Abstract
Antifreeze proteins (AFPs) can produce a difference between the nonequilibrium freezing point and the melting point is termed thermal hysteresis (TH). The TH activity of an antifreeze protein (AFP) depends on the specific AFP, its concentration as well as the presence of co-solutes including low-molecular-mass solutes and/or proteins. We recently identified series of carboxylates and polyols as efficient enhancers for an AFP from the beetle Dendroides canadensis. In this study, we chemically modified DAFP-1 using the arginine-specific reagent 1,2-cyclohexanedione. We demonstrated that 1,2-cyclohexanedione specifically modifies one arginine residue and the modified DAFP-1 loses its enhancing ability completely or partially in the presence of previously identified enhancers. The stronger the enhancement ability of the enhancer on the native DAFP-1, the stronger the enhancement effect of the enhancer has on the modified DAFP-1. The weaker enhancers (e.g., glycerol) completely lose their enhancement effect on the modified DAFP-1 due to their inability to compete with 1,2-cyclohexanedione for the arginine residue. Regeneration of the arginine residue using hydroxylamine fully restored the enhancing ability of DAFP-1. These studies indicated that an arginine residue is critical for the enhancing ability of DAFP-1 and the guanidinium group of the arginine residue is important for its interaction with the enhancers, where the general mechanism of arginine-ligand interaction is borne. This work may initiate a complete mechanistic study of the enhancement effect in AFPs.
Keywords: Beetle antifreeze protein, Antifreeze activity enhancement, Guanidinium-ligand interaction, Molecular mechanism of antifreeze proteins
As an adaptive response to cold environmental conditions, many organisms, such as fish and insects, have developed antifreeze proteins (AFPs) that allow them to survive at subfreezing temperatures. AFPs are characterized by their unique abilities to lower the freezing point of water without affecting the melting point. The gap between the nonequilibrium freezing and melting points is termed thermal hysteresis (TH), and is usually used as a measure of the activity of AFPs (1). The adsorption-inhibition mechanism of AFP function states that AFPs depress the freezing point by absorbing onto the ice crystal surface at preferred growth sites (2-5). The interactions between the AFP and the ice can be one of, or any combination of, hydrophobic interactions, van der Waals interactions (6, 7), and hydrogen bonds (8, 9).
Beetle AFPs typically exhibit high antifreeze activities compared to fish AFPs. Beetle AFPs from Dendroides canadensis (DAFPs) are a family of 13 known AFPs containing varying numbers of 12- or 13-mer repeats with sizes of 7.3 – 16.2 kDa (10). There are sixteen Cys residues well distributed in the seven repeats in the isomer, DAFP-1, all of which are disulfide bridged (Figure 1A). The resulting eight disulfide bonds make DAFP-1 highly stable (11). In addition, there are four Arg and two Lys residues in DAFP-1.
FIGURE 1.
The structures of DAFP-1 from Dendroides canadensis (A) The sequence of DAFP-1 with the location of the disulfide bridges (11), (B) Model of DAFP-1 tertiary structure. The eight disulfide bridges are shown in yellow. The positions of the four arginine residues are labeled. DAFP-1 consists of seven repeats and Arg 9 is located in the first irregular repeat.
The activities of many AFPs can be further enhanced in the presence of certain co-solutes. Both small molecules and macromolecules have been identified as enhancers for DAFP-1 (12-19). Recently, various series of efficient low molecular mass enhancers have been reported, however, the mechanism of enhancement still remains elusive (14, 15, 20, 21).
In searching for highly efficient low molecular mass enhancers, the understanding of the molecular mechanisms of the AFP enhancers and the enhancing function of AFPs are both critically important. In order to understand the molecular mechanism of AFP enhancers, we first correlated the enhancement abilities of the enhancers with their varying physicochemical properties and reported series of polycarboxylates and polyols as efficient AFP enhancers (14, 15). As more highly efficient enhancers have been identified, we have noticed that almost all of the low molecular mass enhancers can interact with the guanidinium group of arginine and/or the ammonium group of lysine through one of, or any combination of the following interactions: ionic interaction, hydrogen bonding, and hydrophobic interaction.
In this study, we examine the role of arginine in the enhancing function of DAFP-1. There are four arginines and two lysines in DAFP-1. Here, we report the modification of arginine in DAFP-1 and the effect of the modification on the enhancing function of DAFP-1 in the presence of various low molecular mass enhancers. This work demonstrates that arginine in DAFP-1 plays a critical role in the enhancing ability of DAFP-1 and provides important information regarding the binding between AFPs and enhancer molecules.
MATERIALS AND METHODS
DAFP-1 3D structure
The starting 3D structure of DAFP-1 was created from a homology model of a Tenebrio molitor AFP, TmAFP (PDB Code: 1EZG) that has considerable sequence homology to DAFP-1, using the 3D-JIGSAW (22-24) online structure prediction tool. This structure was immersed in a 69 Å, pre-equilibrated, cubic box of water molecules described by the F3C (25) water model. The water molecules that overlapped the protein were removed, and 1 Na+ counter ion added to achieve charge neutrality. The system was then simulated for 1.5 ns of NPT dynamics at 1 bar and 300 K, after initial energy minimization and heating, using the LAMMPS (26) MD code, with the protein described by the AMBER99 (27) forcefield. Snapshots of the system were saved every 10 ps, with the final snapshot taken as the equilibrated 3D structure (28).
Materials
1,2-Cyclohexanedione, hydroxylamine hydrochloride, and all the tested enhancer compounds were purchased from Sigma-Aldrich (St. Louis, MO) at ACS grade or better. All the chemicals were used without additional purification. Solvents and chemicals for the HPLC experiments were purchased at HPLC grade from Sigma-Aldrich (St. Louis, MO). All the aqueous solutions were prepared using Milli-Q water produced from a Synergy water system (Millipore Co.) with a minimum resistivity of 18 MΩ·cm. The protein samples were in 50 mM sodium borate buffer, pH 9.0 and all the samples were filtered through a 0.2 μm filter before use unless otherwise indicated.
