Abstract
In Nature, bacteria rarely exist as single, isolated entities, but rather as communities comprised of many other species including higher host organisms. To survive in these competitive environments, microorganisms have developed elaborate tactics such as the formation of biofilms and the production of antimicrobial toxins. Recently, it was discovered that the Gram-negative bacterium Pseudomonas aeruginosa, an opportunistic human pathogen, produces an antibiotic, 3-(1-hydroxydecylidene)-5-(2-hydroxyethyl)pyrrolidine-2,4-dione (C12-TA), derived from one of its quorum sensing molecules. Here, we present a comprehensive study of the expanded spectrum of C12-TA antibacterial activity against microbial competitors encountered by P. aeruginosa in Nature as well as significant human pathogens. The mechanism of action of C12-TA was also elucidated and C12-TA was found to dissipate both the membrane potential and pH gradient of Gram-positive bacteria, correlating well with cell death. Notably, in stark contrast to its parent molecule 3-oxo-dodecanoyl homoserine lactone (3-oxo-C12-HSL), neither activation of cellular stress pathways nor cytotoxicity was observed in human cells treated with C12-TA. Our results suggest that the QS machinery of P. aeruginosa has evolved for a dual-function, both to signal others of the same species, and also to defend against both host immunity and competing bacteria. Because of the broad-spectrum antibacterial activity, established mode of action, lack of rapid resistance development, and tolerance by human cells, the C12-TA scaffold may also serve as a new lead compound for the development of antimicrobial therapeutics.
Introduction
To exist in a world with limited resources, microorganisms have developed elegant survival mechanisms that often act at the expense of other competitors, including bacteria. These tactics encompass highly specialized iron uptake systems, the formation of sessile communities known as biofilms, and the production of toxins against both microbial organisms and eukaryotic hosts. By harnessing the conflict that occurs among these microorganisms, however, a plethora of new antimicrobial therapeutics has been discovered via the exploitation of the chemical agents used in this bacterial warfare. In fact, natural products directly from bacterial sources or their semisynthetic derivatives account for the majority of currently employed antibiotics, and, as a result of the rich environmental diversity inhabited by microorganisms, comprise a vast structural and mechanistic landscape.1 One class of natural products that has attracted considerable attention is the tetramic acids and their derivatives, owing to their structural complexity and diversity as well as their broad spectrum of biological activity.2
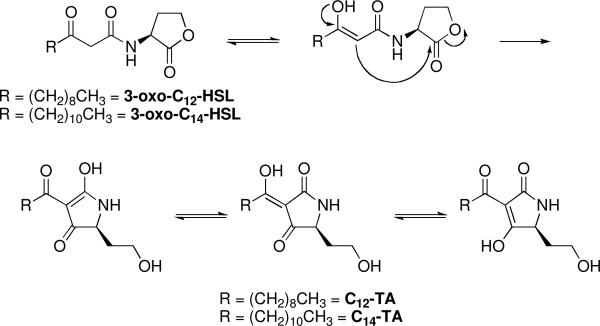
Tetramic acids (TAs) are a class of compounds containing a 2,4-pyrrolidinedione ring system with biological activities ranging from antibacterial and antiviral to mycotoxic, fungicidal, as well as anti-cancer properties.2 Recently, during our efforts to generate monoclonal antibodies against bacterial acyl homoserine lactones, we uncovered the conversion of 3-oxo-dodecaonyl homoserine lactone (3-oxo-C12-HSL), an intercellular signaling molecule used by P. aeruginosa, to the tetramic acid 3-(1-hydroxydecylidene)-5-(2-hydroxyethyl)pyrrolidine-2,4-dione (C12-TA) through an irreversible, nonenzymatic Claisen-like reaction (Figure 1).3,4 Both this tetramic acid and its parent homoserine lactone exhibited antibacterial activity against selected Gram-positive strains, with C12-TA exhibiting significantly more potent antibacterial activity than 3-oxo-C12-HSL.4
Figure 1.
Conversion of 3-oxo-AHLs to the corresponding TA compounds.
P. aeruginosa is a common environmental Gram-negative bacterium that has become an opportunistic human pathogen because of the ability to take advantage of weaknesses in the host immune system. The intercellular communication mediated by 3-oxo-C12-HSL, a process known as quorum sensing (QS), is crucial to the pathogenicity of P. aeruginosa, controlling such functions as virulence factor production, iron acquisition, and biofilm formation.5,6 However, recent reports have suggested that this signal imparts benefits beyond communication, as mutants arise during chronic P. aeruginosa infections that exhibit a loss of function in the gene encoding the 3-oxo-C12-HSL receptor, while the HSL synthase remains active.5,7-12 With this in mind, we hypothesized that 3-oxo-C12-HSL, via the action of C12-TA, might be used by P. aeruginosa as a strategy to hamper encroachment by competing bacteria;4 herein, we define the antimicrobial properties of tetramic acids to include several relevant human pathogens and known natural competitors of P. aeruginosa. We also demonstrate the failure of S. aureus to develop resistance to C12-TA after 20 passages, as well as the lack of cytotoxicity against human cells. Moreover, the mode of action of C12-TA was elucidated by focusing on four Gram-positive bacteria: Bacillus cereus, Bacillus subtilis, Enterococcus faecalis, and Staphylococcus aureus, each showing varying degrees of susceptibility towards C12-TA.
