Abstract
We have identified isoforms of dystrophin and utrophin, a dystrophin homologue, expressed in astrocytes and examined their expression patterns during dibutyryl-cAMP (dBcAMP)-induced morphological differentiation of astrocytes. Immunoblot and immunocytochemical analyses showed that full-length-type dystrophin (427 kDa), utrophin (395 kDa), and Dp71 (75 kDa), a small-type dystrophin isoform, were coexpressed in cultured nondifferentiated rat brain astrocytes and were found to be located in the cell membrane. During morphological differentiation of the astrocytes induced by 1 mM dBcAMP, the amount of Dp71 markedly increased, whereas that of dystrophin and utrophin decreased. Northern blot analyses revealed that dBcAMP regulates the mRNA levels of Dp71 and dystrophin but not that of utrophin. dBcAMP slightly increased the amount of the β-dystroglycan responsible for anchoring dystrophin isoforms and utrophin to the cell membrane. Immunocytochemical analyses showed that most utrophin was observed in the cytoplasmic area during astrocyte differentiation, whereas Dp71 was found along the cell membrane of the differentiated astrocytes. These findings suggest that most of the dystrophin/utrophin-dystroglycan complex on cell membrane in cultured astrocytes was replaced by the Dp71-dystroglycan complex during morphological differentiation. The cell biological roles of Dp71 are discussed.
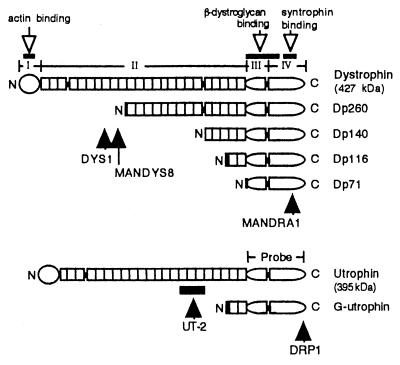
Dystrophin is a 427-kDa protein composed of four functional domains: (i) the NH2-terminal actin-binding domain, (ii) the large rod domain, (iii) the cysteine-rich domain, and (iv) the COOH-terminal domain binding to β-dystroglycan, syntrophins, and dystrobrevin (A0) (see Fig. 1). Dystrophin associates with the muscle sarcolemma via a transmembranous component (β-dystroglycan), which in turn binds to α-dystroglycan. The latter tightly binds to laminin. These findings indicate that dystrophin and dystroglycans form cross-bridges between the extracellular matrix and the subsarcolemmal cytoskeletal network (dystrophin axis), including actin filaments (for reviews see refs. 1 and 2).
Figure 1.
Schematic representations of dystrophin isoforms and utrophin. N and C indicate NH2 and COOH termini of the proteins. Four dystrophin domains, i.e., an actin-binding domain, rod domain, cysteine-rich domain, and a COOH-terminal domain, are indicated as I, II, III, and IV, respectively. Open and solid arrowheads indicate the functional (protein-binding) domains and epitopes recognized by the antibodies DYS1, MANDYS8, MANDRA1, DRP1, and UT-2, respectively. The solid box for utrophin indicates the region corresponding to a recombinant fragment used as an antigen. The region encoded by the cDNA probe used for Northern blotting is indicated as Probe.
Utrophin (395 kDa) is an autosomal homologue of dystrophin encoded by a 13-kb transcript (3). This protein is expressed in a variety of tissues (4), whereas dystrophin is primarily expressed in muscle. The NH2- and COOH-terminal regions of utrophin share considerable amino acid sequence homology with the actin binding and dystroglycan-binding domains of dystrophin, respectively (see Fig. 1) (5). This structural conservation suggests that utrophin forms a part of a similar cross-bridge to the dystrophin axis (1). The “utrophin axis” may mechanically protect the cell membrane, as does the dystrophin axis. In nonmuscle cells, in addition to utrophin, short dystrophin isoforms having molecular masses of 260 kDa (Dp260), 140 kDa (Dp140), 116 kDa (Dp116), and 70–80 kDa (Dp71 or apodystrophin-1) are translated from transcripts derived from the 3′ end of the dystrophin gene (2, 6–9). These isoforms commonly contain the domains involved in binding β-dystroglycan and syntrophins, but lack the domain for actin binding (see Fig. 1). Thus, the small isoforms of dystrophin do not fix the actin cytoskeleton to extracellular matrix and must play different roles from those of full-length-type dystrophin and utrophin.
In the brain, full-length-type as well as the short-type isoforms of dystrophin are expressed (4, 10–12). Full-length utrophin and its short-type isoform (G-utrophin; 113 kDa) are also transcribed from the utrophin gene locus (13). In this study we focused on astrocytes, which are the most numerous cell type in the central nervous system, and analyzed the expression of the dystrophin and utrophin isoforms. Treatment of astrocytes in vitro with dibutyryl-cAMP (dBcAMP) causes dramatic morphological change accompanied by the reorganization of the actin cytoskeleton (14). We show that full-length-type dystrophin, utrophin, and Dp71 were coexpressed in cultured rat brain astrocytes and that their expression concomitantly changes during dBcAMP-induced morphological differentiation of astrocytes. We also indicate changes in the distribution patterns of these molecules on differentiation.
