Abstract
Few Discoveries have had the impact of RNAi. Most researchers who use small RNAs to control gene expression in mammalian cells assume that mRNA will be the target. Recent studies, however, have suggested that small RNAs can also target chromosomal DNA.
Keywords: Promoter-Targeted RNA, non-coding RNA
Introduction
Small duplex RNAs have become ubiquitous tools for controlling gene expression.[1] When introduced into cells or expressed inside cells, duplex RNAs can target mRNA and reduce gene expression. Gene silencing by small RNAs is robust, potent, and readily achievable by laboratories with modest expertise working with mammalian cells. Few discoveries have had such a large and immediate impact on the conduct of experimental science.
The common assumption for RNA-mediated gene silencing in mammalian cells is that small RNAs target mRNA. In 2004, however, two papers appeared which challenged this common wisdom.[2,3] These papers reported that small RNAs could target promoter DNA, induce DNA methylation, and reduce gene expression.
While these findings were intriguing, acceptance of the possibility that RNAs might mediate promoter function has been slow. One problem was that one[3] of the initial two papers was subsequently retracted.[4] On a fundamental scientific level, there was no mechanistic framework for understanding the results. mRNA and chromosomal DNA are very different, how could duplex RNAs recognize both of them? On a practical level, experiments that involve introduction of nucleic acids into cells are notoriously artifact-prone.[5] Sceptics could easily dismiss initial reports by assuming that effects were due to interactions between the introduced RNAs and nontarget molecules (also known as off-target effects, these can be caused by unintended interactions with proteins, RNA, or DNA).
In late 2006, the excitement and the scepticism caused by promoter-targeted RNAs was further increased by a report that these RNAs could also activate gene expression,[6] a capability that would substantially expand the experimental and therapeutic value of duplex RNA. This claim, however, fit even less well into the established theoretical framework for RNAi.[7]
How do promoter-targeted RNAs function?
Prior to these initial reports on modulating gene expression by promoter-targeted RNAs, our laboratory had been working on recognition of chromosomal DNA by synthetic peptide nucleic acid oligomers (PNAs) (Figure 1) complementary to transcription start sites.[8] Recognition of chromosomal DNA by PNAs would necessarily follow much different rules than recognition of DNA by duplex RNA because PNAs are single stranded and have an unnatural amide backbone. Nevertheless, it was straightforward for us to use our established experimental systems to investigate the potential for duplex RNA to target chromosomal DNA.
Figure 1.
Chemical structure of RNA compared to structures of synthetic nucleic acid mimics locked nucleic acid (LNA)[11] and peptide nucleic acid (PNA)[8] that are known to recognize duplex DNA inside cells. Antigene PNAs and LNAs are introduced into cells as single strands, while antigene RNAs are duplexes. PNAs and LNAs provide useful benchmarks for evaluating the activities of agRNAs and considering potential mechanisms of action.
We designed duplex RNAs to target the promoter of the progesterone receptor (PR) gene.[9] We refer to these RNAs as antigene RNAs (agRNAs) to distinguish them from duplex RNAs that are designed to target mRNA and to conform to traditional chemical nomenclature for synthetic oligomers intended to interact with chromosomal DNA.
We chose to target PR because we had already shown that its expression could be reduced by single-stranded PNAs[8] and its promoter region was well-characterized.[10] This last point is critical for experiments with RNAs that target DNA. Experimenters must be confident that the transcription start site has been accurately determined and that observed effects are not due to interactions with mRNA. Recently, we have also shown that single-stranded locked nucleic acids (LNAs) can recognize chromosomal DNA inside cells.[11] This observation reinforces the suggestion that the PR promoter is susceptible to binding by synthetic oligomers and a productive model system for examining agents designed to modulate gene expression.
Our experiments revealed a dramatic reduction of PR mRNA levels and protein expression upon addition of agRNAs.[9] In contrast to experiments with PNAs or LNAs, that required extensive optimization, the first experiments with agRNAs revealed potent inhibition and subsequent experiments have demonstrated that the results are easy to reproduce. This robustness hints at the possibility that the activity of our agRNAs reflects natural mechanisms for promoter-targeted RNAs. Extensive control experiments suggested that our agRNAs were interacting with their intended target sequences, supporting the conclusion that we were investigating a novel phenomenon.