RESULTS
C12-TA is cytotoxic against bacterial competitors and clinically relevant pathogens
To elucidate the antibacterial activity of 3-oxo-AHL derived tetramic acids, we examined the activity of C12- and C14-TA against several bacteria, both Gram-positive and -negative species, potentially encountered by P. aeruginosa in natural environments as well as several important human pathogens. Although the C14-AHL is not one of the major signals of P. aeruginosa, we included C14-TA for comparison, and in each case, the minimum inhibitory concentration (MIC) of C14-TA is slightly less than that of C12-TA, likely due to the increased hydrophobicity of the longer acyl chain allowing for better membrane partitioning or cell penetration. Especially noteworthy is the activity of C12- and C14-TA against Bacillus anthracis and the community acquired methicillin resistant strain of Staphylococcus aureus (a USA-300 clone). Additionally, both C12- and C14-TA exhibit potent activity against Mycobacterium tuberculosis, the causative agent of tuberculosis (Table 1). As a result, these data give credence to our proposition of C12-TA as a new antibiotic scaffold for future drug development efforts. M. tuberculosis and Corynebacterium diphtheriae represent members of the phylum Actinobacteria, whereas the other Gram-positive bacteria previously examined are members of the phylum Firmicutes. Members of these two phyla differ based on their DNA composition, but all share a common feature in the absence of an outer membrane. This fundamental difference between Gram-negative and –positive bacteria is likely the origin of the selective cytotoxicity of C12-TA. Gram- negative bacteria possess an outer membrane composed of lipopolysaccharide (LPS) or -oligosaccharide (LOS), phospholipids, and proteins, providing a permeability barrier that is absent in Gram-positive bacteria, in which the bacterial cell wall is composed of a thicker peptidoglycan layer.4
Table 1.
Antibacterial Activity of C12- and C14-TA
| Bacteria | C12-TA EC50, μg/mL (μM) | C12-TA MIC, μg/mL | C14-TA MIC, μg/mL |
|---|---|---|---|
| Gram-positive | |||
| B. anthracis Ames strain | n.d. | 54 | n.d. |
| B. cereus PCI 213 | 2.4 (8.3) | 12.5 | 3.12 |
| B. subtilis Marburg | n.d. | 25 | 3.12 |
| C. diphtheriae 48255 | 8.9 (30.1) | 25 | 6.25 |
| E. faecalis NCTC 775 | 10.0 (33.7) | 50 | 6.25 |
| L. plantarum NCIMB 8826 | n.d. | 25 | 12.5 |
| M. tuberculosis H37Rv | 0.71 (2.38) | 5 | 2.5 |
| S. aureus USA-300 | n.d. | 25 | 6.25 |
| S. aureus Wood 46 | 8.0 (26.7) | 25 | 6.25 |
| Gram-negative | |||
| B. cepacia UCB 717 | >30 (>100) | n.d. | n.d. |
| E. coli K12 | >30 (>100) | n.d. | n.d. |
| E. coli D22 | 10.7 (35.9) | 50 | 25 |
| E. coli Δimp | 5.7 (19.2) | 25 | 12.5 |
| H. influenzae AMC 36-A-1 | 23.4 (78.5) | n.d. | n.d. |
| P. aeruginosa PAO-1 | >30 (>100) | n.d. | n.d. |
| V. cholerae N16961 | 11.2 (37.7) | >100 | >100 |
| V. cholerae TP | 9.1 (30.4) | 100 | 100 |
| V. harveyi BB120 | 21.6 (72.6) | 100 | 6.25 |
n.d. = not determined
Although there is a general trend of insensitivity of Gram-negative strains to C12-TA, our studies revealed that Vibrio spp., including two strains of the important human pathogen V. cholerae,13 are in fact susceptible to C12-TA. The reasoning for this may lie in the difference in LPS glycosylation pattern in the outer membrane of Vibrio spp. compared to other Gram-negative bacteria. The outer membrane of V. cholerae has also been shown to be particularly permeable to hydrophobic compounds compared to other Gram-negative strains.14 Likewise, Haemophilus influenzae, which has an outer membrane composed of lipooligosaccharide units rather than the longer LPS units of other Gram-negative bacteria,15 was found to be sensitive to C12-TA.
To uncover how the outer membrane modulates Gram-negative bacterial resistance towards C12-TA, we examined two LPS defective E. coli strains: D22 and a Δimp (increased membrane permeability gene) mutant.16,17 The D22 strain, which possesses an envA mutation resulting in impaired lipid A biosynthesis, and the Δimp mutant, which exhibits abnormal assembly of LPS at the cell surface, both have compromised outer membranes as a result of these defects in LPS biogenesis.18,19 Indeed, these two strains have been used to monitor the effects of detergents and other hydrophobic organic compounds normally ineffective against Gram-negative bacteria.20,21 We reasoned that they would be sensitive to the antibacterial effects of C12- and C14-TA, and this was found to be the case. Because of the differential activity between Gram-positive and -negative strains, as well as its effectiveness against Vibrio spp. and membrane-defective E. coli strains, we predicted the bacterial membrane as the target of antibacterial activity of C12-TA.
C12-TA interferes with the bacterial proton gradient and membrane potential
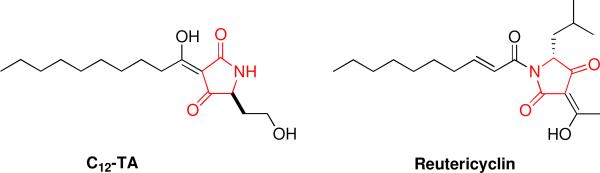
Naturally occurring tetramic acid derivatives are of great interest because of their broad spectrum of antibacterial activity. Interestingly, a structurally related tetramic acid antibiotic, reutericyclin (Figure 2), has been shown to act as an ionophore against Gram-positive bacteria.22,23 Reutericyclin selectively imports protons into Lactobacilli spp., thereby eliminating the proton motive force (PMF) of the cell through dissipation of the transmembrane proton gradient (ΔpH), ultimately culminating in cell death. The PMF is vital to cell survival, as it is used to generate energy for functions such as ATP synthesis and solute transport. Bacteria ultimately use two electrochemical gradients to maintain the PMF: the membrane potential (ΔΨ) and the proton gradient (ΔpH), and, accordingly, we evaluated the mechanism of action of C12-TA by assessing its effects on these two membrane gradients.