Protein Expression and Purification
DAFP-1 was expressed and purified as described previously (14). In brief, DAFP-1 was expressed as a fusion protein with thioredoxin, His, and S tags in Escherichia coli Origami B cells. The cells were harvested by centrifugation at 4 °C and then were disrupted by passing through a French press (Thermo Fisher) at 1250 psi two to three times. The crude protein was purified using Ni-NTA agarose (Qiagen). The tags were cleaved using enterokinase (New England Biolabs) and removed using Ni-NTA agarose. The cleaved protein was finally purified using ÄKTA Purifier 10 (GE Healthcare) with a Sephacryl S-100 gel filtration column (GE Healthcare). The identity of the purified protein was confirmed by SDS-PAGE, HPLC and Matix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. The protein concentration was determined by UV-Vis spectroscopy (Cary 100 Bio, Varian) using the absorption at 280 nm (ε280 = 5.47 × 103 M−1 cm−1) (29).
Reaction of DAFP-1 with Arginine-Modifying Agents and the Reversal of the Reaction with hydroxylamine
In a typical reaction, a tenfold molar excess of freshly made 1,2-cyclohexanedione to arginine was incubated with DAFP-1 in 0.10 M borate buffer, pH 9.0, in the dark for as long as 3 h at 37 °C. The sample pH was 9.0 and was adjusted using 0.20 M NaOH if necessary. DAFP-1 treated under the same conditions without 1,2-cyclohexanedione was used as a control. Aliquots were taken every 1 h for TH activity measurement and for mass spectrometry to determine the extent of modification. The excess 1,2-cyclohexanedione was removed by a PD-10 desalting column (GE Healthcare) equilibrated with 0.10 M sodium borate buffer, pH 9.0. In the modified DAFP-1, the conjugated π system of 1, 2-cyclohexanedione is destroyed. The UV absorption properties at 280 nm are mainly derived from the Tyr residues and to a small extent on the amount of disulfide bonds. The extinction coefficient of the modified DAFP-1 is the same as that of the native DAFP-1 (ε280 = 5.47 × 103 M−1 cm−1). The concentration of the modified DAFP-1 was determined using a Cary 100 Bio UV-Vis spectrophotometer. In order to test whether hydroxylamine would reverse this modification, the modified DAFP-1 was incubated with 0.60 M hydroxylamine, pH 7.0, for 6 h in the dark at 37°C under N2. After the excess hydroxylamine and other small molecules were removed, the arginine regenerated DAFP-1 was concentrated and its concentration was determined as above.
MALDI Mass Spectrometry
Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectra were obtained on a PerSeptive Biosystems/Voyager-DE MALDI-TOF mass spectrometer at the Stanford PAN facility. Calibrations of the mass spectrometer against external mass standards were carried out before the sample analysis. Matrix, sinapinic acid, was prepared as a saturated solution in 1:1 (v/v) water:acetonitrile with 0.1% trifluoroacetic acid (TFA). After being desalted using Ziptip C18 resin (Millipore), the sample was dissolved in double-distilled H2O-TFA at pH 3.0. Then 0.5 μL of the sample solution was mixed with an equal volume of the saturated matrix solution in a MALDI sample plate and air dried before analysis.
Quantitative Analysis of the Modification Reaction of DAFP-1
HPLC chromatograms were recorded on a computer connected to the HPLC system (Dionex, Sunnyvale, CA) consisting of an Ultimate 3000 pump and an Ultimate 3000 Photodiode Array Detector. The samples were injected on a BioSuite™ 125, 4 μm UHR SEC HPLC column, 4.6 × 300 mm (Waters, Milford, MA). The aqueous buffer for SEC consisted of 0.10 M sodium borate, 0.10 M Na2SO4, pH 8.50. All the buffers and samples were filtered through a 0.1 μm filter and were vacuum degassed before use. The flow rate was 0.50 mL/min at ambient temperature. The reaction mixtures were prepared by incubating equal volumes of 1.160 mM 1,2-cyclohexanedione and 0.290 mM DAFP-1; and 1.910 mM 1,2-cyclohexanedione and 0.139 mM DAFP-1 as described above for 1 h. Then the elution profiles of the reaction mixtures, the DAFP-1 control, and 1,2-cyclohexanedione were procured. The consumed amount of 1,2-cyclohexanedione obtained from the HPLC elution profiles was used to estimate the stoichiometry of the reaction.
Thermal Hysteresis Activity Measurements
The determinations of the thermal hysteresis (TH) activities followed the known procedure using differential scanning calorimetry (DSC) except that 0.05 M sodium borate buffer, pH 9.0, was used in this study (15). The measurements were performed on a DSC 823e (Mettler Toledo, OH) with an HSS7 high sensitivity sensor and a Julabo FT900 intracooler chiller (Julabo Company, Allentown, PA). The protein concentrations were 320 μM in all the samples. The enhancing abilities of glycerol, trehalose, and sodium citrate were assessed on DAFP-1 (the control), the Arg-modified DAFP-1, and the arginine regenerated DAFP-1. In addition, the following previously identified enhancers (ethanol, ethylene glycol, threitol, glucose, sodium acetate, sodium glutarate (GTA), sodium tricarboxylate (TCA), sodium ethylenediaminetetraacetate (EDTA), sodium diethylenetriaminepentaacetate (DTPA), sodium triethylenetetramine-N,N,N′,N″,N‴,N‴-hexaacetate (TTHA), were tested for the ability to enhance the TH of both DAFP-1 (the control) and the Arg-modified DAFP-1. The tested concentrations of the enhancers were varied from 0.00 M to 1.5 M (depends on the solubility). Each combination of DAFP-1 and enhancer were made up three times and the TH of a given sample was measured at least twice. The TH values were presented as means and the standard deviations were 0.02 °C.
Circular Dichroism (CD) Spectroscopy
CD spectra were measured on a Jasco 810 spectropolarimeter at ambient temperature using a quartz cuvette with a path length of 0.1 cm. Protein samples were prepared at a concentration of 15 μM in 50 mM sodium borate buffer, pH 9.0. The buffer contributions were subtracted from the sample spectra. The far-UV CD spectrum (190 - 260 nm) of each sample was reported as an average of three scans in molar ellipticity.
RESULTS
DAFP-1 3D Structure
There is no higher order structure available for DAFP-1 to view the arginines. A model of DAFP was proposed, while it is not identified which DAFP isoform was modeled or shows the arginine residues (3). Therefore, a model of the tertiary structure of DAFP-1 (Figure 1B) was built here and was subsequently used to locate arginine side chains with respect to the remainder of DAFP-1 structure. The model structure of DAFP-1 shows that the side chains of all the arginine residues to some extent point away from the protein surface (Figure 1B). Arg9 is located in the only irregular repeat, repeat one. The side chain of Arg25 reaches too far away from the surface to form any contacts with other residues in DAFP-1 and those of Arg30 and Arg54 have similar conformations to each other.