MATERIALS AND METHODS
Cell Culture.
Astrocytes were obtained from the cerebrum of a Wistar rat embryo (18-day embryo) by using the procedure of Hama et al. (15). Astrocytes were cultured in DMEM (GIBCO/BRL) supplemented with 10% fetal calf serum. The cells were used for stimulation with dBcAMP after the third passage. At confluency, about 95% of the cells in the cultures were found to be positive for glial fibrillary acidic protein (GFAP), a differentiation marker of astrocytes (16), by immunostaining with anti-GFAP antibody (GA-5; Sigma). The medium was then changed to DMEM supplemented with 1 mM dBcAMP and 1% horse serum (HS), and cells were cultured for 1–6 days.
Reverse Transcription (RT)–PCR.
Poly(A)-rich RNA was prepared by using a QuickPrep mRNA purification kit (Pharmacia) from rat astrocytes cultured in the absence of dBcAMP.
PCR amplification of Dp71 cDNA.
RT reactions were performed with 10 ng of the poly(A)-rich RNA using a 26-mer oligonucleotide reverse primer complementary to a sequence in the 3′-untranslated region of mouse dystrophin mRNA [5′- TGTCTAATCCTCTTTGTTGTACGAAT-3′, nucleotide position 11408–11383 (17)]. The resulting single-stranded DNA was used as a template for PCR amplification. The amplification was carried out using LA-Taq (Takara Shuzo, Kyoto) for 30 cycles, each cycle consisting of 94°C for 1 min and 65°C for 5 min, the RT primer that was used for the above single-strand cDNA synthesis, and a forward primer corresponding to a sequence in the first exon of the rat Dp71 mRNA [5′-GCCCTCCCCTGACTGCCTGTGAAATC-3′ nucleotide position 56–81 (18)]. The amplification products (2 kbp) contained a sequence identical to that of the first exon for rat Dp71 (18).
PCR amplification of utrophin cDNA.
RT reactions were performed with 300 ng of the poly(A)-rich RNA using a 24-mer oligonucleotide reverse primer complementary to a sequence in the 3′-region of human utrophin mRNA. [5′-AAACGTGCTGTGAATCTGCTCCAT-3′, nucleotide position 10257–10234 (5)]. The resulting single-stranded DNA was used as template for PCR amplification. The amplification was carried out under the same conditions described above except that a forward primer corresponding to nucleotide position 8476–8499 of human utrophin mRNA (5′-AAAGTGCCCTATTACATCAACCAT-3′) was used. The amplified utrophin fragment (1.8 kbp) was cloned into the pCRII (Invitrogen) vector (pCR-UTR) and was confirmed to encode rat utrophin by DNA sequencing of its 5′-terminal region.
Northern Blotting.
Five micrograms of poly(A)-rich RNA from rat cultured astrocytes was separated by formaldehyde/1.1% agarose gel electrophoresis and transferred onto Hybond N membranes (Amersham). Rat Dp71 PCR products (2 kbp) and an EcoRI fragment (2 kbp utrophin fragment) from pCR-UTR were used as probes. These probes were labeled with [α-32P]dCTP by the random-priming method. The membranes in the hybridization solution (ExpressHyb; CLONTECH) were hybridized with the probes under high stringency conditions according to the manufacturer’s protocol, and washed in 0.1× SSC containing 0.1% SDS at 50°C. The hybridization signals were detected by use of a BAS2000 image analyzer (Fujifilm).
Antibodies.
Rabbit antiserum to utrophin, UT-2, was raised against a recombinant human utrophin fragment [aa position 1768–2078 (5)], which shows very low amino acid sequence identity (20%) with human dystrophin. A utrophin cDNA fragment (0.9 kbp) was amplified from a human skeletal muscle cDNA library (CLONTECH) by PCR using the following oligonucleotide primer set in which each primer contains a BamHI site (underlined): 5′-GGATCCACTGGTGTGCCTTTCTCTGAC-3′ (nucleotide position 5302–5322); 5′-GGATCCTTGGCTGCCTATCCTTCACTT-3′ (nucleotide position 6232–6212). The amplification was carried out using LA-Taq for 35 cycles, each cycle consisting of 94°C for 1 min, 55°C for 1 min, and 72°C for 3 min. The amplified utrophin DNA fragment was subcloned into the pCRII vector and the clone (pCR-UTH) was confirmed to be human utrophin cDNA by sequencing. Then, the BamHI fragment of pCR-UTH was ligated into the pGEX2TK vector (Pharmacia). Recombinant protein was expressed as a fusion protein with glutathione S-transferase (GST) in Escherichia coli and purified from the soluble fraction of cell lysates on a glutathione-Sepharose column as described by Suzuki et al. (19). GST was removed from the recombinant proteins by digestion with thrombin. The anti-utrophin antiserum UT-2 stained a 400-kDa protein band on an immunoblot of the rat lung homogenate, but not any other band on that of rat skeletal muscle homogenate, indicating that UT-2 is specific for utrophin.
