs [2, 5]. We examined this as a potential mechanism of TNIP1 repression by expressing HA tagged TNIP1 in Cos-7 cells and measuring the endogenous HDAC activity in the fraction immunoprecipitated with HA antibody. We found no increase in activity compared to control levels (Fig. 2C) despite western detection of HDAC1-3, 5 and 6 in Cos-7 lysates (not shown). Moreover, when we coexpressed Flag tagged HDAC1 with HA tagged TNIP1, we were unable to detect any coimmunoprecipitation (Fig. 2D). Similar results were obtained for other class 1 and 2 HDACs (not shown). Together, these results argue against association between TNIP1 and HDACs and suggest that TNIP1 repression of RAR activity may be HDAC-independent.
TNIP1 does not interact with RXRα and transfer of RARα AF-2 domain to it does not confer interaction
Some NRs, e.g., TRα, RARα, RXRα, and ERα, share ligand-dependent interaction with the same NR box-containing coregulator, e.g., TRAP220, because, in part, of conservation in the helix 12 AF-2 sequence (φφXEφφ, φ= hydrophobic amino acid, E=glutamic acid, X=any amino acid) and despite amino acid sequence variation throughout the rest of the receptor LBD [20]. Like the PPARα used to isolate TNIP1, RARs are group 1 NRs but RXRα, the heterodimer partner of both PPARs and RARs, is a group 2 NR [10]. We compared RARα and RXRα interaction with TNIP1 in mammalian two-hybrid assays. RARα interacted with TNIP1 equally well whether liganded with ATRA or 9-cis-RA. However, in the presence of 9-cis-RA, we saw minimal RXRα association with TNIP1, relatively 40-fold less than holo-RARα and TNIP1 (Fig. 3A) suggesting some selectivity by TNIP1 for interaction with retinoid receptors. Since the AF-2 of retinoid receptors is conserved (PLIQEMLE for RARα versus TFLMEMLE for RXRα) but not identical, the inability of RXRα to interact with TNIP1 was addressed by replacing its AF-2 domain with that of the efficiently interacting RARα. This chimeric receptor approach was used previously to establish the role of the RAR-related orphan receptor α (RORα) AF-2 domain with corepressor hairless (hr), where substitution of RARα AF-2 with that of RORα enabled RARα to associate with hr [21]. For RXRα, however, replacement of its AF-2 domain with that of RARα, necessary for RARα's interaction with TNIP1 (Fig. 1B), was insufficient to enable RXRα-TNIP1 association. Notable, in the context of the full-length receptor activating transcription via an RXR binding site, RXR functionality was reduced (Fig. 3B) to empty vector control levels supporting previous interpretations that interplay of AF-2 with other LBD helices supports interaction with NR box coregulators.
Fig. 3.
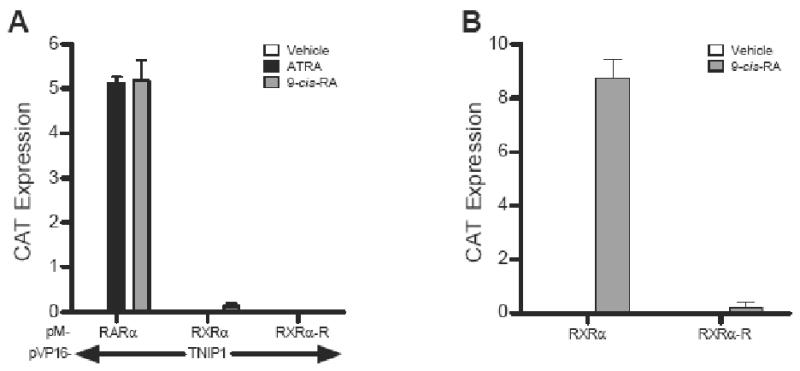
RXRα does not interact with TNIP1 and introduction of RARα AF-2 domain does not confer interaction. (A) M2H assays showing interaction of RARα with TNIP1 compared to wild-type RXRα and RXRα with its AF-2 replaced by that of RARα (RXRα-R). (B) Comparison of RXRα and RXRα-R activation of RXRE-tk-CAT in Cos-7 cells presented as difference in reporter activation between receptor constructs and empty vector.