Arginine-Modified DAFP-1 by Reaction with 1,2-Cyclohexanedione
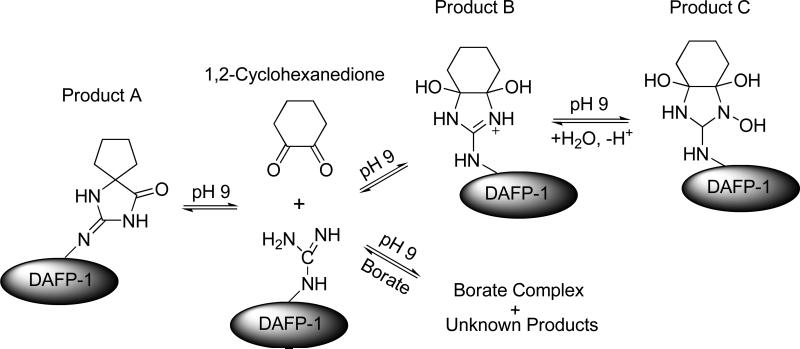
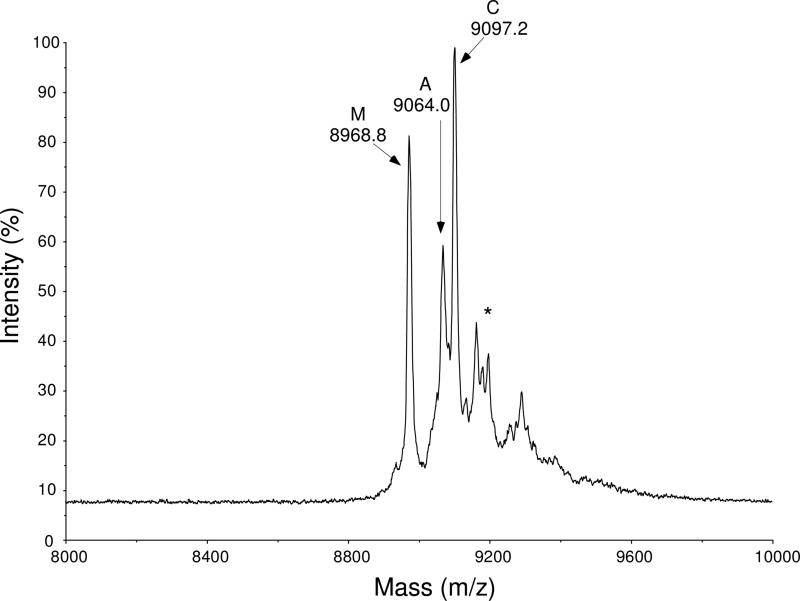
The arginine modification of DAFP-1 was performed by the reaction of 1,2-cyclohexanedione and DAFP-1 at pH 9.0 in 0.10 M sodium borate as sketched in Figure 2. To test the kinetics of the modification reaction, aliquots taken after 1 h, 2 h, and 3 h of incubation were analyzed by HPLC and MALDI-TOF mass spectrometry. Nearly identical results were obtained from all three aliquots, which suggests the reaction can be completed in one hour. Thus, all the modified DAFP-1 samples used for enhancing ability tests were then prepared by one hour incubation with 1,2-cyclohexanedione. Figure 3 shows the MALDI-TOF mass spectrum of DAFP-1 incubated with 1,2-cyclohexanedione for 1 h. It is known that 1,2-cyclohexanedione attacks only arginine residues in a protein under the mild conditions we used (30, 31). As revealed in Figure 3, there is no lysine modification in DAFP-1 and only one of the four arginines in DAFP-1 was modified although a large excess of 1,2-cyclohexanedione was used in the reaction. The calculated molecular mass of DAFP-1 is 8968.8 Da. The two peaks at 9064.0 and 9097.8 are due to one arginine modified DAFP-1, associated with the 1:1 products A and C, respectively, while the peak at 8968.8 is associated with the DAFP-1 fragment dissociated from the modified DAFP-1 (Figures 2 and 3).
FIGURE 2.
The reaction scheme of the key arginine in DAFP-1 with 1,2-cyclohexanedione at pH 9.0 in 0.10 M sodium borate buffer.
FIGURE 3.
MALDI-TOF mass spectrum of arginine modified DAFP-1 products. A and C represent products A and C in Figure 2, respectively. M represents the molecular fragment of the products. The asterisk (*) indicates the borate complexes of product C.
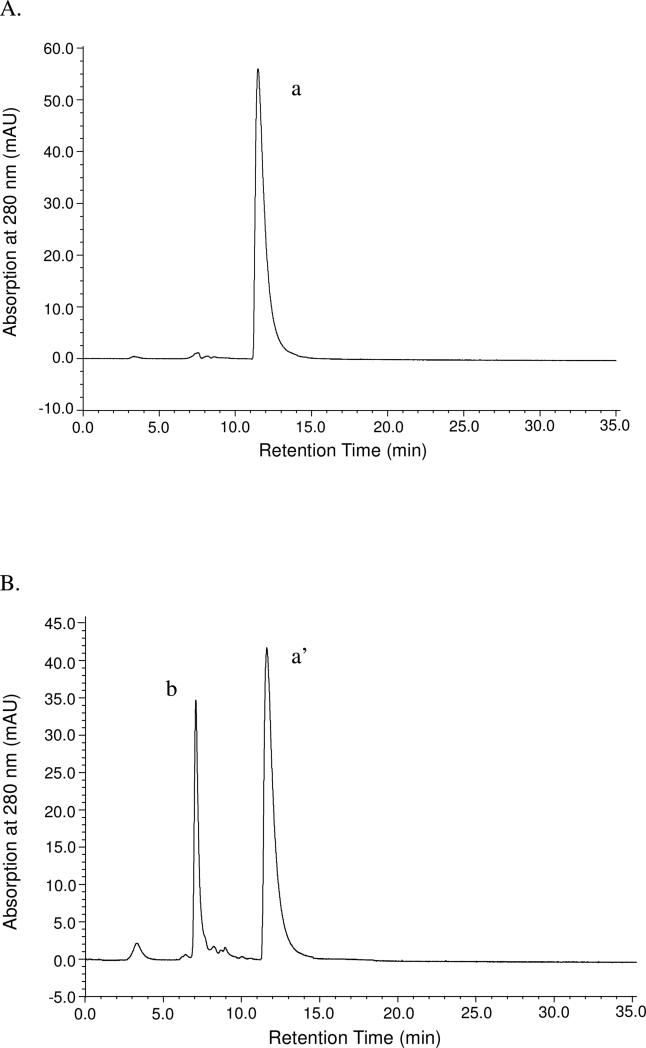
Furthermore, the stoichiometry of the reaction between 1,2-cyclohexanedione and DAFP-1 was estimated to be 1:1 by analyzing the decrease of the UV absorption of 1,2-cyclohexanedione in the size exclusion HPLC profiles obtained with varied reactant molar ratios. A typical example is presented in Figure 4. The elution peak of 1,2-cyclohexanedione was decreased by exactly one fourth in the reaction mixture of DAFP-1 and 1,2-cyclohexanedione with a molar ratio of 1:4 (Figure 4B). Another special example is shown in the supporting materials, in which 9.55 pmol 1,2-cyclohexanedione (Figure S1 A and B) and 0.695 pmol DAFP-1 (Figure S1 B) were used. Similar results were obtained though this non-integral molar ratio of DAFP-1 and 1,2-cyclohexanedione with the amount of 1,2-cyclohexanedione more than 10 times in excess of that of DAFP-1 used in the reaction (Figure S1). These results suggest that one arginine residue in DAFP-1 is modified under these conditions.
