Abstract
The eukaryotic replisome is a crucial determinant of genome stability, but its structure is still poorly understood. We found previously that many regulatory proteins assemble around the MCM2-7 helicase at yeast replication forks to form the replisome progression complex (RPC), which might link MCM2-7 to other replisome components. Here, we show that the RPC associates with DNA polymerase α that primes each Okazaki fragment during lagging strand synthesis. Our data indicate that a complex of the GINS and Ctf4 components of the RPC is crucial to couple MCM2-7 to DNA polymerase α. Others have found recently that the Mrc1 subunit of RPCs binds DNA polymerase epsilon, which synthesises the leading strand at DNA replication forks. We show that cells lacking both Ctf4 and Mrc1 experience chronic activation of the DNA damage checkpoint during chromosome replication and do not complete the cell cycle. These findings indicate that coupling MCM2-7 to replicative polymerases is an important feature of the regulation of chromosome replication in eukaryotes, and highlight a key role for Ctf4 in this process.
Keywords: Ctf4, DNA polymerase α, GINS, MCM2-7, replisome progression complex
Introduction
The replication of large chromosomes that are compacted into chromatin is a major challenge for the proliferation of eukaryotic cells. The presence of single strand DNA at replication forks makes them vulnerable to attack by nucleases, and defects in chromosome replication are a major source of genome instability. To combat these dangers and preserve the integrity of the chromosomes, eukaryotic cells have evolved mechanisms that regulate the progression of DNA replication forks, monitor defects in DNA synthesis, and deal with problems when they occur (DePamphilis, 2006; Branzei and Foiani, 2007; Tourriere and Pasero, 2007; Aguilera and Gomez-Gonzalez, 2008).
Chromosome replication in eukaryotic cells is a highly complex process that is still understood much less well than in prokaryotes. One reason for the greater complexity in eukaryotes is that DNA synthesis is coupled to other processes at replication forks, such as the establishment of cohesion between sister chromatids and the duplication of complex epigenetic patterns of histone modifications throughout each chromosome. Our current understanding of eukaryotic DNA replication forks was inspired by earlier work with bacteria, in which chromosome replication is understood in much greater detail. In Escherichia coli, progression of DNA replication forks is dependent on the DnaB DNA helicase that unwinds the parental DNA duplex. The DnaB helicase is connected physically to a subset of the other replication factors to form a larger assembly called the replisome (Yao and O'Donnell, 2008). In addition to DnaB, the replisome includes the DNA polymerase that works on the leading and lagging strand templates, the primase that initiates each new DNA molecule, and the γ-complex that loads the β-clamp that serves as a processivity factor for the DNA polymerase. Interactions between the components of the replisome serve to ensure that unwinding by DnaB is coordinated with efficient DNA synthesis on the leading and lagging strand. This minimises the period during which single strand DNA is exposed at forks, as well as influencing the rate at which forks move (Pomerantz and O'Donnell, 2007).
The replisome in eukaryotic cells has been characterised in much less detail, and indeed the identities of the replicative helicase and the leading and lagging strand polymerases were only determined relatively recently (Kunkel and Burgers, 2008; Stillman, 2008). Each new DNA molecule is initiated by the DNA polymerase α complex, which contains both primase and DNA polymerase subunits. The leading strand is then extended by DNA polymerase epsilon (Pursell et al, 2007), whereas each Okazaki fragment on the lagging strand is completed by DNA polymerase delta (Nick McElhinny et al, 2008). Progression of eukaryotic DNA replication forks is dependent on the activity of the Mcm2-7 DNA helicase, the activity of which is regulated very carefully in vivo so that each origin fires once and a single copy of the genome is produced during each cell cycle (Blow and Dutta, 2005; Sclafani and Holzen, 2007). It seems likely that MCM2-7 will be coupled physically to replisome components such as DNA polymerases and primase in an analogous manner to the situation in E. coli, to regulate fork progression and help preserve genome stability. For example, work with yeast shows that depletion of deoxynucleotides slows the progression of MCM2-7 in addition to slowing the movement of DNA polymerases (Aparicio et al, 1997; Katou et al, 2003). But the nature of such links between MCM2-7 and other replisome components is poorly understood.