We screened through a large number of agRNAs and identified several capable of inhibiting PR gene expression.[9] In the course of these experiments, we also identified RNAs that reproducibly caused a 25–50% increase in PR gene expression. While such relatively small changes were far from definitive, they did suggest that some small RNAs might be able to act like protein transcription factors and enhance gene expression. Up to this point our experiments had been carried out in a cell line with high expression of our target gene. We reasoned that using a cell line that expressed low levels of PR would allow increases in gene expression to be observed more easily.
After testing agRNAs in a cell line with low background PR expression it became clear that they had the potential to increase gene expression many-fold above background.[12] We identified several agRNAs capable of activating PR expression while multiple mismatch and scrambled control duplex RNAs did not affect gene expression. These data suggested that agRNAs could activate gene expression.
Unlike traditional siRNAs, agRNAs are not complementary to mRNA. What molecules might agRNAs be associating with to produce such varied effects on gene expression? One hypothesis is that agRNAs bind directly to chromosomal DNA (Figure 2A). While conceptually simple, there is no known cellular machinery for promoting this recognition. An alternative explanation is that the agRNAs bind to nascent RNA transcripts that overlap gene promoters (Figure 2B).
Figure 2.
Possible mechanisms for agRNA mechanism. A) agRNAs could directly recognize chromosomal DNA. B). agRNAs could recognize ncRNA transcripts originating within the target gene promoter. Argonaute family proteins (Ago) are known to mediate agRNA function. Other, unidentified proteins could bridge a ncRNA and promoter DNA to affect transcription and gene expression.
These hypotheses were first described by investigations using yeast as a model system.[13] RNAi proteins had been implicated in heterochromatin formation in fission yeast through the RNA-initiation of transcriptional gene silencing (RITS) complex formation.[14] The yeast RITS complex is comprised of Chp1, Tas3, and Ago1. Of these proteins, only Ago1 has clear mammalian homologues. Mammalian cells contain four Ago genes (Ago1 – Ago4). Of these genes, Ago2 has been described as the catalytic engine for RNAi while the functions of Ago1, Ago3, and Ago4 remain unclear.[15,16]
We investigated the potential involvement of Ago proteins in gene regulation by agRNAs in mammalian cells. We observed that Ago2, and to a lesser extent Ago1, appeared to be necessary for silencing by agRNAs.[17] Ago1 involvement was reported for gene silencing by Rossi and co-workers,[18] while Li and coworkers implicated Ago2 in RNA-mediated gene activation.[6] The Ago proteins are known to promote RNA-mediated recognition of RNA, further suggesting a model in which agRNAs bind nascent RNA transcripts rather than chromosomal DNA.
While there were no reports of RNA being produced from the PR gene promoter, recent studies had revealed that most of the genome is transcribed.[19–22] Complex networks of noncoding RNAs (ncRNAs) overlap gene promoters as well as other regions of the genome. These ncRNAs are transcribed in both the sense and antisense direction relative to the genes in which they overlap. These findings led us to search for potential ncRNAs that overlapped the PR gene promoter.
Using RT-PCR we detected RNA species originating from the PR gene promoter and 5′-RACE identified a ncRNA transcribed through the promoter in the antisense direction relative to the PR gene.[23] We were unable to detect any RNA species being transcribed upstream from the previously determined[10] transcription start site for PR mRNA in the sense direction relative to the gene. The antisense transcript was, therefore, the only known candidate for recognition by our agRNAs. Having identified a candidate RNA target for our agRNAs, we used a battery of techniques to investigate the possible mechanistic role of this ncRNA in our observations.
Our experiments revealed several lines of evidence implicating the antisense transcript in the action of agRNAs: 1) We developed a biotin-avidin purification assay in which the agRNA is biotinylated on either strand of the duplex and transfected into cells. The bound nucleic acid is isolated using avidin coated beads and analyzed using RT-PCR. We observed association of the agRNA with the antisense ncRNA transcript and no association with chromosomal DNA; 2) We found that agRNAs could induce association of Ago proteins with the ncRNA transcript using RNA immunoprecipitation; 3) reducing the levels of the antisense ncRNA reversed the activity of activating agRNAs.