FIGURE 4.
UV absorption detected size exclusion HPLC profiles of (A) 1,2-cyclohexanedione (1.160 mM, 8 μL, the retention time tR = 11.5 min); and (B) the reaction mixture of DAFP-1 (0.290 mM, 8 μL, tR = 7.1 min) and 1,2-cyclohexanedione (1.160 mM, 8 μL, tR = 11.5 min). The molar ratio of DAFP-1 to 1,2-cyclohexanedione in the reaction mixture was 1:4. The ratio of the area under peak a (Aa = 38.9 mAu · min) to the area under peak a’ (Aa’ = 29.0 mAu · min) is 4:3 indicating only one-fourth of the original amount of 1,2-cyclohexanedione was consumed.
Thermal Hysteresis Characterization
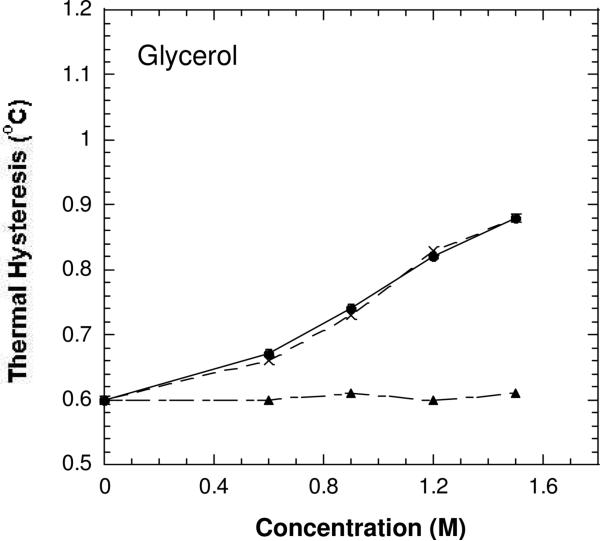
In the control, in which DAFP-1 was treated the same as the treated DAFP-1 except that 1,2-cyclohexanedione was not added, the TH activities in the absence and in the presence of low-molecular-mass enhancers were in agreement with those previously reported for DAFP-1 (14, 15). The TH activity of Arg-modified DAFP-1 alone was the same as those of the control or the native DAFP-1, while the efficiencies of the antifreeze enhancement in the Arg-modified DAFP-1 in the presence of different enhancers were decreased to different extents or completely abolished (Figures 5-7 and S2-S4). The ability of the modified DAFP-1 to be enhanced was completely lost in the presence of weak enhancers. The results further support that the modification reaction was complete within one hour and there was little unmodified DAFP-1 in the solution. To test the completion of the reaction of the modified DAFP-1 with enhancers, the modified DAFP-1 and three representative enhancers (glycerol as a weak enhancer, trehalose as a medium strong enhancer, and citrate as a stronger enhancer) were incubated at room temperature for different time periods (0.5 h, 1 h, and 2h, respectively) before thermal hysteresis activity measurements. Little changes in the TH activities of the samples with different incubation time were observed indicating the equilibrium was reached within 30 minutes at ambient temperature (data not shown). Thus the incubation time of 0.5 h was used for the modified DAFP-1 with the enhancers for all the measurements in this study.
FIGURE 5.
The antifreeze activities of DAFP-1 (•), the Arg-modified DAFP-1 (▲), and the Arg-modified DAFP-1 after treatment with NH2OH (×) in the presence of varying concentrations of glycerol in 50 mM borate buffer pH 9.0.
FIGURE 7.
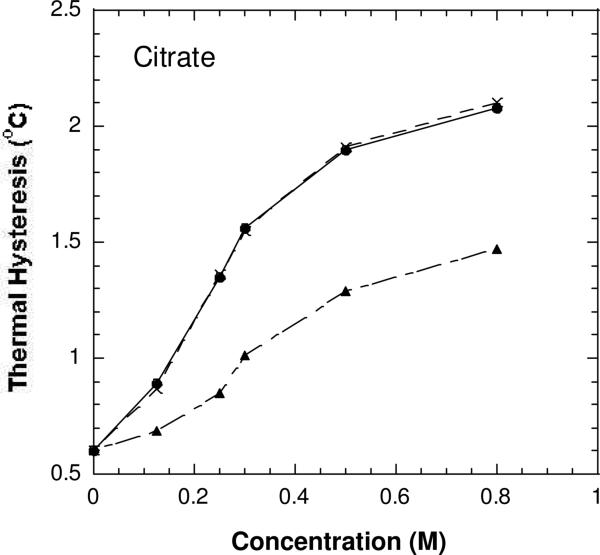
The antifreeze activities of DAFP-1 (•), the Arg-modified DAFP-1 (▲), and the Arg-modified DAFP-1 treated with NH2OH (×) in the presence of varying concentrations of sodium citrate in 50 mM borate buffer pH 9.0.
