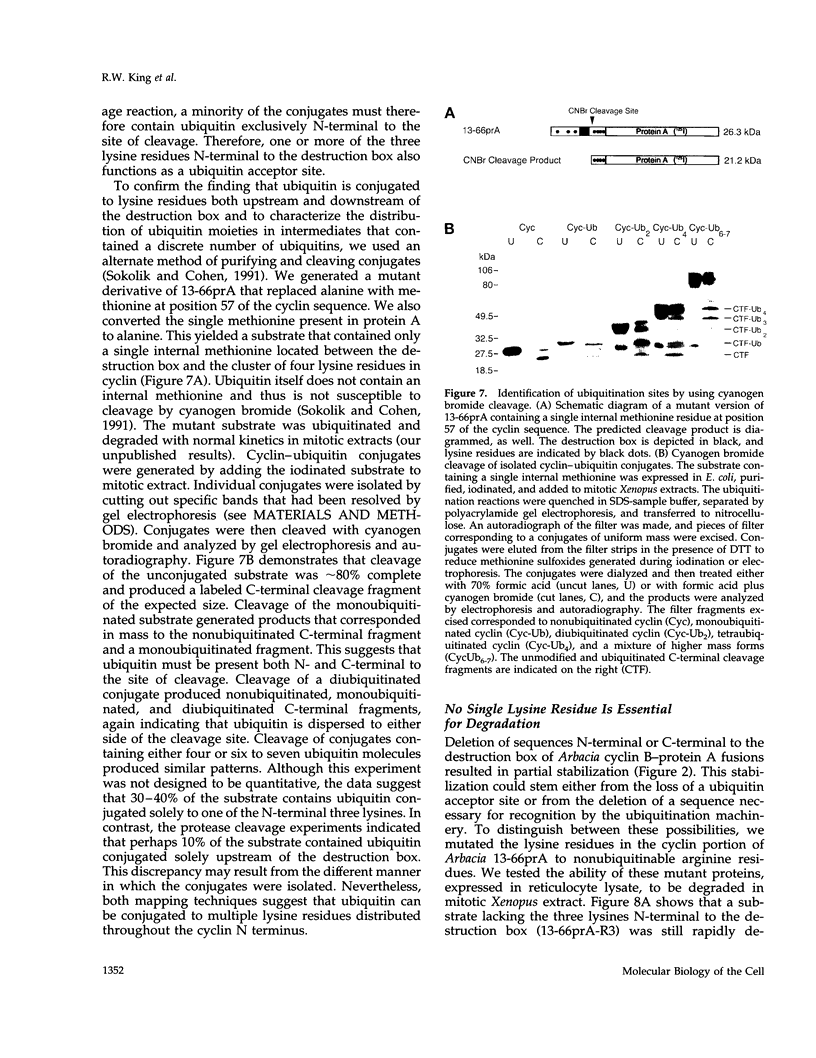
Abstract
Mitotic cyclins are abruptly degraded at the end of mitosis by a cell-cycle-regulated ubiquitin-dependent proteolytic system. To understand how cyclin is recognized for ubiquitin conjugation, we have performed a mutagenic analysis of the destruction signal of mitotic cyclins. We demonstrate that an N-terminal cyclin B segment as short as 27 residues, containing the 9-amino-acid destruction box, is sufficient to destabilize a heterologous protein in mitotic Xenopus extracts. Each of the three highly conserved residues of the cyclin B destruction box is essential for ubiquitination and subsequent degradation. Although an intact destruction box is essential for the degradation of both A- and B-type cyclins, we find that the Xenopus cyclin A1 destruction box cannot functionally substitute for its B-type counterpart, because it does not contain the highly conserved asparagine necessary for cyclin B proteolysis. Physical analysis of ubiquitinated cyclin B intermediates demonstrates that multiple lysine residues function as ubiquitin acceptor sites, and mutagenic studies indicate that no single lysine residue is essential for cyclin B degradation. This study defines the key residues of the destruction box that target cyclin for ubiquitination and suggests there are important differences in the way in which A- and B-type cyclins are recognized by the cyclin ubiquitination machinery.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amon A., Irniger S., Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994 Jul 1;77(7):1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Aristarkhov A., Eytan E., Moghe A., Admon A., Hershko A., Ruderman J. V. E2-C, a cyclin-selective ubiquitin carrier protein required for the destruction of mitotic cyclins. Proc Natl Acad Sci U S A. 1996 Apr 30;93(9):4294–4299. doi: 10.1073/pnas.93.9.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A., Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989 Mar 24;56(6):1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- Beers E. P., Callis J. Utility of polyhistidine-tagged ubiquitin in the purification of ubiquitin-protein conjugates and as an affinity ligand for the purification of ubiquitin-specific hydrolases. J Biol Chem. 1993 Oct 15;268(29):21645–21649. [PubMed] [Google Scholar]
- Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989 Mar 24;243(4898):1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Deshaies R. J. The self-destructive personality of a cell cycle in transition. Curr Opin Cell Biol. 1995 Dec;7(6):781–789. doi: 10.1016/0955-0674(95)80061-1. [DOI] [PubMed] [Google Scholar]
- Evans T., Rosenthal E. T., Youngblom J., Distel D., Hunt T. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983 Jun;33(2):389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- Galan J. M., Volland C., Urban-Grimal D., Haguenauer-Tsapis R. The yeast plasma membrane uracil permease is stabilized against stress induced degradation by a point mutation in a cyclin-like "destruction box". Biochem Biophys Res Commun. 1994 Jun 15;201(2):769–775. doi: 10.1006/bbrc.1994.1767. [DOI] [PubMed] [Google Scholar]
- Gallant P., Nigg E. A. Cyclin B2 undergoes cell cycle-dependent nuclear translocation and, when expressed as a non-destructible mutant, causes mitotic arrest in HeLa cells. J Cell Biol. 1992 Apr;117(1):213–224. doi: 10.1083/jcb.117.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant P., Nigg E. A. Identification of a novel vertebrate cyclin: cyclin B3 shares properties with both A- and B-type cyclins. EMBO J. 1994 Feb 1;13(3):595–605. doi: 10.1002/j.1460-2075.1994.tb06297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiara J. B., Richardson H. E., Sugimoto K., Henze M., Lew D. J., Wittenberg C., Reed S. I. A cyclin B homolog in S. cerevisiae: chronic activation of the Cdc28 protein kinase by cyclin prevents exit from mitosis. Cell. 1991 Apr 5;65(1):163–174. doi: 10.1016/0092-8674(91)90417-w. [DOI] [PubMed] [Google Scholar]
- Glotzer M. Cell cycle. The only way out of mitosis. Curr Biol. 1995 Sep 1;5(9):970–972. doi: 10.1016/s0960-9822(95)00190-4. [DOI] [PubMed] [Google Scholar]