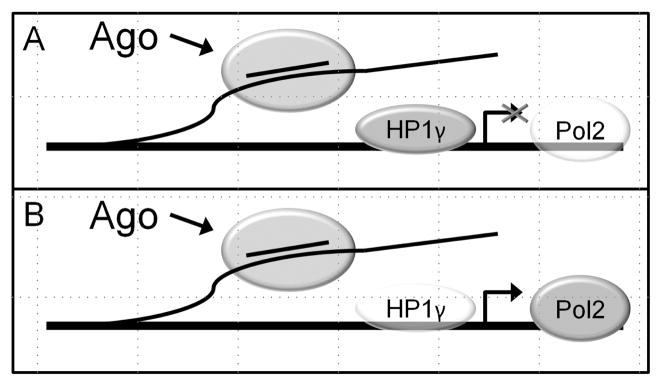
Taken together these data suggest a model in which the agRNA recognizes the antisense ncRNA in close proximity to the chromosome and transcriptionally modulates gene expression (Figure 3). Supporting this hypothesis, chromatin immunoprecipitation for RNA Pol II reveals increased and decreased transcription for activating and silencing agRNAs, respectively.
Figure 3.
Interactions of duplex RNAs at the PR promoter during gene silencing or gene activation. A) Silencing agRNAs recruit both AGO and HP1γ to the target gene promoter in cells that normally express a high level of PR. This results in decreased levels of RNA polymerase II on the target gene. B) Activating agRNAs recruit AGO to the target gene promoter and increase RNA polymerase II levels on the target gene in cells that normally express a low level of PR.
Our studies[23] and those of others[24,25] have suggested that ncRNAs transcribed through gene promoters can be utilized to regulate gene transcription using synthetic small duplex RNA, but why are noncoding RNAs transcribed at gene promoters? What is the natural function of these ncRNAs?
The noncoding RNA world
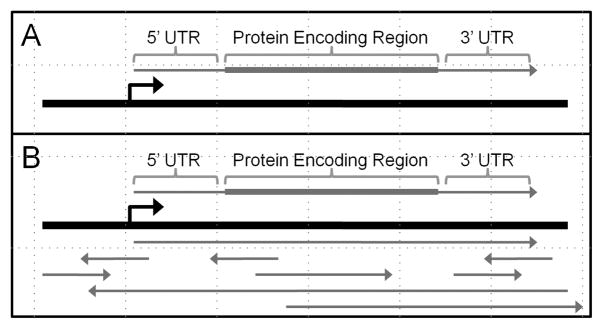
To begin to answer these questions, it is necessary to understand that transcription is much more complicated than has been appreciated previously. Traditionally, transcription of a gene is pictured as synthesis of an mRNA (Figure 4A). Slightly over 1 % of the human gene is transcribed into mRNA and this RNA has been the focus of most experimental studies. Recent studies have made it clear, however, that greater than 80 % of the genome is transcribed into RNA.[19–22] Of special significance for the mechanism of agRNAs, a substantial fraction of all genes have ncRNA transcripts that overlap their promoters.[26,27]
Figure 4.
Models of RNA expression. A) Traditional model: an mRNA is synthesized and translated into protein. B) Emerging model: Many overlapping RNAs are synthesized alongside the protein-encoding mRNA. The overall effect of these RNAs on transcription and translation of mRNA are only beginning to be understood.
Instead of picturing a simple mRNA being transcribed, it is more accurate to envision the mRNA being just one of many overlapping RNAs (Figure 4B). Our understanding of the function of these RNAs, or whether most have any function at all, is in its infancy. Several recent reports, however, have investigated various functional aspects of ncRNAs and their involvement in regulating gene transcription (Figure 5).
Figure 5.