Figure 2.
Structures of C12-TA and reutericyclin. The 2,4-pyrrolidinedione ring system characteristic of TA compounds is shown in red.
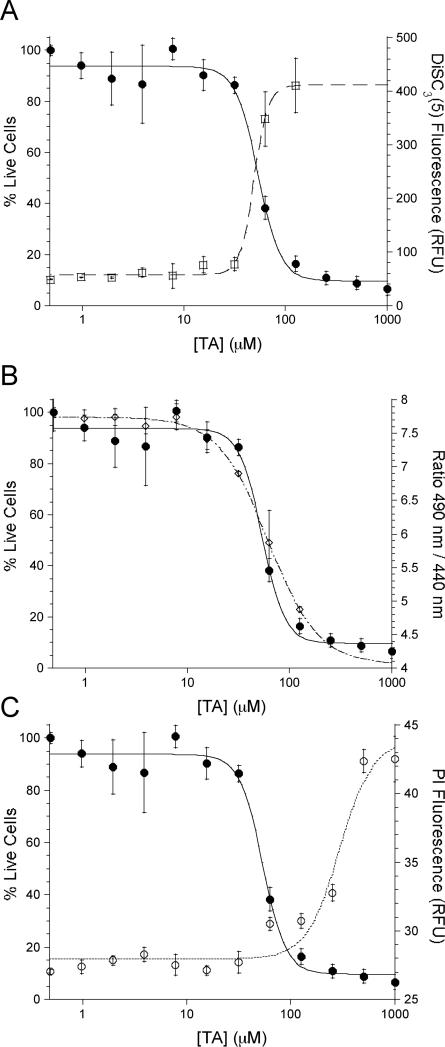
The capacity of C12-TA to disrupt the ΔΨ was measured using the fluorescent probe DiSC3(5). The cationic nature of DiSC3(5) allows the dye to accumulate inside cells with polarized membranes, where its fluorescence is quenched by aggregation with other DiSC3(5) molecules. Upon depolarization of the membrane, the probe is released and fluorescence is restored; that is, an increase in emission corresponds to a depolarization of the membrane. Treatment of DiSC3(5)-loaded B. cereus with C12-TA resulted in a dose-dependent increase in fluorescence with an EC50 value of 49.0 ± 2.0 μM (Figure 3A). To correlate membrane depolarization with cytotoxicity, the effects of C12-TA on cultures of B. cereus (OD600 = 0.5) were determined and dose-dependent cytotoxicity was observed with an EC50 of 53.6 ± 3.7 μM (Figure 3A). ΔΨ is maintained due in large part to the maintenance of K+ gradients across the cell membrane,24 and indeed, when B. cereus was incubated with C12-TA and varying concentrations of KCl in HEPES buffer, the extent of depolarization varied with the concentration of K+. When performed in K+-free HEPES buffer, ΔΨ was still diminished, implicating the transport of other ions across the membrane by C12-TA (data not shown).
Figure 3.
Comparison between death of B. cereus and the membrane effects elicited by C12-TA. (A.) Correlation between cell death and the dissipation of ΔΨ (□) and (B.) ΔpH (◇). (C.) Cell death (•,Y1) does not occur at concentrations needed for membrane permeabilization (○, Y2).
Another ion used in the maintenance of the PMF is the proton, which is responsible for the chemical gradient across the membrane (ΔpH). The effect of C12-TA on the transmembrane ΔpH was measured using the fluorescent probe cFDA-SE, which is internalized by the bacteria and becomes sensitive to fluctuations in pH. Figure 3B shows the decrease of the internal pH (open symbols), and thus the ΔpH of B. cereus, with increasing concentrations of C12-TA (EC50 = 61.6 ± 8.7 μM). This value also corresponds closely to the EC50 for cell death and implicates the loss of this gradient in the death of the cell. Similar results also were observed for three other Gram-positive bacteria: B. subtilis, Enterococcus faecalis, and S. aureus (Table 2). In the case of S. aureus and E. faecalis, the effect on ΔpH was observed at lower concentrations than that required for cell death, which may be indicative of a lower contribution of the ΔpH to the overall PMF under these growth conditions.
Table 2.
Activity of C12-TA in Gram-positive bacteria. All EC50 values are reported in μM.
| Bacteria | Death | ΔpH | Δψ | Membrane Permeation |
|---|---|---|---|---|
| B. cereus PCI 213 | 53.6±3.7 | 61.6±8.7 | 49.0±2.0 | 293±48.0 |
| B. subtilis Marburg | 64.5±1.9 | 60.3±16.2 | 23.9±5.1 | 110±12.6 |
| E. faecalis NCTC 775 | 116±26.2 | 17.5±2.8 | 121±6.3 | ~875a |
| S. aureus Wood 46 | 294±33.5 | 15.0±3.0 | 186±6.2 | ~575a |
Insufficient data to calculate an accurate EC50
It is worth noting that the EC50 values reported for the PMF disruption are higher than the EC50 values reported for growth inhibition in Table 1. This increase in values can be attributed to the difference in the two assays, in that the values in Table 1 represent growth inhibition, whereas the values measured in the mechanistic studies represent the concentration needed to kill bacterial cultures at an OD600 of 0.5. Thus, the lethal concentrations needed to kill dense cultures were evaluated for a more direct comparison of cell death to the dissipation of ΔpH and ΔΨ. Furthermore, the concentrations of C12-TA needed for dissipation of ΔpH and ΔΨ correlate well with the MIC values, suggesting membrane depolarization as the mechanism of action, rather than an artifact of artificially high antibiotic concentrations.