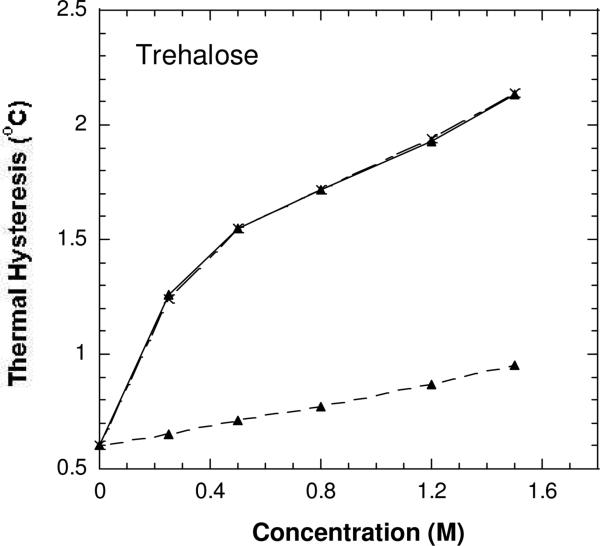
The TH activities of the modified DAFP-1 in the presence of the weak enhancers (e.g., glycerol) were nearly identical to that of the native DAFP-1 alone (Figures 5 and S2-S3). The ability of DAFP-1 to be enhanced by these weak enhancers was completely abolished after the arginine residue was modified in DAFP-1. In contrast, in the presence of relatively stronger enhancers, such as trehalose, citrate, and EDTA, the ability to be enhanced can be partially regained in the modified DAFP-1, indicating that ability of the modified DAFP-1 to be enhanced appears to be positively associated with the enhancement abilities of these enhancers (Figures 6-7 and S2-S4). This suggests that these stronger enhancers can compete with the 1,2-cyclohexanedione for the side chain of the arginine residue in DAFP-1 (a proposed scheme is shown in Figure 8), and that they form complexes with DAFP-1. The peak of the complex of DAFP-1 and citrate (as a stronger enhancer) was indeed observed in the MALDI-TOF mass spectrum (Figure S5), suggesting the formation of a complex of DAFP-1 and a stronger enhancer. The arginine modification reaction by 1,2-cyclohexanedione is usually reversible, and the arginine can be regenerated if treated by hydroxylamine (30). As expected, the enhancing ability of the protein samples was nearly fully recovered in the presence of the previously tested enhancers after the modified DAFP-1 was treated in 0.5 M hydroxylamine, pH 7.0 for six hours (Figures 5-7). These results suggest that the arginine residue does not account for the antifreeze activity of DAFP-1, but is critical for the enhancing ability of DAFP-1.
FIGURE 6.
The antifreeze activities of DAFP-1 (•), the Arg-modified DAFP-1 (▲), and the Arg-modified DAFP-1 treated with NH2OH (×) in the presence of varying concentrations of trehalose in 50 mM borate buffer pH 9.0.
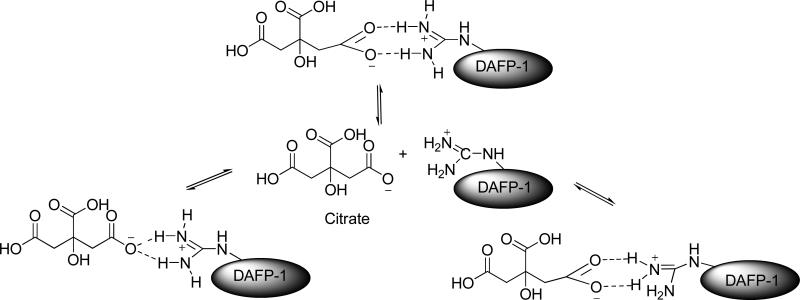
FIGURE 8.
The proposed binding of arginine in DAFP-1 with citrate.
CD Spectra of DAFP-1 and Modified DAFP-1
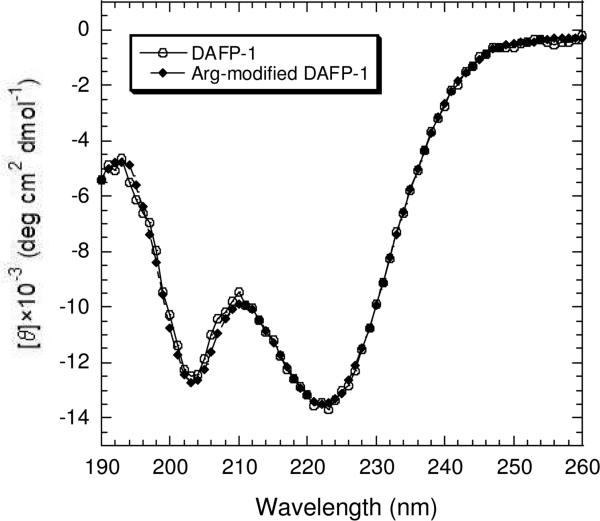
CD spectra (Figure 9) were obtained for both DAFP-1 and the Arg-modified DAFP-1 to determine whether the secondary structure of the native protein was retained in the modified DAFP-1. As earlier reported with the protein isolated from D. canadensis hemolymph (32), the far-UV CD spectrum of DAFP-1 showed negative ellipiticity near 195 nm and two minima around 203 and 222 nm, respectively, with a much broader 222-nm band (Figure 9). These characteristics were also seen in the CD spectra of the expressed DAFP-1 used in this study indicating that it also consists primarily of turns and short β-sheet structures (32). Furthermore, there were no apparent differences in the CD spectra of DAFP-1 and Arg-modified DAFP-1 (Figure 9), indicating that the modification of DAFP-1 does not introduce major changes in the protein structure.
FIGURE 9.
CD spectra of DAFP-1 and the Arg-modified DAFP-1 in 50 mM borate buffer, pH 9.0. The protein concentrations were 15 μM. The experiments were performed at ambient temperature.