Different Mechanisms of Gene Regulation by ncRNAs. A) Transcription of an upstream ncRNA alters chromatin and allows transcription of subsequent ncRNAs. The end result is increased transcription of the associated gene. B) Protein binding to a ncRNA produced from the gene promoter undergoes allosteric modifications that result in local transcriptional silencing. C) Transcriptional silencing complexes bind to the ncRNA in close proximity to the chromosome and are transferred to the DNA, resulting in transcriptional silencing. D) Transcriptional silencing complexes bind to the ncRNA and are recruited to distant genomic locations by unclear mechanisms.
One group has found that transcription through a gene’s promoter modifies the chromatin in a manner that prepares the gene for transcriptional activation.[28] Using the fission yeast Schizosaccharomyces pombe as a model system, Hirota et al were able to demonstrate that a cascade of four ncRNAs transcribed through the promoter of the fbp1+ gene prepare the locus for activation (Figure 5A). The authors report that transcription of the most upstream ncRNA prepares the chromatin for binding of factors that induce transcription of the following ncRNA within the cascade. This ‘stepwise’ process results in robust activation of fbp1+ during glucose starvation. Removal of the ncRNAs via insertion of a premature transcription terminator sequence within the locus was sufficient to prevent chromatin remodelling and transcriptional activation. This report highlights at least one functional importance of transcription within gene promoters.
Additional reports have focused on the functional roles of the actual ncRNA molecules. Wang et al. showed in human cells that the cyclin D1 gene promoter produces a ncRNA in response to DNA damage signals.[29] The nascent ncRNA recruits protein factors that repress the cyclin D1 gene (Figure 5B). A consensus sequence within the ncRNA serves as a ligand for the RNA-binding protein TLS (for translocated in liposarcoma). Upon binding, it is suggested that an allosteric modification in TLS results in an inhibitory effect on the histone acetyltransferases CBP (CREB-binding protein) and p300, thus silencing transcription of the cyclin D1 gene. The authors propose a general model in which induced ncRNAs can serve as ligands for co-regulatory proteins. The binding of the ncRNA to the co-regulator would result in activation (or inactivation) of the protein causing changes in transcription.
Interestingly, regulation of transcription by ncRNAs does not seem to be restricted to gene promoters. The idea of ncRNAs acting as ligands for co-regulators has been extended to X-chromosome inactivation (XCI). It has long been known that expression of a ncRNA from the X-chromosome, termed Xist, is responsible for the initiation of XCI.[30,31] Zhao et al. have demonstrated that a short ncRNA expressed from within Xist functions to recruit the Polycomb complex, PRC2, to the X chromosome (Figure 5C).[32] They report that depletion of either PRC2 or the ncRNA results in failure to initiate XCI.
The reports described above have all studied the function of ncRNAs acting in cis with respect to their own genomic origin. However, there are some examples of ncRNAs that silence transcription by acting in trans (Figure 5D). HOTAIR is a long ncRNA transcribed from the human HOXC locus (on chromosome 12) that represses transcription of the HOXD locus (on chromosome 2).[33] Rinn et al. have reported that depletion of the HOTAIR transcript using multiple siRNAs results in significant increases in expression of the HOXD cluster while having no effect on the HOXC cluster itself. The authors also suggest involvement of the same Polycomb complex (PRC2) that was reported to be involved in ncRNA-mediated X-chromosome inactivation.
Another example of a ncRNA acting in trans to silence transcription is the Air ncRNA. The Air transcript is expressed from paternally inherited chromosomes and is involved in imprinting (or silencing) of the proximal genes on the chromosome.[34] The Air transcript does not affect expression of the same locus on the maternally inherited chromosome. A recent report from Nagano et al. showed that the ncRNA physically interacts with the promoter of one of the imprinted genes, Slc22a3.[35] Air is considered to work in trans because the gene and the ncRNA are positioned more than 100kb apart and are transcribed in divergent orientation. The report suggests that the Air ncRNA recruits the histone methyltransferase G9a to the Slc22a3 promoter. However, evidence also suggests that the Air ncRNA uses different mechanisms to imprint other genes in the locus.
Conclusions
It is becoming apparent that ncRNAs play important roles in diverse cellular processes and the pool of interesting ncRNAs is increasing at a rapid pace. However, because ncRNAs inherently have no coding potential it remains difficult to accurately predict their function. Much more work will be required before we can understand how these RNAs contribute to regulating cellular processes.