Based on our experimental data, the hydrophobicity of C12-TA, and the knowledge that lipid substituted antibiotics are known to target the membranes of Gram-positive bacteria,25 we reasoned that our observations may be due to a detergent-like general disintegration of the membrane as the underlying mode of action to account for C12-TA cytotoxicity. The ability of C12-TA to permeabilize the cell membrane was measured by incubating log phase cultures with propidium iodide (PI) and C12-TA. PI, which is unable to penetrate intact membranes, will enter cells with permeabilized membranes and bind DNA to emit a fluorescent signal. Fluorescence was observed in B. cereus with an EC50 of 293 ± 48.0 μM C12-TA (Figure 3C): the high concentration needed to observe PI fluorescence indicates that membrane permeabilization is not involved in cell death, but is instead more likely a secondary effect of bacterial death. This reasoning was corroborated by measuring the ability of C12-TA to kill similarly dense cultures of B. cereus, which occurred with an EC50 of 53.6 ± 3.7 μM (Figure 3).
If the mechanism is indeed the nonspecific shuttling of ions across the bacterial membrane, then there should be no difference in the MIC values of the natural (S)-C12-TA and the unnatural R form. Towards this end, we synthesized (R)-C12-TA as described previously4 and evaluated its antibacterial activity against B. cereus, S. aureus, and C. diphtheriae. For all three species examined, there was no difference between the MIC values of (R)-C12-TA and (S)-C12-TA. This finding supports a nonspecific interaction, such as that suggested by the membrane depolarization data, rather than interaction of the TA compounds with an unknown specific receptor.
S. aureus does not develop resistance to C12-TA
To study the development of resistance to C12-TA, we monitored S. aureus growth in the presence of subinhibitory concentrations of C12-TA. These studies focused on S. aureus due to its relevance in human disease and the fact that it is a known competitor of P. aeruginosa in the lungs of CF patients. Furthermore, S. aureus is the pathogen most commonly associated with antibacterial resistance, and to date it has developed resistance to every antibiotic used to treat its infections.26 Cultures of S. aureus were incubated for 24 h in the presence of varying concentrations of C12-TA, and the culture just below the MIC was taken and used for subculturing in the fresh growth medium the same range of concentrations of C12-TA as before. The MIC was recorded after each passage, and, after 20 passages, there was no observed increased in the MIC of C12-TA or C14-TA (Table 3).
Table 3.
Onset of resistance of S. aureus Wood 46 to C12- and C14-TA.
| Passage | C12-TA MIC (μg/mL) | C14-TA MIC (μg/mL) |
|---|---|---|
| 0 | 25 | 6.25 |
| 20 | 25 | 6.25 |
To gain further insight into the activity of C12-TA against clinically relevant scenarios of bacterial infection, we also measured the capacity of C12-TA to disrupt biofilms of S. aureus. However, C12-TA did not exhibit activity in this regard (Figure S1).
C12- and C14-TA are not toxic to human cells
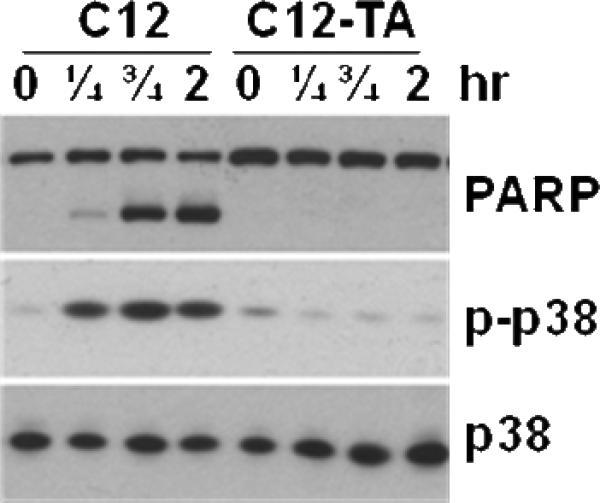
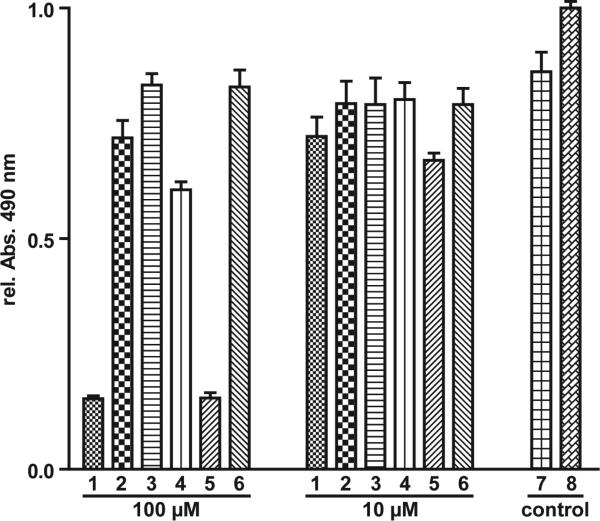
Collectively, these microbiological data suggest the use of tetramic acid-derived compounds as therapeutic agents against bacterial infections. As an initial step in this direction, the tetramic acids were evaluated for the ability to induce apoptosis, as well as general cytotoxic effects, in bone marrow-derived macrophage (BMDM) cells. We have previously shown that, upon 3-oxo-C12-HSL stimulation, macrophages activate several intracellular signaling events, including pro- and anti-apoptotic pathways. For example, activation of the anti-apoptotic p38 mitogen-activated protein kinase (MAPK) is induced via the phosphorylation of p38 kinase domain.27,28 In turn, the pro-apoptotic effects of 3-oxo-C12-HSL are mediated through the activation of caspase-9 and caspase-3, and subsequent cleavage of poly(ADP-ribose) polymerase (PARP), a biochemical marker indicative of apoptosis.27,29 To address whether C12-TA has the same or a similar effect on mammalian cells, we investigated its effect on p38 phosphorylation (p-p38) and PARP cleavage in BMDM. In contrast to 3-oxo-C12-HSL, C12-TA failed to induce p-p38 and PARP cleavage, providing strong evidence that the conversion of 3-oxo-C12-HSL to C12-TA results in functionally distinctive biological activities of these molecules (Figure 4). Thus, it is very likely that 3-oxo-C12-HSL possesses the ability to act as an agonist for both eukaryotic and prokaryotic cells, whereas action of C12-TA is confined to bacterial cells. To evaluate the toxicity of C12-TA against human cells, BMDM cells were treated with C12- and C14-TA for 22 h, and under both conditions the cells exhibited similar viability to the DMSO control even at concentrations up to 100 μM (Figure 5). However, both 3-oxo-C12- and 3-oxo-C14-HSL were found to be toxic, which is consistent with our previous reports.27
Figure 4.