DISCUSSION
Both small molecules and proteins have been reported to be able to enhance the TH activity of AFPs (12-18, 20). It has been demonstrated that the TH activity enhancement effects of the protein enhancers is due to their ability to bind to DAFPs (21, 33). Despite persistent studies, the enhancer binding sites of small molecular mass enhancers in AFPs are essentially unknown (14, 15, 20, 21). In this study, by using a chemically modified DAFP-1, we demonstrated that an arginine residue in DAFP-1 is critical for the TH enhancing ability of DAFP-1 by small enhancers. The mass spectrum shows the product peaks with one arginine modification, indicating that the modification is only for arginine and only one of the four arginine residues in DAFP-1 was modified (Figure 3). The modification of arginine by 1,2-cyclohexanedione is well known to be reversed by hydroxylamine (30), and because the enhancing ability of the modified DAFP-1 was fully restored by hydroxylamine treatment (Figures 6-8), this also indicates that the modification only affects an arginine residue. The analytical HPLC results provide further support that the stoichiometry of the reaction between 1,2-cyclohexanedione and DAFP-1 is 1:1 (Figure 4). DAFP-1 consists of seven tandem 12- or 13-residue repeats. All the sixteen Cys residues in DAFP-1 are disulfide bridged and form seven intra-repeat and one inter-repeat disulfide bonds. The inter-repeat disulfide bond links repeat one and repeat two. The four arginines, Arg9, 25, 30 and 54, are located in repeat one, two, three, and five, respectively. Arginine-ligand interactions are known to exist in aqueous phase only when there are hydrophobic interactions and/or hydrogen bonds with other amino acids (37). The simple ion pairs between guanidinium cations and ligands are not stable in aqueous solutions, since water can compete for both donor and acceptor sites for the ligand solvation and decrease the stability of the ion pairs significantly. The side chain of Arg25 reaches too far away from the surface to form any contacts with other residues in DAFP-1, which is makes it less possible for the Arg-enhancer pair to be stabilized. Thus Arg25 has the least possibility to be the key Arg for the enhancing ability of DAFP-1. The side chains of Arg30 and Arg54 have similar conformations (Figure 1). Due to the repeat structure of DAFP-1, Arg30 and Arg54 are located in a similar environment. Since only one Arg residue was involved in the enhancement molecular mechanism, the similarities between Arg30 and Arg54 make them less possible to be the key Arg. Arg9 is located in the irregular repeat, repeat one, next to the only inter-repeat disulfide bond at the N-terminus (Figure 1) and may have the highest possibility of being the critical arginine residue for the enhancing ability of DAFP-1. The model structure of DAFP-1 gives us some hints on which arginine plays a major role in the enhancing ability of DAFP-1. Studies are underway to identify the key arginine and the assisting residues.
Guanidine is one of the most basic forms of neutral nitrogen compounds, with a pKa of 13.5 in water at room temperature (34). This results from the stability of the conjugate acid (guanidinium) in water, where the positive charge on guanidinium can be distributed evenly among the three nitrogen atoms by resonance (35, 36). When the guanidinium moiety of the arginine residue was chemically modified with 1,2-cyclohexanedione, the modified arginine residue lost the resonance stability in guanidinium and its ability to bind to anions was greatly reduced. Furthermore, in contrast to a free arginine residue, the resulting adducts have a five or six atom ring. The increased steric hindrance of the modified arginine greatly reduces the possibility of the formation of interactions such as hydrogen bonds and/or ionic interactions among guanidinium and the ligand (or enhancer). As a result the modified arginine residue lost its ability to bind to the enhancer molecules, completely in the case of weak enhancers or partially with strong enhancers.
The 1,2-cyclohexanedione modification of arginine is a reversible reaction in borate buffer at pH 9.0 (30). In the presence of enhancer molecules that can bind to the side chain of arginine, the competition for arginine between the enhancer molecules and 1,2-cyclohexanedione will occur until a chemical equilibrium is finally reached (Figures 2 and 8). In this study, the competition for the guanidinium group in the arginine residue in DAFP-1 between a strong enhancer and 1,2-cyclohexanedione was suggested by the partial retrieval of the enhancing ability of the modified DAFP-1 in the presence of a strong enhancer (Figure 8). The stronger the enhancer presented, the more efficient was the competition of the enhancer with the arginine residue in DAFP-1, and the greater the enhancement effect on the modified DAFP-1 (Figures 5-7). The weaker enhancers (e.g., glycerol), however, were not able to compete for the arginine residue, resulting in the complete loss of their enhancement abilities. In the case of citrate, the simple ion pairs between the guanidinium cation and citrate are not very stable in aqueous solution as water molecules compete for the formation of the ion pairs by solvating the individual ions. The solvates of the guanidinium cation and citrate greatly decrease the stability of their ion pairs, leading to the break-up of the ion pairs. Other amino acids in DAFP-1 may assist the formation of the ion pairs between the guanidinium cation and citrate by interacting with citrate through hydrogen bonds, van der Waals contacts and/or hydrophobic interactions to stabilize the products. The product peak of the 1:1 complex of DAFP-1 and citric acid was observed in the MALDI-TOF mass spectrum (Figure S5).
The identification of the participation of arginine in the recognition of enhancers in DAFP-1 may initiate a more complete mechanistic study of the enhancement effect in AFPs. Simple anions (e.g., Cl− and Br−) that have only ionic interaction with the guanidinium cation are usually very weak enhancers. Likewise, simple organic molecules (e.g., ethanol) having weak hydrogen-bonding and hydrophobic interactions with the arginine and DAFP-1 are also weak enhancers, while carbohydrates having multiple hydrogen bonds and hydrophobic interactions with the arginine and DAFP-1 are better enhancers. Only those molecules that can interact with the key factors in DAFP-1 more extensively by all the possible ways (namely, ionic binding, hydrogen-bonding, and hydrophobic interaction) function as efficient enhancers (e.g., EDTA and citrate) (15). This requirement is consistent with known arginine-ligand interactions (37-39).
The guanidinium cation is a good molecular recognition motif for oxoanion binding in nature (37, 40). The great basicity of the guanidine functional group allows it to play a key roles in recognition and catalysis (41, 42), while the guanidinium group of Arg is widely used as a mediator for noncovalent binding in the biological world, such as oxoanionic enzyme substrates and the base pairing in nucleic acids (40). In addition, guanidinium can form hydrogen bonds to polyhydroxy compounds (43) and the guanidinium-oxoanion ionic pair can assist drug transport across the cell membrane (44, 45). The results presented here bear on the general mechanism of arginine-ligand interactions, though the biological function of this interaction is novel – the enhancing function of small molecular mass solutes on DAFP-1 activity. We believe that 1) any good anion that pairs to the guanidinium cation in nature and has strong hydrogen bonding and hydrophobic interactions with AFPs is an excellent candidate to be a DAFP-1 enhancer; 2) a similar molecular mechanism is involved in the enhancement of the other isomers of DAFP having arginines; and 3) other potential key amino acid interaction pairs exist in other AFPs that lack arginine residues, such as certain of the D. canadensis DAFP isoforms and AFPs from Tenebrio molitor. Furthermore, uncovering the details of the binding is the key step to the design of super efficient enhancers for DAFP-1, by which we may finally make DAFP-1 and its enhancers more useful in applications. It is also important to address whether and how other residues recognize and orient water and/or ice interactions that help DAFP-1 bind tightly to the surface of the ice crystal and finally increase the TH activity.