In addition to the antiserum against utrophin (UT-2) described above, the antibodies used were MANDRA1 (20) (Sigma), MANDYS8 (21) (Sigma), NCL-DYS1 (Dy4/6D3) (NovoCastra, Newcastle, U.K.) (22) (NovoCastra), NCL-DRP1 (NovoCastra), NCL-43-DAG (anti-β-dystroglycan) (NovoCastra), PA3a (rabbit anti-β-dystroglycan antibody) (6), and GA-5 (anti-GFAP).
Immunoblotting and Quantitative Analysis.
Astrocytes cultured on a 3.5-cm dish were washed three times with Tris-buffered saline (pH 7.4) and then lysed in 400 μl of preheated solution A containing 100 mM Tris⋅HCl (pH 6.8), 1% SDS, 0.5% 2-mercaptoethanol. The cell lysates were electrophoresed on SDS/PAGE (4.5–15% polyacrylamide gel) and were transferred to a polyvinylidene difluoride (PVDF) membrane (Immobilon; Millipore). The PVDF membrane was sequentially treated with primary antibodies and horseradish peroxidase-conjugated secondary antibody. Immunoreactive protein bands were detected with the enhanced chemiluminescence (ECL) system and Hyperfilm ECL (Amersham).
Astrocytes cultured in DMEM supplemented with 1% HS and 1 mM dBcAMP at day 0, 1, 2, 4, 6, and 6-day culture in control medium (+1% HS without dBcAMP) were lysed in preheated solution A. Protein concentrations of the lysates were determined by protein assay (Bio-Rad). Equal amounts of total protein isolated from cells at each time point were used for immunoblotting. Utrophin and β-dystroglycan were detected with UT-2 and NCL-43DAG as 400-kDa and 43-kDa bands, respectively. Dystrophin and Dp71 were detected with MANDRA1 as 400-kDa and 75-kDa bands, respectively. These immunoreactive bands were quantified on BioImage Gel Print 2000i/VGA and Advanced Quantifier (BioImage, Ann Arbor, MI). For quantification of the dystrophin, utrophin, and β-dystroglycan bands, 8 μg of lysate at each time point was analyzed. For Dp71 quantification, 2 μg of the lysates was analyzed. The analysis was performed within the range of proportionality of the Hyperfilm ECL.
Immunofluorescence Microscopy.
Coverslip cultures of rat brain astrocytes were fixed and permeabilized in cold acetone (−20°C) for 5 min. The cells were incubated overnight with primary antibodies at 4°C. The reacted antibodies were detected with fluorescein isothiocyanate (FITC)-conjugated or tetramethylrhodamine isothiocyanate (TRITC)-conjugated secondary antibodies. Confocal laser scanning microscopy was performed using a Axioplan 2 microscope (Zeiss) on MRC-1024 (Bio-Rad) (see Fig. 6 j–o) and LSM 510 system (Zeiss) (see Fig. 6 a–i).
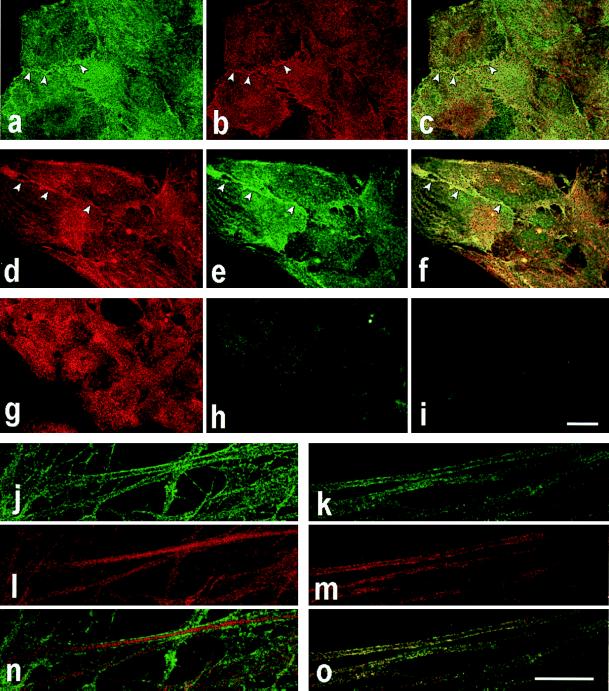
Figure 6.
Immunofluorescence staining of dystrophin, Dp71, and utrophin in cultured astrocytes. The rat brain astrocytes, cultured in DMEM supplemented with 1% HS for 8 days in the absence (a–i) and presence (j–o) of 1 mM dBcAMP, were stained. The undifferentiated polygonal cells were double-stained with anti-β-dystroglycan (NCL-43DAG) (a) and anti-utrophin (UT-2) (b), anti-β-dystroglycan (PA3a) (d) and anti-COOH-terminal region of dystrophin (MANDRA1) (e), anti-β-dystroglycan (PA3a) (g), and anti-full-length dystrophin (MANDYS8) (h). c and f show the merged images. Polyclonal (b, d, and g) and monoclonal (a, e, and h) antibodies are visualized with TRITC-labeled anti-rabbit (red) and FITC-labeled anti-mouse (green) secondary antibodies, respectively. i is the control of e and h, which was incubated with only FITC-labeled anti-mouse secondary antibody. Arrowheads indicate regions of cell-to-cell contact. Processes of the differentiated cells were double-stained with NCL-43DAG (j), and UT-2 (l), PA3a (k), and MANDRA1 (m). n and o show the merged images. NCL-43DAG and PA3a were visualized with FITC-labeled (green) anti-mouse and anti-rabbit antibody, respectively. UT-2 and MANDRA1 were visualized with TRITC-labeled (red) anti-rabbit and anti-mouse secondary antibody, respectively. (Bars = 20 μm.)