- Glotzer M., Murray A. W., Kirschner M. W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991 Jan 10;349(6305):132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ganoth D., Pehrson J., Palazzo R. E., Cohen L. H. Methylated ubiquitin inhibits cyclin degradation in clam embryo extracts. J Biol Chem. 1991 Sep 5;266(25):16376–16379. [PubMed] [Google Scholar]
- Hershko A., Heller H. Occurrence of a polyubiquitin structure in ubiquitin-protein conjugates. Biochem Biophys Res Commun. 1985 May 16;128(3):1079–1086. doi: 10.1016/0006-291x(85)91050-2. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M., Ellison M. J., Chau V., Varshavsky A. The short-lived MAT alpha 2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway S. L., Glotzer M., King R. W., Murray A. W. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 1993 Jul 2;73(7):1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- Hunt T., Luca F. C., Ruderman J. V. The requirements for protein synthesis and degradation, and the control of destruction of cyclins A and B in the meiotic and mitotic cell cycles of the clam embryo. J Cell Biol. 1992 Feb;116(3):707–724. doi: 10.1083/jcb.116.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S., Piatti S., Michaelis C., Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995 Apr 21;81(2):269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Izumi T., Maller J. L. Phosphorylation of Xenopus cyclins B1 and B2 is not required for cell cycle transitions. Mol Cell Biol. 1991 Aug;11(8):3860–3867. doi: 10.1128/mcb.11.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplon T., Jacquet M. The cellular content of Cdc25p, the Ras exchange factor in Saccharomyces cerevisiae, is regulated by destabilization through a cyclin destruction box. J Biol Chem. 1995 Sep 1;270(35):20742–20747. doi: 10.1074/jbc.270.35.20742. [DOI] [PubMed] [Google Scholar]
- King R. W., Jackson P. K., Kirschner M. W. Mitosis in transition. Cell. 1994 Nov 18;79(4):563–571. doi: 10.1016/0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- King R. W., Peters J. M., Tugendreich S., Rolfe M., Hieter P., Kirschner M. W. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995 Apr 21;81(2):279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Klotzbücher A., Stewart E., Harrison D., Hunt T. The 'destruction box' of cyclin A allows B-type cyclins to be ubiquitinated, but not efficiently destroyed. EMBO J. 1996 Jun 17;15(12):3053–3064. [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H., Stewart E., Poon R., Adamczewski J. P., Gannon J., Hunt T. Identification of the domains in cyclin A required for binding to, and activation of, p34cdc2 and p32cdk2 protein kinase subunits. Mol Biol Cell. 1992 Nov;3(11):1279–1294. doi: 10.1091/mbc.3.11.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Meyer A. N., Donoghue D. J. Requirement for phosphorylation of cyclin B1 for Xenopus oocyte maturation. Mol Biol Cell. 1995 Sep;6(9):1111–1124. doi: 10.1091/mbc.6.9.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorca T., Devault A., Colas P., Van Loon A., Fesquet D., Lazaro J. B., Dorée M. Cyclin A-Cys41 does not undergo cell cycle-dependent degradation in Xenopus extracts. FEBS Lett. 1992 Jul 13;306(1):90–93. doi: 10.1016/0014-5793(92)80844-7. [DOI] [PubMed] [Google Scholar]
- Luca F. C., Shibuya E. K., Dohrmann C. E., Ruderman J. V. Both cyclin A delta 60 and B delta 97 are stable and arrest cells in M-phase, but only cyclin B delta 97 turns on cyclin destruction. EMBO J. 1991 Dec;10(13):4311–4320. doi: 10.1002/j.1460-2075.1991.tb05009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q., Michaelis C., Weeks G. Overexpression of a truncated cyclin B gene arrests Dictyostelium cell division during mitosis. J Cell Sci. 1994 Nov;107(Pt 11):3105–3114. doi: 10.1242/jcs.107.11.3105. [DOI] [PubMed] [Google Scholar]