BMDM cells were treated with 50 μM 3-oxo-C12-HSL (C12) or C12-TA as indicated, and protein extracts were analyzed by Western blot for PARP cleavage and p38 phosphorylation (p-p38).
Figure 5.
Viability of BMDM cells in the presence of C12-TA and related compounds after 22 hour incubation. (Legend: 1, 3-oxo-C12-HSL; 2, C12-TA; 3, (R)-3-oxo-C12-HSL; 4, (R)-C12-TA; 5, 3-oxo-C14-HSL; 6, C14-TA; 7, 1% DMSO; 8, control)
DISCUSSION
The past decade has seen a marked increase in the understanding of bacterial QS systems and the various classes of QS compounds along with their effects on bacterial population behavior have been investigated extensively.30-32 Although AHL-regulated QS in P. aeruginosa has become one of the best-studied systems, the capacity of 3-oxo-C12-HSL to undergo a spontaneous Claisen-like cyclization to form the tetramic acid C12-TA, a compound with antibacterial activity against Gram-positive and certain Gram-negative bacteria, was only recently discovered.4 The nonenzymatic, base-catalyzed conversion of 3-oxo-C12-HSL to C12-TA is also reminiscent of other bacterially produced antimicrobial agents, particularly the production of the microcin E492m by Klebsiella pneumoniae. The active form of this toxin is formed by an enzyme independent, base-catalyzed rearrangement of an enzymatically produced precursor.33 In light of these reports, the cytotoxic activity, and the resistance of P. aeruginosa to its own C12-TA, we envision a role for C12-TA both in microbial warfare as well as a scaffold for future medicinal chemistry efforts. This hypothesis is supported by clinical data, as isolation of S. aureus and H. influenzae, but not B. cepacia, occurs less frequently as incidences of P. aeruginosa increase, suggesting a competition for survival among these pathogens in the lungs of CF patients.34 The selective activity of C12-TA may provide one plausible explanation for these observations and the corresponding success of P. aeruginosa in its infections of CF patients.
The conversion of 3-oxo-C12-HSL to C12-TA and the antibacterial activity of C12-TA are indicative of a role for AHL-based QS beyond that of strictly cell-to-cell communication.35,36 Several reports have described the role of 3-oxo-C12-HSL in the modulation of several host responses including immunomodulation and the induction of apoptosis.27,37,38 These effects have led to a hypothesized role of 3-oxo-C12-HSL in the establishment of P. aeruginosa infections by modulating the responses of the host. In a similar fashion, 3-oxo-C12-HSL, through the action of C12-TA, may also aid in the establishment of infection by inhibiting the growth of competing bacteria. Similar studies have suggested the role of P. aeruginosa in the selection of antibiotic resistant strains of S. aureus through the function of a 2-alkyl-4-quinolone, a class of molecules also involved in the interspecies communication of P. aeruginosa.39 As such, it is evident that P. aeruginosa possesses a host of mechanisms to mediate interspecies competition. From our results we believe that C12-TA has a place in this arsenal, and, accordingly, it is likely that QS systems play a larger role in bacterial communities than was previously recognized. Indeed, it is a plausible scenario in which P. aeruginosa produces one molecule, 3-oxo-C12-HSL, to mediate competition with both host cells and bacterial competitors in that 3-oxo-C12-HSL exerts its effects primarily on host cells and the rearranged product C12-TA primarily acting on bacterial competitors.