Other Procedures.
SDS/PAGE and protein transfer to the PVDF membrane were performed as described by Laemmli (23) and Kyhse-Anderson (24), respectively. The DNA sequencing was performed using an Applied Biosystems cycle-sequencing system (Perkin–Elmer).
RESULTS
Identification of Isoforms of Dystrophin and Utrophin Expressed in Cultured Rat Astrocytes.
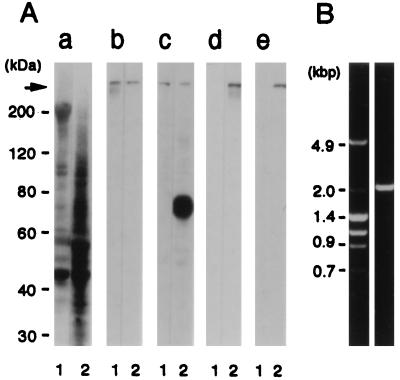
We examined the expression of dystrophin isoforms in cultured rat cerebral astrocytes by immunoblot analyses using two mAbs specific for dystrophin. The mAbs MANDRA1 and DYS1 were raised against the COOH-terminal (20) and mid-rod regions (22) of recombinant dystrophin protein, respectively (Fig. 1). On a blot of the homogenate of rat astrocytes, both antibodies stained a protein band, which migrated to the same position as that of the rat skeletal muscle dystrophin (427 kDa; Fig. 2A). This 427-kDa band was also detected with another mAb, MANDYS8 (21), which recognizes an epitope in the mid-rod region of dystrophin molecule (data not shown). In addition to the 427-kDa band, MANDRA1 detected a predominant 75-kDa band only on a blot of the astrocyte homogenate, suggesting that the protein band represented Dp71, a small-type isoform of dystrophin (6–9). To confirm that the 75-kDa protein is not a degradation product of full-length dystrophin but a translational product from the Dp71 transcript, RT–PCR analysis was performed to search for Dp71 mRNA. Template cDNA for the PCR was synthesized from rat cultured astrocyte RNA using a reverse primer complementary to a sequence in the 3′-untranslated region of mouse dystrophin mRNA. The PCR using the same reverse primer and a forward primer corresponding to a sequence in the first exon specific for the rat Dp71 mRNA revealed an expected 2-kb product (Fig. 2B). The product was identified as rat Dp71 cDNA by direct sequencing of its 5′- and 3′-terminal regions. We examined the expression of utrophin using the mAb DRP1 and rabbit antiserum (UT-2) specific for utrophin. Two anti-utrophin antibodies, DRP1 and UT-2, were raised against the last 11 amino acids at the COOH-terminus and a recombinant protein fragment (36 kDa) of the rod region of utrophin, respectively (Fig. 1). Both antibodies reacted only to a 400-kDa protein band on a blot of the astrocyte homogenate but not on that of skeletal muscle homogenate (Fig. 2A d and e). Taken together with the results from immunoblotting using anti-dystrophin antibodies, these results indicate that cultured rat brain astrocytes express two dystrophin isoforms–i.e., full-length dystrophin and Dp71, and full-length utrophin.
Figure 2.
Expression of dystrophin and utrophin isoforms in rat cultured astrocytes. (A) Twenty-four micrograms of total protein from rat cultured astrocytes (lanes 2) and eight micrograms of rat skeletal muscle homogenate (lanes 1) were separated on a SDS/polyacrylamide gel and transferred to PVDF membranes. The membranes were stained with Coomassie brilliant blue (panel a), DYS1 (panel b), MANDRA1 (panel c), UT-2 (panel d), and DRP1 (panel e). Arrow indicates the position of the 400-kDa band. (B) Detection of rat astrocyte Dp71 transcripts by RT–PCR. A reverse primer containing a sequence complementary to a sequence in the 3′-untranslated region of mouse dystrophin mRNA was used to reverse transcribe RNA isolated from cultured rat astrocytes. The single-strand cDNA was amplified by PCR using the same reverse primer and a forward primer located within the unique exon 1 of rat Dp71 mRNA (right lane). Size markers are indicated on the left lane.
Effect of dBcAMP on Expression of Dp71, Dystrophin, and Utrophin in Rat Cultured Astrocytes.