Supplementary Material
ACKNOWLEDGMENT
We thank the Protein and Nucleic Acid Facility at Stanford University, Stanford, CA for MALDI-TOF mass spectrometry. VMD was developed by the Theoretical and Computational Biophysics Group in the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana-Champaign.
Abbreviations
- AFP
Antifreeze protein
- TH
Thermal hysteresis activity
- DSC
Differential scanning calorimetry
- DAFP
Dendroides canadensis antifreeze protein
- CHD
1,2-cyclohexanedione
- EDTA
Ethylenediaminetetraacetate
- DTPA
Diethylenetriaminepentaacetate
- TTHA
Triethylenetetramine-N,N,N′,N″,N‴,N‴-hexaacetate
- TCA
Tricarboxylate
- GTA
Glutarate
Footnotes
This study was supported by the National Institutes of Health Grant 1SC3GM086249-01 and the NIHRIMI program at California State University, Los Angeles (P20 MD001824-01).
BRIEFS. Role of Arginine in an Antifreeze Protein from the Beetle Dendroides canadensis in Molecular Recognition of Enhancers.
SUPPORTING INFORMATION AVAILABLE
HPLC profiles of the modification reaction of DAFP-1, the antifreeze activities of DAFP-1and the Arg-modified DAFP-1 in the presence of various enhancers, MALDI-TOF mass spectrum of DAFP-1 incubated with citrate. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.DeVries AL. Glycoproteins as biological antifreeze agents in Antarctic fishes. Science. 1971;172:1152–1155. doi: 10.1126/science.172.3988.1152. [DOI] [PubMed] [Google Scholar]
- 2.Raymond JA, DeVries AL. Adsorption inhibition as a mechanism of freezing resistance in polar fishes. Proc. Natl. Acad. Sci. U.S.A. 1977;74:2589–2593. doi: 10.1073/pnas.74.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia ZC, Davies PL. Antifreeze proteins: An unusual receptor-ligand interaction. Trends Biochem. Sci. 2002;27:101–106. doi: 10.1016/s0968-0004(01)02028-x. [DOI] [PubMed] [Google Scholar]
- 4.Raymond JA, Wilson P, DeVries AL. Inhibition of growth of nonbasal planes in ice by fish antifreezes. Proc. Natl. Acad. Sci. U.S.A. 1989;86:881–885. doi: 10.1073/pnas.86.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knight CA, Cheng CC, DeVries AL. Adsorption of alpha-helical antifreeze peptides on specific ice crystal surface planes. Biophys. J. 1991;59:409–418. doi: 10.1016/S0006-3495(91)82234-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng A, Merz KM., Jr. Ice-binding mechanism of winter flounder antifreeze proteins. Biophys. J. 1997;73:2851–2873. doi: 10.1016/S0006-3495(97)78315-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haymet ADJ, Ward LG, Harding MM, Knight CA. Valine substituted winter flounder ‘antifreeze’: Preservation of ice growth hysteresis. FEBS Lett. 1998;430:301–306. doi: 10.1016/s0014-5793(98)00652-8. [DOI] [PubMed] [Google Scholar]
- 8.DeVries AL, Cheng C-HC. In: Water and life: Comparative analysis of water relationships at the organismic, cellular and molecular level. Somero GN, Osmond CB, editors. Springer-Verlag; Berlin, Heidelberg: 1992. pp. 301–315. [Google Scholar]
- 9.Sicheri F, Yang DSC. Ice-binding structure and mechanism of an antifreeze protein from winter flounder. Nature. 1995;375:427–431. doi: 10.1038/375427a0. [DOI] [PubMed] [Google Scholar]
- 10.Andorfer CA, Duman JG. Isolation and characterization of cDNA clones encoding antifreeze proteins of the pyrochroid beetle Dendroides canadensis. J. Insect Physiol. 2000;46:365–372. doi: 10.1016/s0022-1910(99)00189-4. [DOI] [PubMed] [Google Scholar]
- 11.Li N, Chibber BAK, Castellino FJ, Duman JG. Mapping of disulfide bridges in antifreeze proteins from overwintering larvae of the beetle Dendroides canadensis. Biochemistry. 1998;37:6343–6350. doi: 10.1021/bi972853i. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Duman JG. A thaumatin-like protein from larvae of the beetle Dendroides canadensis enhances the activity of antifreeze proteins. Biochemistry. 2006;45:1278–1284. doi: 10.1021/bi051680r. [DOI] [PubMed] [Google Scholar]
- 13.Li N, Andorfer C, Duman J. Enhancement of insect antifreeze protein activity by solutes of low molecular mass. J. Exp. Biol. 1998;201:2243–2251. doi: 10.1242/jeb.201.15.2243. [DOI] [PubMed] [Google Scholar]
- 14.Amornwittawat N, Wang S, Banatlao J, Chung M, Velasco E, Duman JG, Wen X. Effects of polyhydroxy compounds on beetle antifreeze protein activity. Biochim. Biophys. Acta. 2009;1794:341–346. doi: 10.1016/j.bbapap.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amornwittawat N, Wang S, Duman JG, Wen X. Polycarboxylates enhance beetle antifreeze protein activity. Biochim. Biophys. Acta. 2008;1784:1942–1948. doi: 10.1016/j.bbapap.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miura K, Ohgiya S, Hoshino T, Nemoto N, Suetake T, Miura A, Spyracopoulos L, Kondo H, Tsuda S. NMR analysis of type III antifreeze protein intramolecular dimer. Structural basis for enhanced activity. J. Biol. Chem. 2001;276:1304–1310. doi: 10.1074/jbc.M007902200. [DOI] [PubMed] [Google Scholar]
- 17.Takamichi M, Nishimiya Y, Miura A, Tsuda S. Fully active QEA isoform confers thermal hysteresis activity on a defective SP isoform of type III antifreeze protein. FEBS J. 2009;276:1471–1479. doi: 10.1111/j.1742-4658.2009.06887.x. [DOI] [PubMed] [Google Scholar]