Given the broad spectrum of cytotoxic effects, C12-TA also represents a potential lead compound for the development of antimicrobial therapeutics. Indeed, medicinal chemistry efforts have recently been directed towards the development of N-substituted tetramic acids as antibiotics.40,41 While these TA derivatives exhibited excellent potency against many Gram-positive bacteria, they exhibited no activity against Gram-negative pathogens, moderate activity against M. tuberculosis, and toxic effects against mammalian cells, all of which are in contrast with the activity of C12- and C14-TA. Despite the somewhat modest antibacterial activity of C12-TA, it is likely that little evolutionary pressure exists to optimize the C12-TA structure based on the fact that it may be used by P. aeruginosa to gain a competitive advantage, rather than the complete elimination of competitors. P. aeruginosa does not necessarily stand to gain from optimization of the C12-TA structure, as it is derived from an autoinducer that has been optimized for P. aeruginosa communication and possibly for interaction with host innate immunity. This is further supported by the observation that there was no decrease in the efficacy of C12- or C14-TA against S. aureus in a resistance development assay. Another important consideration in the identification of lead compounds is the capacity of the molecule to exist in biological settings, and indeed, C12-TA has been detected in cultures of P. aeruginosa.4 Thus, C12-TA may represent the balance of activity and biocompatibility characteristic of small molecule natural product drug targets.42 Based on these studies of C12-TA, which upon further elucidation of its antibacterial activity implicates its role in the survival of P. aeruginosa, the discovery of new AHL-derived TAs may represent an exciting new avenue in the study of interspecies bacterial interactions as a whole, as all organisms that produce 3-oxo-AHLs for communication purposes may also benefit from the conversion to TA-derived molecules.
Experimental
Antibacterial activity of C12-TA
The following bacterial strains were purchased from the American Type Culture Collection (ATCC) unless otherwise stated: Bacillus cereus ATCC 11778, B. anthracis Ames strain (Dr. R. Ulrich), B. subtilis ATCC 6051, Enterococcus faecalis ATCC 29212, Staphylococcus aureus ATCC 25923, S. aureus USA-300 clone (Network on Antimicrobial Resistance in Staphylococcus aureus, NARSA), L. plantarum ATCC BAA-793, Corynebacterium diphtheriae ATCC 13812, M. tuberculosis ATCC 27294 (Dr. C. Barry), V. cholerae TP (Prof. D. Bartlett), V. cholerae ATCC 39315, V. harveyi ATCC BAA-1116, H. influenzae ATCC 49401, B. cepacia ATCC 25416, E. coli D22, E. coli Δimp (Prof. F. Romesberg), E. coli ATCC 25404. A bacterial colony was picked and grown overnight in the growth medium and at the temperature recommended by ATCC. For MIC determinations, all bacteria were grown in Mueller Hinton broth. On the next day, the culture was diluted to ~1 × 104 CFU/mL. Aliquots (198 μ) were added into a 96-well microtiter plate containing the test compound dissolved in a DMSO/water solution for a final DMSO concentration of 0.05%. The microtiter plate was sealed (BreatheEasy, Research Products International) and incubated on a shaker at the appropriate temperature overnight. The next day, OD600 was measured by using a ThermoMax plate reader (Molecular Devices). EC50 values were determined by nonlinear curve-fitting using Kaleidagraph 3.6.2 (Synergy Software), and MIC values were determined by visual examination of the wells for turbidity. Reported values are the average of a minimum of three replicates.
Bactericidal Assay
The following bacterial strains were purchased from the American Type Culture Collection (ATCC): Bacillus cereus ATCC 11778, Bacillus subtilis ATCC 6051, Enterococcus faecalis ATCC 29212, and Staphylococcus aureus ATCC 25923. Bactericidal activity was assessed using the BacTiter-Glo™ Microbial Cell Viability Assay kit available from Promega (Madison, WI). Briefly, cells of an overnight culture were harvested and resuspended to an OD600 of 0.4 in fresh growth medium. The cells were then incubated with varying concentrations of C12-TA in 96 well plate format for 1 hour (100 μL per well), at which time 100 μL of BacTiter-Glo™ Reagent was added to each well. Luminescence was measured using a SpectraMax Gemini EM (Molecular Devices, Sunnyvale, CA) plate reader and values were plotted and analyzed using the program Kaleidagraph (Synergy Software, Reading, PA).
Dissipation of the transmembrane potential (ΔΨ)
Perturbations of the ΔΨ were measured using the cationic fluorescent probe 3,3’-dipropylthiadicarbocyanine iodide (DiSC3(5)) from Molecular Probes (Eugene, OR) as described previously.43 Cells were cultured to an OD600 of 0.4 and washed twice with potassium phosphate buffer (pH 7.0) containing 10 mM glucose. The cells were resuspended in the same buffer containing 5 μM DiSC3(5) and incubated for 15 minutes, or until a steady signal was achieved, at the appropriate temperature. After this time, the suspension was transferred to a 96 well plate (200 μL per well) containing 2 μL (in DMSO) of the test compound in each well. Fluorescence was measured (λexc 643 nm, λem 665 nm) over 20 minutes using a SpectraMax Gemini EM (Molecular Devices) fluorescence plate reader.
Dissipation of the transmembrane proton gradient (ΔpH)
Internal pH was monitored according to the procedure developed by Breeuwer et al.44 In short, cells of an overnight culture were washed and resuspended in 50 mM HEPES buffer (pH 8.0) to an OD600 of 0.4. Carboxyfluorescein diacetate succinimidyl ester (cFDA-SE, final concentration 1 μM) was added to the suspension, followed by incubation for 10 minutes at the appropriate temperature. The cells were washed and resuspended in 50 mM potassium phosphate buffer (pH 7.0) and 10 mM glucose was added to eliminate non-conjugated cFDA-SE. After incubation for 30 min at the appropriate temperature, cells were washed twice with potassium phosphate buffer (pH 6.5) containing 10 mM glucose. The cells were resuspended in the same buffer and transferred to a 96 well plate containing the test compound, as in the ΔΨ assay. Assays were performed and fluorescence was measured over 20 minutes using the fluorescence plate reader with excitations of 440 nm and 490 nm and an emission of 525 nm.44 The ratio of signals (490-to-440) was plotted as a function of concentration and analyzed using Kaleidagraph.