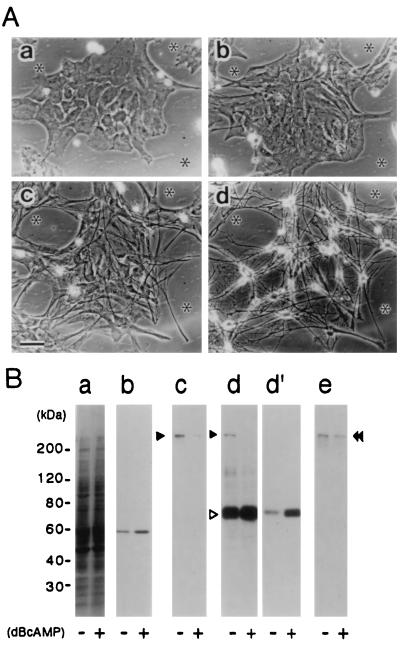
Elevation of intracellular cAMP levels in cultured astrocytes following treatment with dBcAMP causes transformation of the flat cell type into the stellate cell type (14, 25). The morphological transformation is accompanied by an increase in GFAP, suggesting that the dBcAMP-induced morphological transformation reflects the transition from immature astroblasts to more differentiated, mature cell types. As shown in Fig. 3A, in the presence of 1 mM dBcAMP and 1% HS, the flat polygonal astrocytes showed expansion and migration within 1–2 days. Almost all cells changed from flat type into fibrous cell type within 4 days. At day 6, the cells changed into process-bearing stellate shape, having small cell bodies and fine processes. The expression of GFAP in the dBcAMP-treated astrocytes (6-day culture) was approximately 2-fold higher than the basal level in control cells (Fig. 3B panel b).
Figure 3.
Time course of morphological differentiation and the expression of dystrophin isoforms in dBcAMP-treated astrocytes. (A) dBcAMP-induced morphological differentiation of rat astrocytes. Serial phase-contrast micrographs of rat brain astrocytes cultured in DMEM supplemented with 1 mM dBcAMP and 1% HS at day 0 (a), 2 (b), 4 (c), and 6 (d). Asterisks in each micrograph indicate the same positions on a culture dish. (Bar = 50 μm.) (B) Effect of dBcAMP on expression of dystrophin isoforms in astrocytes. Twenty micrograms of total proteins from astrocytes cultured for 6 days in the presence (+) or absence (−) of 1 mM dBcAMP were analyzed by immunoblotting (panels a, c, d, and e). The membranes were stained with Coomassie brilliant blue (panel a), anti-GFAP (G5A) (panel b), DYS1 (panel c), MANDRA1 (panels d and d′), and anti-utrophin rabbit antiserum (panel e). For panels b and d′, 0.2 and 2 μg of the total proteins were transferred to the membranes, respectively. Solid and open arrowheads indicate the bands representing full-length dystrophin and Dp71, respectively. Double arrowheads indicate the band representing utrophin.
Using this in vitro model for the differentiation of astrocytes, we examined the changes in the expression of full-length dystrophin, Dp71, and utrophin. Immunoblot analysis with DYS1 and MANDRA1 revealed a marked decrease in the expression of full-length dystrophin following treatment of the astrocytes with dBcAMP (Fig. 3B panels c and d). Utrophin expression was also reduced to some extent by this treatment, although the level of reduction was lower than that of the reduction of full-length dystrophin expression (Fig. 3B panel e). In contrast, the dBcAMP treatment induced a marked increase in the expression of Dp71 (Fig. 3B panels d and d′).
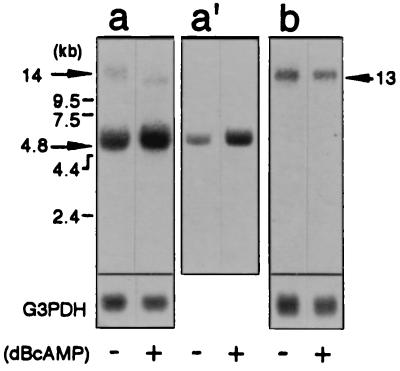
To elucidate whether dBcAMP affects the mRNA levels of full-length dystrophin, Dp71, and utrophin, we performed Northern blot analyses using cDNA probes of rat Dp71 and the COOH-terminal region (70 kDa) of utrophin (see Fig. 1). The Dp71 cDNA probe hybridized strongly to a 4.8-kb band on a blot of an RNA sample from astrocytes cultured in the absence of dBcAMP (Fig. 4). The probe also weakly hybridized to a band of 14 kb. On the other hand, the utrophin probe hybridized to a 13-kb band on a blot of RNA from the astrocytes cultured in the absence of dBcAMP. The sizes of the bands, 4.8 kb, 14 kb, and 13 kb, corresponded to those reported for Dp71 (4, 12), full-length dystrophin (10), and utrophin (3) mRNAs, respectively. Following culture in the presence of dBcAMP for 6 days, the level of 4.8-kb transcript in astrocytes increased (≈2-fold), whereas that of the 14-kb transcript was reduced to an undetectable level (Fig. 4 panels a and a′). These expression patterns of Dp71 and dystrophin gene transcripts coincided well with the results from immunoblot analyses (Fig. 3). Although the level of utrophin expression was reduced at protein level, its mRNA level was only slightly affected by the dBcAMP treatment (Fig. 4 panel b). It is noted that a weak 12-kb band hybridizing with the Dp71 cDNA probe was detected on a blot of RNA from the dBcAMP-treated astrocytes. The size of this band did not correspond to that of any of the other reported dystrophin isoforms.
Figure 4.