RARα helices 5-9 dictate its preferential interaction with TNIP1
Coexpression of TNIP1 with RARα in the mammalian two-hybrid system resulted in higher reporter readout than with RARγ suggesting better interaction (Fig. 1C). Subtype selectivity among RARs for interaction with coregulators was observed previously with the corepressor SMRT [22, 23] where LBD helical chimeras derived from RARα and RARβ were able to confer upon β the better interaction with SMRT exhibited by α. We took this approach to test if the greater interaction of RARα with TNIP1 could be mapped to specific helices within its LBD and generated a series of RARα/RARγ chimeric receptors by replacing portions of RARγ LBD with the corresponding helices of RARα (Fig. 4A). Receptor surfaces used for coregulator interaction are formed by varying three-dimensional arrangements of LBD helices 3-12 [24, 25], therefore the chimera constructs were made in the context of helices 3-12. Chimeras wherein RARα helices 3-5 (αγ1) and 10-carboxyl terminus (αγ3) replaced the corresponding regions in the RARγ LBD (Fig. 4A) did not confer increased interaction. In contrast, the αγ2 chimera where helices 5-9 of RARγ LBD are replaced by the corresponding region of RARα was able to increase the level of reporter activation in the mammalian two-hybrid assay (Fig. 4B). Parallel with the two-hybrid constructs, each chimera was made in the context of a full-length receptor and confirmed to activate an RAR-driven reporter gene to a similar degree as intact RARγ (Fig. 4C). Thus all the chimeras are capable of the corepressor/coactivator exchange required for NR transactivation, i.e. they are coregulator competent. Readout in the two-hybrid assays with the chimeras, then, is likely reflective of the helices within the construct rather than overall LBD functionality suggesting RARα helices 5-9 dictate its preferential interaction with TNIP1.
Fig. 4.
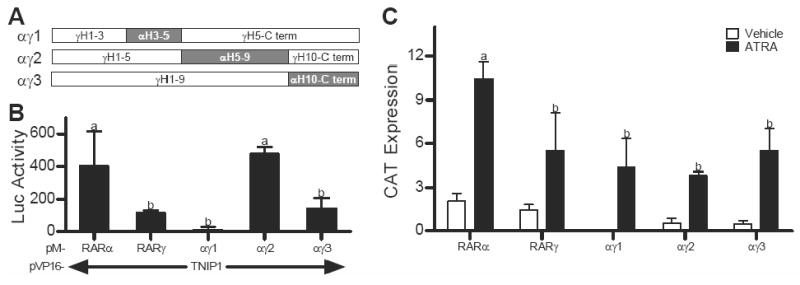
Replacement of helices 5-9 of RARγ ligand binding domain (LBD) with those of RARα increases RARγ-TNIP1 interaction. (A) Schematic of LBD helices in RARα/RARγ chimera constructs with RARγ as open and RARα replacements as shaded rectangles, C term: carboxyl terminus. (B) M2H assays in ATRA treated Cos-7 cells showing interaction of RARα, RARγ or αγ chimeras with TNIP1 presented as difference in reporter activation between TNIP1 constructs and empty vector control. (C) Activation of TRE-tk-CAT reporter by RARα, RARγ, or RARα/RARγ chimeras in Cos-7 cells presented as difference in reporter activation between receptor constructs and empty vector control. RXRα was cotransfected with all RAR constructs. In (B) and (C) bars with different letter indicate significantly different values.