Membrane permeation assay
Membrane permeation was assayed by determination of the permeability of the cell membranes to propidium iodide (PI). Cells of an overnight culture were washed twice with 50 mM potassium phosphate buffer (pH 6.5) and resuspended in the same buffer containing 13 μM PI. Membrane permeation was then monitored by the measurement of fluorescence (λexc 488 / λem 617) upon incubation of cells with varying concentrations of C12-TA in 96 well plate format, as in the previous two assays.
Determination of resistant bacteria
Culture tubes containing BHI and C12-TA were inoculated with an overnight culture of S. aureus to obtain 1 × 104 cfu/mL. The concentrations of C12-TA ranged from three doubling dilutions above and below the previously measured MIC. The cultures were incubated for 24 h at 37°C, and the culture incubated with a concentration of TA one dilution step below the MIC was used as the inoculum for the next transfer (1 × 104 cfu/mL). The process was then repeated for 19 times for a total of 20 passages, with MIC measured as before.
Mammalian cell studies
Bone marrow-derived macrophages (BMDM) were prepared from C57BL/6 mice by using standard protocols. BMDM were cultured in 70% growth medium [Dulbecco's modified Eagle's medium (4.5 g/liter glucose) supplemented with 10% fetal bovine serum (HyClone), L-glutamine, pyruvate, penicillin/streptomycin and nonessential amino-acids] and 30% L929 condition medium as described.27
Cytotox Assay
Cytotoxicity of test compounds was assessed using bone marrow-derived macrophages (BMDM) and a solution assay based on the reduction of MTS into formazan (Promega) according to the manufacturer's protocol. BMDM were cultured according to standard techniques in 70% growth medium (Dulbecco's modified Eagle medium (4.5 g/L glucose) supplemented with 10% fetal bovine serum, 100 mM HEPES, 5 mM sodium Pyruvate, L-glutamine, penicillin/streptomycin, and nonessential amino acids) and 30 % L929 conditioned medium. In brief, cells were allowed to recover for 24 hours after transfer to the assay plate and medium was refreshed prior to the assay. Compounds were added directly from DMSO stocks to a final DMSO concentration of 1% and cells were incubated at 37°C, 5% CO2 for 24 hours before addition of the MTS substrate. Absorbance was measured after 3 hours incubation at 490 nm and data were normalized to untreated controls. All experiments have been performed in triplicate.
Supplementary Material
Acknowledgement
We thank Professor Michael Gänzle of the University of Alberta, Edmonton, AB, Canada for generously providing us with reutericyclin, and Prof. Floyd Romesberg for providing the mutant E. coli strains. The following isolate was obtained through the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) Program: Staphylococcus aureus USA300 (NRS384);supported under NIAID/NIH Contract No. HHSN272200700055C We would also like to thank Prof. Bastiaan P. Krom of the University of Groningen for critical reading of the manuscript. This work was supported by the National Institutes of Health (AI079503 to KDJ, AI080715 to GFK, AI079436 to VVK), the Skaggs Institute for Chemical Biology, and a Sanofi-Aventis Graduate Fellowship (C.A.L.), and in part by the Intramural Research program of the NIAID, NIH.
Footnotes
Supporting Information Synthetic procedures, spectral data, and biofilm protocols. This information is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Clardy J, Fischbach MA, Walsh CT. Nat Biotechnol. 2006;24:1541–50. doi: 10.1038/nbt1266. [DOI] [PubMed] [Google Scholar]
- 2.Royles BJL. Chem Rev. 1995;95:1981–2001. [Google Scholar]
- 3.Kaufmann GF, Sartorio R, Lee SH, Mee JM, Altobell LJ, 3rd, Kujawa DP, Jeffries E, Clapham B, Meijler MM, Janda KD. Journal of the American Chemical Society. 2006;128:2802–3. doi: 10.1021/ja0578698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaufmann GF, Sartorio R, Lee SH, Rogers CJ, Meijler MM, Moss JA, Clapham B, Brogan AP, Dickerson TJ, Janda KD. Proc Natl Acad Sci U S A. 2005;102:309–14. doi: 10.1073/pnas.0408639102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heurlier K, Denervaud V, Haas D. Int J Med Microbiol. 2006;296:93–102. doi: 10.1016/j.ijmm.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 6.Geske GD, O'Neill JC, Blackwell HE. Chem Soc Rev. 2008;37:1432–47. doi: 10.1039/b703021p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabrol S, Olliver A, Pier GB, Andremont A, Ruimy R. J Bacteriol. 2003;185:7222–30. doi: 10.1128/JB.185.24.7222-7230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Favre-Bonte S, Chamot E, Kohler T, Romand JA, van Delden C. BMC microbiology. 2007;7:33. doi: 10.1186/1471-2180-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee B, Haagensen JA, Ciofu O, Andersen JB, Hoiby N, Molin S. J Clin Microbiol. 2005;43:5247–55. doi: 10.1128/JCM.43.10.5247-5255.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaber JA, Carty NL, McDonald NA, Graham ED, Cheluvappa R, Griswold JA, Hamood AN. Journal of medical microbiology. 2004;53:841–53. doi: 10.1099/jmm.0.45617-0. [DOI] [PubMed] [Google Scholar]