The effect of dBcAMP on dystrophin, Dp71, and utrophin mRNA expression in cultured astrocytes. Five micrograms of poly(A)-rich RNA from rat cultured astrocytes was separated by electrophoresis, transferred to a nylon membrane, and hybridized with 32P-labeled rat Dp71 (panels a and a′) PCR products and utrophin (panel b) cDNA. The hybridization signals (arrows) were visualized following autoradiography for 18 hr (panels a and b) and 2 hr (panel a′) by use of a BAS2000 image analyzer. RNA samples: astrocytes cultured for 6 days in the absence (−) and presence (+) of 1 mM dBcAMP. The filters used for panels a and a′ were the same. The membrane was rehybridized with a 32P-labeled human glyceraldehyde-3-phosphate dehydrogenase (G3PDH) cDNA probe to correct for the amount of RNA loaded in each lane.
Time Course of the Expression of Dystrophin, Dp71, Utrophin, and β-Dystroglycan During dBcAMP-Induced Astrocyte Differentiation.
Immunoblot analyses revealed that changes in the expression pattern of β-dystroglycan, as well as those of dystrophin isoforms and utrophin, took place as the morphological differentiation progressed (Figs. 3 and 5).
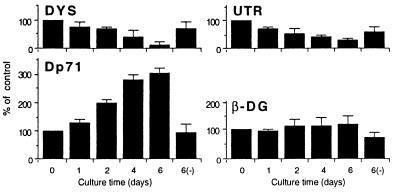
Figure 5.
The expression of dystrophin isoforms, utrophin and β-dystroglycan, during dBcAMP-induced astrocyte differentiation. The astrocytes cultured in DMEM supplemented with 1 mM dBcAMP and 1% HS at day 0, 1, 2, 4, and 6 were lysed in SDS solution. These lysates were used for quantitative analysis of full-length dystrophin (DYS), utrophin (UTR), Dp71, and β-dystroglycan (β-DG) as described. The protein level (mean ± SD from three independent experiments) at each time point was expressed as a percentage of the amount of protein compared with that on day 0 of culture. The astrocytes cultured for 6 days in control medium (+1% HS without dBcAMP) are indicated as 6(−).
The astrocytes 6 days after the treatment showed expression levels of utrophin and dystrophin that had decreased to 50% and 20%, respectively, of those of the day 0 culture. In contrast, the expression of Dp71 markedly increased to approximately 3-fold of the control level by 4–6 days of culture. The expression level of β-dystroglycan increased slightly (1.4 times the level of the control).
Distribution of Dystrophin, Utrophin, and Dp71 in Polygonal- and Stellate-Shaped Cultured Astrocytes.
Previous studies suggested that Dp71 and utrophin are localized beneath the plasma membrane as a result of anchoring to β-dystroglycan, as in the case of full-length dystrophin (26–30).
We examined the expression and distribution of full-length dystrophin, utrophin, and Dp71 in cultured rat astrocytes by confocal laser scanning microscopy. The antibodies against the rod region of utrophin (UT-2) and COOH-terminal of dystrophin (MANDRA1) stained all over its surface, especially the cell-to-cell contact region of undifferentiated polygonal astrocytes (Fig. 6 b and e), and the staining pattern was similar to the pattern with anti-β-dystroglycan (Fig. 6 a and d). The merged images indicated that most of utrophin and dystrophin/Dp71 colocalized with β-dystroglycan (Fig. 6 c and f). The MANDYS8, which recognizes only full-length dystrophin (see Fig. 1), weakly stained the astrocytes (Fig. 6h). Because the MANDYS8 clearly stained sarcolemma of skeletal muscle cells at the same level as that with MANDRA1 (data not shown), the weak signal of MANDYS8 indicated a very low level expression of full-length dystrophin in astrocytes. This is compatible with the finding of immunoblot analysis (Fig. 3B panels d and d′). This also indicated that most of the fluorescence signal of MANDRA1 presented the distribution of Dp71 (Fig. 6e). In differentiated astrocytes by dBcAMP treatment, MANDRA1 tended to stain the edges along foot processes and the staining was mostly overlapped with the signal of anti-β-dystroglycan antibody (Fig. 6 k, m, and o), whereas the prominent signal of UT-2 antibody was observed throughout the processes including the cytoplasmic area (Fig. 6 j, l, and n). Fluorescence signals for full-length dystrophin were not detected when the differentiated cells were stained with MANDYS8 (data not shown).
DISCUSSION
In this paper we have shown that a full-length dystrophin, utrophin, and a Dp71 dystrophin isoform are coexpressed in primary cultures of rat brain astrocytes. In primary undifferentiated polygonal astrocytes, full-length dystrophin, utrophin, and Dp71 were mostly localized all over the cell surface and the region of cell-to-cell contact where β-dystroglycan is located. Because the dystrophin, utrophin, and Dp71 bind to β-dystroglycan, it is likely that dystrophin and utrophin axes are formed to associate the cell membrane with submembraneous cytoskeletal networks in undifferentiated astrocytes.
On morphological differentiation induced by dBcAMP, Dp71 increased by about three times and was mostly present on the cell membrane where β-dystroglycan was localized. However, full-length dystrophin was greatly reduced and utrophin substantially decreased. Furthermore, utrophin was mostly found in cytoplasmic regions, suggesting that the amount of utrophin present on the cell membrane is greatly reduced. Indeed, there was less yellow area showing colocalization of utrophin and β-dystroglycan on the membranes of differentiated cells (Fig. 6n) compared with the area showing Dp71 and β-dystroglycan (Fig. 6o).