Given the wide tissue distribution of TNIP1 [7, 8], and non-uniform tissue distribution of different RAR subtypes [26] we expect its effects on RARs will vary widely from tissue to tissue depending largely on which RAR subtype is predominantly expressed. For instance, altered TNIP1 levels may have limited effect on NR function if its preferred target receptors are minimally present highlighting the importance of characterizing NR-coregulator selectivity. While NR selectivity has been examined for numerous coregulators [2, 5], differences between receptor subtypes in coregulator interaction have received a limited study. Along these lines, Privalsky and coworkers used a receptor chimera approach similar to the one used here and demonstrated the selectivity of RAR subtypes for interaction with the traditional corepressor SMRT centered on amino acid differences within the receptors' LBD helix 3 and, to a lesser extent, the carboxyl-terminal F domain and enabled interaction with RARα but not RARβ or γ. The RAR LBD sequence differences presumably affected the tertiary structure of the receptors' C-termini allowing for RARα but not for β and γ access to the corepressor binding [22, 23, 27, 28]. In contrast, our results point to helices 5-9 in RARs as being responsible for the differing interaction of this receptor's subtypes with TNIP1. This is a novel finding as no previous role for this region has been reported for interaction with coregulators.
In summary, we show that TNIP1 interacts with liganded RARs but acts as a corepressor of their activity. TNIP1 repression is mediated partly through interference with coactivator recruitment but appears HDAC-independent. While the receptor AF-2 domain is required for association with TNIP1, it is not sufficient by itself to confer interaction. We further note a preference of TNIP1 for RARα subtype over RARγ and map the region responsible for this preference to helices 5-9 of the receptor LBD – a region with no previously known roles in NR-coregulator interaction.
Acknowledgments
This work was supported by the National Institutes of Health, NIAMS, AR048860. Pre-doctoral fellowships from Boehringer-Ingelheim Pharmaceuticals and the American Foundation for Pharmaceutical Education (IG) provided partial support. We thank A.M. Flores for generating some of the plasmid constructs and J. Ciarlo and R. Wilson for assistance with some of the two-hybrid assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lonard DM, O'Malley BW. Nuclear receptor coregulators: Judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 2.Lazar MA. Nuclear receptor corepressors. Nucl Recept Signal. 2003;1:e001. doi: 10.1621/nrs.01001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lonard DM, O'Malley BW. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125:411–4. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 4.Treuter E, Albrektsen T, Johansson L, et al. A regulatory role for RIP140 in nuclear receptor activation. Mol Endocrinol. 1998;12:864–81. doi: 10.1210/mend.12.6.0123. [DOI] [PubMed] [Google Scholar]
- 5.Gurevich I, Flores AM, Aneskievich BJ. Corepressors of agonist-bound nuclear receptors. Toxicol Appl Pharmacol. 2007;223:288–98. doi: 10.1016/j.taap.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White R, Morganstein D, Christian M, et al. Role of RIP140 in metabolic tissues: Connections to disease. FEBS Lett. 2008;582:39–45. doi: 10.1016/j.febslet.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 7.Fukushi M, Dixon J, Kimura T, et al. Identification and cloning of a novel cellular protein Naf1, nef-associated factor 1, that increases cell surface CD4 expression. FEBS Lett. 1999;442:83–8. doi: 10.1016/s0014-5793(98)01631-7. [DOI] [PubMed] [Google Scholar]
- 8.Gupta K, Ott D, Hope TJ, et al. A human nuclear shuttling protein that interacts with human immunodeficiency virus type 1 matrix is packaged into virions. J Virol. 2000;74:11811–24. doi: 10.1128/jvi.74.24.11811-11824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heyninck K, Kreike MM, Beyaert R. Structure-function analysis of the A20-binding inhibitor of NF-kappa B activation, ABIN-1. FEBS Lett. 2003;536:135–140. doi: 10.1016/s0014-5793(03)00041-3. [DOI] [PubMed] [Google Scholar]
- 10.Germain P, Staels B, Dacquet C, et al. Overview of nomenclature of nuclear receptors. Pharmacol Rev. 2006;58:685–704. doi: 10.1124/pr.58.4.2. [DOI] [PubMed] [Google Scholar]