- 11.Sandoz KM, Mitzimberg SM, Schuster M. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15876–81. doi: 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diggle SP, Griffin AS, Campbell GS, West SA. Nature. 2007;450:411–4. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 13.Purdy A, Rohwer F, Edwards R, Azam F, Bartlett DH. J Bacteriol. 2005;187:2992–3001. doi: 10.1128/JB.187.9.2992-3001.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatterjee A, Chaudhuri S, Saha G, Gupta S, Chowdhury R. J Bacteriol. 2004;186:6809–14. doi: 10.1128/JB.186.20.6809-6814.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilsdorf JR. Infect Immun. 1998;66:5053–9. doi: 10.1128/iai.66.11.5053-5059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Normark S, Boman HG, Matsson E. J Bacteriol. 1969;97:1334–42. doi: 10.1128/jb.97.3.1334-1342.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sampson BA, Misra R, Benson SA. Genetics. 1989;122:491–501. doi: 10.1093/genetics/122.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. Proc Natl Acad Sci U S A. 2008;105:5537–42. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young K, Silver LL, Bramhill D, Cameron P, Eveland SS, Raetz CR, Hyland SA, Anderson MS. J Biol Chem. 1995;270:30384–91. doi: 10.1074/jbc.270.51.30384. [DOI] [PubMed] [Google Scholar]
- 20.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Cell. 2005;121:307–17. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 21.Sperandeo P, Pozzi C, Deho G, Polissi A. Research in microbiology. 2006;157:547–58. doi: 10.1016/j.resmic.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 22.Ganzle MG. Appl Microbiol Biotechnol. 2004;64:326–32. doi: 10.1007/s00253-003-1536-8. [DOI] [PubMed] [Google Scholar]
- 23.Ganzle MG, Vogel RF. Appl Environ Microbiol. 2003;69:1305–7. doi: 10.1128/AEM.69.2.1305-1307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bakker EP, Mangerich WE. J Bacteriol. 1981;147:820–6. doi: 10.1128/jb.147.3.820-826.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong SD, Oberthur M, Losey HC, Anderson JW, Eggert US, Peczuh MW, Walsh CT, Kahne D. J Am Chem Soc. 2002;124:9064–5. doi: 10.1021/ja026342h. [DOI] [PubMed] [Google Scholar]
- 26.Walsh CT, Fischbach MA. Angew Chem Int Ed Engl. 2008;47:5700–2. doi: 10.1002/anie.200801801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kravchenko VV, Kaufmann GF, Mathison JC, Scott DA, Katz AZ, Wood MR, Brogan AP, Lehmann M, Mee JM, Iwata K, Pan Q, Fearns C, Knaus UG, Meijler MM, Janda KD, Ulevitch RJ. J Biol Chem. 2006;281:28822–30. doi: 10.1074/jbc.M606613200. [DOI] [PubMed] [Google Scholar]
- 28.Kravchenko VV, Kaufmann GF, Mathison JC, Scott DA, Katz AZ, Grauer DC, Lehmann M, Meijler MM, Janda KD, Ulevitch RJ. Science. 2008;321:259–63. doi: 10.1126/science.1156499. [DOI] [PubMed] [Google Scholar]
- 29.Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, Gareau Y, Griffin PR, Labelle M, Lazebnik YA, et al. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 30.Deziel E, Lepine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. Proc Natl Acad Sci U S A. 2004;101:1339–44. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lazdunski AM, Ventre I, Sturgis JN. Nat Rev Microbiol. 2004;2:581–92. doi: 10.1038/nrmicro924. [DOI] [PubMed] [Google Scholar]
- 32.Venturi V. FEMS Microbiol Rev. 2006;30:274–91. doi: 10.1111/j.1574-6976.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- 33.Nolan EM, Fischbach MA, Koglin A, Walsh CT. J Am Chem Soc. 2007;129:14336–47. doi: 10.1021/ja074650f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foundation CF. Cystic Fibrosis Foundation; Bethesda, MD: 2005. [Google Scholar]
- 35.Schertzer JW, Boulette ML, Whiteley M. Trends Microbiol. 2009;17:189–95. doi: 10.1016/j.tim.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Cooley M, Chhabra SR, Williams P. Chem Biol. 2008;15:1141–7. doi: 10.1016/j.chembiol.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Telford G, Wheeler D, Williams P, Tomkins PT, Appleby P, Sewell H, Stewart GS, Bycroft BW, Pritchard DI. Infect Immun. 1998;66:36–42. doi: 10.1128/iai.66.1.36-42.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wagner VE, Frelinger JG, Barth RK, Iglewski BH. Trends Microbiol. 2006;14:55–8. doi: 10.1016/j.tim.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Hoffman LR, Deziel E, D'Argenio DA, Lepine F, Emerson J, McNamara S, Gibson RL, Ramsey BW, Miller SI. Proc Natl Acad Sci U S A. 2006;103:19890–5. doi: 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yendapally R, Hurdle JG, Carson EI, Lee RB, Lee RE. J Med Chem. 2008;51:1487–91. doi: 10.1021/jm701356q. [DOI] [PubMed] [Google Scholar]
- 41.Hurdle JG, Yendapally R, Sun D, Lee RE. Antimicrob Agents Chemother. 2009 doi: 10.1128/AAC.00457-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson RM, Danishefsky SJ. J Org Chem. 2006;71:8329–51. doi: 10.1021/jo0610053. [DOI] [PubMed] [Google Scholar]
- 43.Wu M, Maier E, Benz R, Hancock RE. Biochemistry. 1999;38:7235–42. doi: 10.1021/bi9826299. [DOI] [PubMed] [Google Scholar]
- 44.Breeuwer P, Drocourt J, Rombouts FM, Abee T. Appl Environ Microbiol. 1996;62:178–183. doi: 10.1128/aem.62.1.178-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.