These findings suggest that in an undifferentiated state when cell shape is rather stable, cell membranes may be fairly well associated to the membranous cytoskeleton network by dystrophin and utrophin binding to β-dystroglycan. However, when morphological changes take place on differentiation (Fig. 3A), this association may prevent the cell from the new cell shape formation. Because Dp71 binds to β-dystroglycan (29, 30), but does not anchor to the submembraneous actin cytoskeleton, the replacement of utrophin axis to the “Dp71 axis” may allow the free morphological changes of astrocytes. Indeed, this is compatible with the fact that the transformation of astrocytes from polygonal flat type into stellate type is accompanied by changes in organization of the actin cytoskeleton (14).
In vivo, the Dp71 transcript is found in glial cells in the developing central nervous system (12), but not in adult brain (31). Presumably, in vivo, Dp71 expression is necessary for morphologically differentiating astrocytes, but not for the steady state of terminally differentiated astrocytes. In the adult brain, full-length utrophin is observed at the inner plasma face of astrocytic foot processes at the abluminal surface of the basal lamina surrounding the capillary endothelial cells (32). It is likely that the terminally differentiated astrocytes utrophin axis may become dominant, replacing the Dp71 axis to fix the cell structure. However, in vitro, we could only find the higher level of Dp71 expression and the reduced level of utrophin in differentiated astrocytes. This may be because cultured astrocytes cannot progress into the fully matured state by treatment with dBcAMP, i.e., the dBcAMP-treated astrocytes in culture are closer to reactive astrocytes than to matured astrocytes in vivo (33).
Among the three types of full-length dystrophin, i.e., brain-, Purkinje cell-, and muscle-type forms, the muscle-type dystrophin mRNA is predominantly expressed in the primary culture of mouse astrocytes (2, 34). We also confirmed the result by detection of a first exon unique for the muscle-type isoform by RT–PCR (data not shown). Thus, the astrocytic full-length dystrophin detected by immunoblotting may be a translational product from the muscle-type dystrophin transcript. The expression of the dystrophin was very low compared with that of Dp71 in undifferentiated astrocytes (Fig. 2A panel c). The dystrophin was no longer detected in differentiated astrocytes by immunocytochemical staining. This is consistent with the previous report that the full-length dystrophin could not be detected in astrocytes in vivo (11).
Acknowledgments
We thank Satoru Noguchi for reviewing the manuscript. This work was supported in part by Grant 8A-1 from the National Center of Neurology and Psychiatry of the Ministry of Health and Welfare, and by the Ministry of Education, Science and Culture of Japan.
ABBREVIATIONS
- dBcAMP
dibutyryl-cAMP
- RT
reverse transcription
- GFAP
glial fibrillary acidic protein
- HS
horse serum
- PVDF
polyvinylidene difluoride
- FITC
fluorescein isothiocyanate
- TRITC
tetramethylrhodamine isothiocyanate
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB011666).
References
- 1.Ozawa E, Yoshida M, Suzuki A, Mizuno Y, Hagiwara Y, Noguchi S. Hum Mol Genet. 1995;4:1711–1716. doi: 10.1093/hmg/4.suppl_1.1711. [DOI] [PubMed] [Google Scholar]
- 2.Sadoulet-Puccio H M, Kunkel L M. Brain Pathol. 1996;6:25–35. doi: 10.1111/j.1750-3639.1996.tb00780.x. [DOI] [PubMed] [Google Scholar]
- 3.Love D R, Hill D F, Dickson G, Spurr N K, Byth B C, Marsden R F, Walsh F S, Edwards Y H, Davies K E. Nature (London) 1989;339:55–58. doi: 10.1038/339055a0. [DOI] [PubMed] [Google Scholar]
- 4.Blake D J, Love D R, Tinsley J, Morris G E, Turley H, Gatter K, Dickson G, Edwards Y H, Davies K E. Hum Mol Genet. 1992;1:103–109. doi: 10.1093/hmg/1.2.103. [DOI] [PubMed] [Google Scholar]
- 5.Tinsley J M, Blake D J, Roche A, Fairbrother U, Riss J, Byth B C, Knight A E, Kendrick-Jones J, Suthers G K, Love D R, Edwards Y H, Davies K E. Nature (London) 1992;360:591–593. doi: 10.1038/360591a0. [DOI] [PubMed] [Google Scholar]
- 6.Mizuno Y, Yoshida M, Yamamoto H, Hirai S, Ozawa E. J Biochem. 1993;114:936–941. doi: 10.1093/oxfordjournals.jbchem.a124281. [DOI] [PubMed] [Google Scholar]