- Minshull J., Golsteyn R., Hill C. S., Hunt T. The A- and B-type cyclin associated cdc2 kinases in Xenopus turn on and off at different times in the cell cycle. EMBO J. 1990 Sep;9(9):2865–2875. doi: 10.1002/j.1460-2075.1990.tb07476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Murray A. W., Solomon M. J., Kirschner M. W. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989 May 25;339(6222):280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- Pines J., Hunter T. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J Cell Biol. 1991 Oct;115(1):1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmington G., Dalby B., Glover D. M. Expression of N-terminally truncated cyclin B in the Drosophila larval brain leads to mitotic delay at late anaphase. J Cell Sci. 1994 Oct;107(Pt 10):2729–2738. doi: 10.1242/jcs.107.10.2729. [DOI] [PubMed] [Google Scholar]
- Scheffner M., Huibregtse J. M., Vierstra R. D., Howley P. M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993 Nov 5;75(3):495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Schwob E., Böhm T., Mendenhall M. D., Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994 Oct 21;79(2):233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Sigrist S., Jacobs H., Stratmann R., Lehner C. F. Exit from mitosis is regulated by Drosophila fizzy and the sequential destruction of cyclins A, B and B3. EMBO J. 1995 Oct 2;14(19):4827–4838. doi: 10.1002/j.1460-2075.1995.tb00164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolik C. W., Cohen R. E. The structures of ubiquitin conjugates of yeast Iso-2-cytochrome c. J Biol Chem. 1991 May 15;266(14):9100–9107. [PubMed] [Google Scholar]
- Sokolik C. W., Cohen R. E. Ubiquitin conjugation to cytochromes c. Structure of the yeast iso-1 conjugate and possible recognition determinants. J Biol Chem. 1992 Jan 15;267(2):1067–1071. [PubMed] [Google Scholar]
- Stewart E., Kobayashi H., Harrison D., Hunt T. Destruction of Xenopus cyclins A and B2, but not B1, requires binding to p34cdc2. EMBO J. 1994 Feb 1;13(3):584–594. doi: 10.1002/j.1460-2075.1994.tb06296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Sudakin V., Ganoth D., Dahan A., Heller H., Hershko J., Luca F. C., Ruderman J. V., Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995 Feb;6(2):185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U., Amon A., Dowzer C., McGrew J., Byers B., Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 1993 May;12(5):1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treier M., Staszewski L. M., Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994 Sep 9;78(5):787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- Tugendreich S., Tomkiel J., Earnshaw W., Hieter P. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995 Apr 21;81(2):261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- Whitfield W. G., Gonzalez C., Maldonado-Codina G., Glover D. M. The A- and B-type cyclins of Drosophila are accumulated and destroyed in temporally distinct events that define separable phases of the G2-M transition. EMBO J. 1990 Aug;9(8):2563–2572. doi: 10.1002/j.1460-2075.1990.tb07437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Guacci V., Koshland D. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J Cell Biol. 1996 Apr;133(1):85–97. doi: 10.1083/jcb.133.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Guacci V., Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s). J Cell Biol. 1996 Apr;133(1):99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., King R. W., Peters J. M., Kirschner M. W. Identification of a novel ubiquitin-conjugating enzyme involved in mitotic cyclin degradation. Curr Biol. 1996 Apr 1;6(4):455–466. doi: 10.1016/s0960-9822(02)00513-4. [DOI] [PubMed] [Google Scholar]
- Zachariae W., Nasmyth K. TPR proteins required for anaphase progression mediate ubiquitination of mitotic B-type cyclins in yeast. Mol Biol Cell. 1996 May;7(5):791–801. doi: 10.1091/mbc.7.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velden H. M., Lohka M. J. Cell cycle-regulated degradation of Xenopus cyclin B2 requires binding to p34cdc2. Mol Biol Cell. 1994 Jul;5(7):713–724. doi: 10.1091/mbc.5.7.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velden H. M., Lohka M. J. Mitotic arrest caused by the amino terminus of Xenopus cyclin B2. Mol Cell Biol. 1993 Mar;13(3):1480–1488. doi: 10.1128/mcb.13.3.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]