- 18.Duman JG, Serianni AS. The role of endogenous antifreeze protein enhancers in the hemolymph thermal hysteresis activity of the beetle Dendroides canadensis. J. Insect Physiol. 2002;48:103–111. doi: 10.1016/s0022-1910(01)00150-0. [DOI] [PubMed] [Google Scholar]
- 19.Zelent B, Bryan MA, Sharp KA, Vanderkooi JM. Influence of surface groups of proteins on water studied by freezing/thawing hysteresis and infrared spectroscopy. Biophys. Chem. 2009;141:222–230. doi: 10.1016/j.bpc.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Wu DW, Duman JG, Xu L. Enhancement of insect antifreeze protein activity by antibodies. Biochim. Biophys. Acta. 1991;1076:416–420. doi: 10.1016/0167-4838(91)90485-i. [DOI] [PubMed] [Google Scholar]
- 21.Duman JG, Wu DW, Olsen TM, Urrutia ME, Tursman D. Thermal-hysteresis proteins. Adv. Low Temp. Biol. 1993;2:131–182. [Google Scholar]
- 22.Bates PA, Kelley LA, MacCallum RM, Sternberg MJE. Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3DPSSM. Proteins. 2001:39–46. doi: 10.1002/prot.1168. [DOI] [PubMed] [Google Scholar]
- 23.Bates PA, Sternberg MJE. Model building by comparison at CASP3: Using expert knowledge and computer automation. Proteins. 1999:47–54. doi: 10.1002/(sici)1097-0134(1999)37:3+<47::aid-prot7>3.3.co;2-6. [DOI] [PubMed] [Google Scholar]
- 24.Contreras-Moreira B, Bates PA. Domain Fishing: a first step in protein comparative modelling. Bioinformatics. 2002;18:1141–1142. doi: 10.1093/bioinformatics/18.8.1141. [DOI] [PubMed] [Google Scholar]
- 25.Levitt M, Hirshberg M, Sharon R, Laidig KE, Daggett V. Calibration and testing of a water model for simulation of the molecular dynamics of proteins and nucleic acids in solution. J. Phys. Chem. B. 1997;101:5051–5061. [Google Scholar]
- 26.Plimpton S. Fast Parallel Algorithms for Short-Range Molecular-Dynamics. J. Comput. Phys. 1995;117:1–19. [Google Scholar]
- 27.Case DA. Amber and its associated force fields: An update. Abstr. Pap. Am. Chem. Soc. 2002;223:U475–U475. [Google Scholar]
- 28.Humphrey W, Dalke A, Schulten K. VMD -Visual Molecular Dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 29.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 30.Patthy L, Smith EL. Reversible modification of arginine residues. Application to sequence studies by restriction of tryptic hydrolysis to lysine residues. J. Biol. Chem. 1975;250:557–564. [PubMed] [Google Scholar]
- 31.Wilbur DS, Hamlin DK, Meyer DL, Mallett RW, Quinn J, Vessella RL, Press OW. Streptavidin in antibody pretargeting. 3. Comparison of biotin binding and tissue localization of 1,2-cyclohexanedione and succinic anhydride modified recombinant streptavidin. Bioconjug. Chem. 2002;13:611–620. doi: 10.1021/bc015574n. [DOI] [PubMed] [Google Scholar]
- 32.Li N, Kendrick BS, Manning MC, Carpenter JF, Duman JG. Secondary structure of antifreeze proteins from overwintering larvae of the beetle Dendroides canadensis. Arch. Biochem. Biophys. 1998;360:25–32. doi: 10.1006/abbi.1998.0930. [DOI] [PubMed] [Google Scholar]
- 33.Wu DW, Duman JG. Activation of antifreeze proteins from larvae of the beetle Dendroides canadensis. J. Comp. Physiol. B. 1991;161:279–283. [Google Scholar]
- 34.Soon Kim K, Qian L. Synthesis of 4,4-difluoro-L-arginine. Tetrahedron Lett. 1993;34:7195–7196. [Google Scholar]
- 35.Giralt E, Gordo S, Belda I, Pujals S, Martinell M, Salvatella X, Fernández--Carneado J. Understanding Biology Using Peptides. Springer; New York: 2006. pp. 649–650. [Google Scholar]
- 36.Berg JM, Tymoczko JL, Stryer L. Biochemistry. 6th Edition. W. H. Freeman and Company; New York: 2006. [Google Scholar]
- 37.Schmuck C. How to improve guanidinium cations for oxoanion binding in aqueous solution?: The design of artificial peptide receptors. Coord. Chem. Rev. 2006;250:3053–3067. [Google Scholar]
- 38.Fersht AR. The hydrogen bond in molecular recognition. Trends Biochem. Sci. 1987;12:301–304. [Google Scholar]
- 39.Fersht AR, Shi J-P, Knill-Jones J, Lowe DM, Wilkinson AJ, Blow DM, Brick P, Carter P, Waye MMY, Winter G. Hydrogen bonding and biological specificity analysed by protein engineering. Nature. 1985;314:235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- 40.Hannon CL, Anslyn EV. Bioorganic Chemistry Frontiers. Springer Verlag; Berlin: 1993. p. 193. [Google Scholar]
- 41.Cuatrecasas P, Wilchek M, Anfinsen CB. Action of staphylococcal nuclease on synthetic substrates. Biochemistry. 1969;8:2277–2284. doi: 10.1021/bi00834a007. [DOI] [PubMed] [Google Scholar]
- 42.Tucker PW, Hazen EE, Cotton FA. Staphylococcal nuclease reviewed: A prototypic study in contemporary enzymology. Mol. Cell. Biochem. 1979;23:67–86. doi: 10.1007/BF00226229. [DOI] [PubMed] [Google Scholar]
- 43.Li G, Gouzy MFO, Fuhrhop H., Jr. Host-Guest Chemistry: Mimetic approaches to study carbohydrate recognition. Top. Curr. Chem. 2002;218:133–158. [Google Scholar]
- 44.Mitchell DJ, Steinman L, Kim DT, Fathman CG, Rothbard JB. Polyarginine enters cells more efficiently than other polycationic homopolymers. J. Pept. Res. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- 45.Ryser HJP, Hancock R. Histones and basic polyamino acids stimulate the uptake of albumin by tumor cells in culture. Science. 1965;150:501–503. doi: 10.1126/science.150.3695.501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.