- 7.Bar S, Barnea E, Levy Z, Neuman S, Yaffe D, Nudel U. Biochem J. 1990;272:557–560. doi: 10.1042/bj2720557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lederfein D, Levy Z, Augier N, Mornet D, Morris G, Fuchs O, Yaffe D, Nudel U. Proc Natl Acad Sci USA. 1992;89:5346–5350. doi: 10.1073/pnas.89.12.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hugnot J P, Gilgenkrantz H, Vincent N, Chafey P, Morris G E, Monaco A P, Berwald-Netter Y, Koulakoff A, Kaplan J C, Kahn A, Chelly J. Proc Natl Acad Sci USA. 1992;89:7506–7510. doi: 10.1073/pnas.89.16.7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chamberlain J S, Pearlman J A, Yuzny D M, Gibbs R A, Ranier J E, Reeves A A, Caskey T C. Science. 1988;239:1416–1418. doi: 10.1126/science.3347839. [DOI] [PubMed] [Google Scholar]
- 11.Lidov H G W, Byers T J, Kunkel L M. Neuroscience. 1993;54:167–187. doi: 10.1016/0306-4522(93)90392-s. [DOI] [PubMed] [Google Scholar]
- 12.Schofield J N, Blake D J, Simmons C, Morris G E, Tinsley J M, Davies K E, Edwards Y H. Hum Mol Genet. 1994;8:1309–1316. doi: 10.1093/hmg/3.8.1309. [DOI] [PubMed] [Google Scholar]
- 13.Blake D J, Schofield J N, Zuellig R A, Gorecki D C, Phelps S R, Barnard E A, Edwards Y H, Davis K E. Proc Natl Acad Sci USA. 1995;92:3697–3701. doi: 10.1073/pnas.92.9.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciesielski-Treska J, Guerold B, Aunis D. Neuroscience. 1982;7:509–522. doi: 10.1016/0306-4522(82)90284-6. [DOI] [PubMed] [Google Scholar]
- 15.Hama H, Sakurai T, Kasuya Y, Fujiki M, Masaki T, Goto K. Biochem Biophys Res Commun. 1992;186:355–362. doi: 10.1016/s0006-291x(05)80815-0. [DOI] [PubMed] [Google Scholar]
- 16.Le Prince G, Fages C, Rolland B, Nunez J, Tardy M. Glia. 1991;4:322–326. doi: 10.1002/glia.440040310. [DOI] [PubMed] [Google Scholar]
- 17.Koenig M, Hoffman E P, Bertelson C J, Monaco A P, Feener C, Kunkel L M. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 18.Rapaport D, Lederfein D, den Dunnen J T, Grootscholten P M, Van Ommen G-J B, Fuchs O, Nudel U, Yaffe D. Differentiation. 1992;49:187–193. doi: 10.1111/j.1432-0436.1992.tb00666.x. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki A, Yoshida M, Ozawa E. J Cell Biol. 1995;128:373–381. doi: 10.1083/jcb.128.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen T M, Ginjaar I B, Van Ommen G-J B, Morris G E. Biochem J. 1992;288:663–668. doi: 10.1042/bj2880663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sedgwick S G, Nguyen T M, Ellis J M, Crowne H, Morris G E. Nucleic Acids Res. 1991;7:509–522. doi: 10.1093/nar/19.21.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholson L V B, Johnson M A, Davison K, O’Donnell E, Falkous G, Barron M, Harris J B. Acta Neurol Scand. 1992;86:8–14. doi: 10.1111/j.1600-0404.1992.tb08046.x. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Nature (London) 1979;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Kyhse-Anderson J. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- 25.Shain W, Forman D S, Madelian V, Turner J N. J Cell Biol. 1987;105:2307–2314. doi: 10.1083/jcb.105.5.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumura K, Ervasti J M, Ohlendieck K, Kahl S D, Campbell K P. Nature (London) 1992;360:588–591. doi: 10.1038/360588a0. [DOI] [PubMed] [Google Scholar]
- 27.James M, Nguyen T M, Wise C J, Jones G E, Morris G E. Cell Motil Cytoskeleton. 1996;33:163–174. doi: 10.1002/(SICI)1097-0169(1996)33:3<163::AID-CM1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 28.Rapaport D, Greenberg D S, Tal M, Yaffe D, Nudel U. FEBS Lett. 1993;328:197–202. doi: 10.1016/0014-5793(93)80992-4. [DOI] [PubMed] [Google Scholar]
- 29.Jung D, Yang B, Meyer J, Chamberlain J S, Campbell K P. J Biol Chem. 1995;270:27305–27310. doi: 10.1074/jbc.270.45.27305. [DOI] [PubMed] [Google Scholar]
- 30.Rosa G, Cecarini M, Cavaldesi M, Zini M, Petrucci T C. Biochem Biophys Res Commun. 1996;223:272–277. doi: 10.1006/bbrc.1996.0883. [DOI] [PubMed] [Google Scholar]
- 31.Gorecki D C, Barnard E A. NeuroReport. 1995;6:893–896. doi: 10.1097/00001756-199504190-00017. [DOI] [PubMed] [Google Scholar]
- 32.Khurana T S, Watkins S C, Kunkel L M. J Cell Biol. 1992;119:357–366. doi: 10.1083/jcb.119.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fedoroff S, McAuley W A J, Houle J D, Devon R M. J Neurosci Res. 1984;12:15–27. doi: 10.1002/jnr.490120103. [DOI] [PubMed] [Google Scholar]
- 34.Chelly J, Hamard G, Koulakoff A, Kaplan J-C, Kahn A, Berwald-Netter Y. Nature (London) 1990;344:64–65. doi: 10.1038/344064a0. [DOI] [PubMed] [Google Scholar]