- 11.Fisher GJ, Voorhees JJ. Molecular mechanisms of retinoid actions in skin. Faseb J. 1996;10:1002–13. doi: 10.1096/fasebj.10.9.8801161. [DOI] [PubMed] [Google Scholar]
- 12.Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81:1269–304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 13.Aneskievich BJ, Fuchs E. Terminal differentiation in keratinocytes involves positive as well as negative regulation by retinoic acid receptors and retinoid×receptors at retinoid response elements. Mol Cell Biol. 1992;12:4862–71. doi: 10.1128/mcb.12.11.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zelent A, Krust A, Petkovich M, et al. Cloning of murine alpha and beta retinoic acid receptors and a novel receptor gamma predominantly expressed in skin. Nature. 1989;339:714–7. doi: 10.1038/339714a0. [DOI] [PubMed] [Google Scholar]
- 15.Flores AM, Li L, Aneskievich BJ. Isolation and functional analysis of a keratinocyte-derived, ligand-regulated nuclear receptor comodulator. J Invest Dermatol. 2004;123:1092–101. doi: 10.1111/j.0022-202X.2004.23424.x. [DOI] [PubMed] [Google Scholar]
- 16.Tate BF, Allenby G, Janocha R, et al. Distinct binding determinants for 9-cis retinoic acid are located within AF-2 of retinoic acid receptor alpha. Mol Cell Biol. 1994;14:2323–2330. doi: 10.1128/mcb.14.4.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox JE, Burow ME, McLachlan JA, Miller CA., 3rd Detecting ligands and dissecting nuclear receptor-signaling pathways using recombinant strains of the yeast saccharomyces cerevisiae. Nat Protoc. 2008;3:637–645. doi: 10.1038/nprot.2008.33. [DOI] [PubMed] [Google Scholar]
- 18.Zhou J, Oakley RH, Cidlowski JA. DAX-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X-chromosome, gene 1) selectively inhibits transactivation but not transrepression mediated by the glucocorticoid receptor in a LXXLL-dependent manner. Mol Endocrinol. 2008;22:1521–34. doi: 10.1210/me.2007-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delage-Mourroux R, Martini PG, Choi I, et al. Analysis of estrogen receptor interaction with a repressor of estrogen receptor activity (REA) and the regulation of estrogen receptor transcriptional activity by REA. J Biol Chem. 2000;275:35848–56. doi: 10.1074/jbc.M001327200. [DOI] [PubMed] [Google Scholar]
- 20.Yuan CX, Ito M, Fondell JD, et al. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci U S A. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moraitis AN, Giguere V, Thompson CC. Novel mechanism of nuclear receptor corepressor interaction dictated by activation function 2 helix determinants. Mol Cell Biol. 2002;22:6831–41. doi: 10.1128/MCB.22.19.6831-6841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farboud B, Hauksdottir H, Wu Y, Privalsky ML. Isotype-restricted corepressor recruitment: A constitutively closed helix 12 conformation in retinoic acid receptors beta and gamma interferes with corepressor recruitment and prevents transcriptional repression. Mol Cell Biol. 2003;23:2844–58. doi: 10.1128/MCB.23.8.2844-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hauksdottir H, Farboud B, Privalsky ML. Retinoic acid receptors beta and gamma do not repress, but instead activate target gene transcription in both the absence and presence of hormone ligand. Mol Endocrinol. 2003;17:373–85. doi: 10.1210/me.2002-0340. [DOI] [PubMed] [Google Scholar]
- 24.Darimont BD, Wagner RL, Apriletti JW, et al. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–56. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perissi V, Staszewski LM, McInerney EM, et al. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 1999;13:3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dolle P, Ruberte E, Leroy P, et al. Retinoic acid receptors and cellular retinoid binding proteins. I. A systematic study of their differential pattern of transcription during mouse organogenesis. Development. 1990;110:1133–1151. doi: 10.1242/dev.110.4.1133. [DOI] [PubMed] [Google Scholar]
- 27.Wong CW, Privalsky ML. Transcriptional silencing is defined by isoform- and heterodimer-specific interactions between nuclear hormone receptors and corepressors. Mol Cell Biol. 1998;18:5724–5733. doi: 10.1128/mcb.18.10.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farboud B, Privalsky ML. Retinoic acid receptor-alpha is stabilized in a repressive state by its C-terminal, isotype-specific F domain. Mol Endocrinol. 2004;18:2839–53. doi: 10.1210/me.2004-0236. [DOI] [PubMed] [Google Scholar]