Many open mechanistic questions remain unanswered and will likely be a rich area of research for many years. For ncRNAs in general, little is known about their function or how they interact with chromosomal DNA, proteins, or mRNA. For agRNAs, the precise molecular details for how they can activate expression in one context and silence expression in another remain to be elucidated. Perhaps the biggest question is whether endogenous small RNAs, for example microRNAs, possess the ability to recognize gene promoters.
For chemists, the ability to either activate or inhibit gene expression using synthetic agRNAs that are complementary to gene promoters hints that noncoding RNAs provide a large and virtually untapped reservoir of molecular targets. It is likely that we have glimpsed only a small fraction of the noncoding RNA world. Chemical approaches will contribute to defining the structure of these RNAs, their biological functions, and the potential for using them to manipulate the expression of genes.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (NIGMS 77253 to DRC), The Robert A. Welch Foundation (I-1244), and an NIH Pharmacological Sciences Training Grant (GM07062 to STY).
References
- 1.Watts JK, Deleavey GF, Damha MJ. Chemically modified siRNA, tools and applications. Drug Discovery Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional silencing in human cells. Science. 2004;305:1289–92. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 3.Kawasaki H, Taira K. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature. 2004;431:211–7. doi: 10.1038/nature02889. [DOI] [PubMed] [Google Scholar]
- 4.Taira K. Retraction: Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature. 2006;441:1176. doi: 10.1038/nature04952. [DOI] [PubMed] [Google Scholar]
- 5.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li L, Okino ST, Zhao H, Pookot D, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Check E. RNA interference: Hitting the switch. Nature. 2007;448:855–858. doi: 10.1038/448855a. [DOI] [PubMed] [Google Scholar]
- 8.Janowski BA, Kaihatsu K, Huffman KE, Schwartz JC, Ram R, Hardy DB, Mendelson CR, Corey DR. Inhibiting transcription of chromosomal DNA using antigene peptide nucleic acids. Nat Chem Biol. 2005;1:210–215. doi: 10.1038/nchembio724. [DOI] [PubMed] [Google Scholar]
- 9.Janowski BA, Huffman KE, Schwartz JC, Ram R, Hardy DB, Shames DS, Minna JD, Corey DR. Inhibition of gene expression at transcription start sites using antigene RNAs (agRNAs) Nat Chem Biol. 2005;1:216–222. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 10.Kastner P, Krust A, Turcotte B, Stropp U, Tora L, Gronemeyer H, Chambon P. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990;9:1603–14. doi: 10.1002/j.1460-2075.1990.tb08280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beane R, Gabillet S, Montallier C, Arar K, Corey DR. Recognition of chromosomal DNA by locked nucleic acids. Biochemistry. 2008;47:13147–13149. doi: 10.1021/bi801930p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 13.Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi-and heterochromatin dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 14.Verdel A, Jia S, Gerber S, Sugiyama T, Bygi S, Greal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–6. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 17.Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR. Involvement of Ago1 and Ago2 in mammalian transcriptional silencing. Nat Struc Mol Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 18.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struc Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 19.RIKEN Group and FANTOM. Consortium Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 20.The Encode Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the Encode pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gingeras TR. Origin of phenotypes: genes and transcripts. Genome Res. 2007;17:682–690. doi: 10.1101/gr.6525007. [DOI] [PubMed] [Google Scholar]
- 22.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 23.Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Corey DR. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15:842–8. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional silencing in human cells. Proc Natl Acad Sci. 2007;104:12422–14227. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008 doi: 10.1371/journal.pgen.1000258. ePub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, Nemzer S, Pinner E, Walach S, Bernstein J, Savitsky K, Rotman G. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–385. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 27.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;302:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirota K, Mioshi T, Kogou K, Hoffman CS, Shibata T, Ohta K. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–4. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Arai S, Song X, Reichart D, Du K, Pascual GP, Tempst G, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–30. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 192;71:527–42. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 31.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–7. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 32.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wutz A, Barlow DP. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–9. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 35.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]