Previous work has shown that the MCM2-7 helicase associates with many other factors during the process of chromosome replication. These include Cdc45 and the four-protein GINS complex that are also important for fork progression (Tercero et al, 2000; Kanemaki et al, 2003; Kubota et al, 2003; Takayama et al, 2003; Pacek and Walter, 2004), and that form the ‘CMG' complex (Cdc45-MCM-GINS) or ‘unwindosome' together with MCM2-7 (Moyer et al, 2006; Pacek et al, 2006; Aparicio et al, 2009). Other factors associating with MCM2-7 in budding yeast include the checkpoint mediator Mrc1, the Tof1–Csm3 complex that is required for forks to pause at protein–DNA barriers, the histone chaperone FACT, the type I topoisomerase Top1, and Mcm10 and Ctf4 that are known to bind DNA polymerase α (Homesley et al, 2000; Katou et al, 2003; Ricke and Bielinsky, 2004; Nedelcheva et al, 2005; Gambus et al, 2006). By purifying GINS from budding yeast cell extracts, we found that all of the above proteins form a very large assembly called the replisome progression complex or RPC (Gambus et al, 2006). The RPC only exists during S-phase, occurs on chromatin, requires the prior loading of MCM2-7 at origins during G1-phase, and persists for as long as DNA replication forks are still present when nucleotide production is inhibited (Gambus et al, 2006). It thus appears that the RPC represents the fraction of MCM2-7 helicase that is present at DNA replication forks and is therefore likely to be a key component of the replisome.
The consequences of RPC formation are poorly understood but are likely to include activation of the MCM2-7 helicase by recruitment of other factors required for fork progression. The Cdc45 protein is required in addition to MCM2-7 for the unwinding of the parental DNA duplex at forks (Tercero et al, 2000; Pacek and Walter, 2004), and GINS is also important for fork progression and is required to preserve a stable interaction between MCM2-7 and Cdc45 (Kanemaki et al, 2003; Gambus et al, 2006). A second reason for RPC assembly could be to connect the MCM2-7 helicase to other components of the replisome such as DNA polymerases. Consistent with this view, recent work has shown that Mrc1 associates with DNA polymerase epsilon throughout the cell cycle and is a key determinant of the rate of progression of DNA replication forks (Szyjka et al, 2005; Tourriere et al, 2005; Hodgson et al, 2007; Lou et al, 2008). In addition, interaction between Mcm10 and Ctf4 has been shown to be important for recruitment of Pol α to chromatin during initiation of chromosome replication in extracts of Xenopus eggs (Zhu et al, 2007). But the subsequent contribution of Mcm10 and Ctf4 at forks to the stable link between DNA polymerase α and the MCM2-7 helicase was not addressed.
Here, we provide direct evidence that the RPC is associated at forks with DNA polymerase α. Our data indicate that a complex of GINS and Ctf4 has an important function in coupling MCM2-7 to Pol α. Ctf4 and Mrc1 are crucial for the normal regulation of chromosome replication and their combined absence leads to chronic activation of the DNA damage checkpoint and cell cycle arrest.
Results
The RPC associates with DNA polymerase α
As a first step towards characterising how the RPC might interact with other components of the replisome, we wanted to isolate the complex from budding yeast under very mild conditions, and then use mass spectrometry to identify any associated proteins in an unbiased manner. Originally, we purified RPCs from cell extracts that contained 300 or 700 mM potassium acetate, as the RPC is stable even at very high concentrations of salt (Gambus et al, 2006). It seemed likely, however, that the interaction of the RPC with other replisome components might not survive these rather harsh conditions. We repeated the purification, therefore, using a cell extract containing 50 mM potassium acetate, to try and preserve weaker ionic interactions between the RPC and other replication factors that might normally occur under physiological conditions. We found previously that GINS could still interact in the presence of 50 mM potassium acetate with all RPC components except Mcm10, which associates preferentially with the RPC in the presence of higher salt concentrations, perhaps through hydrophobic interactions (Gambus et al, 2006).
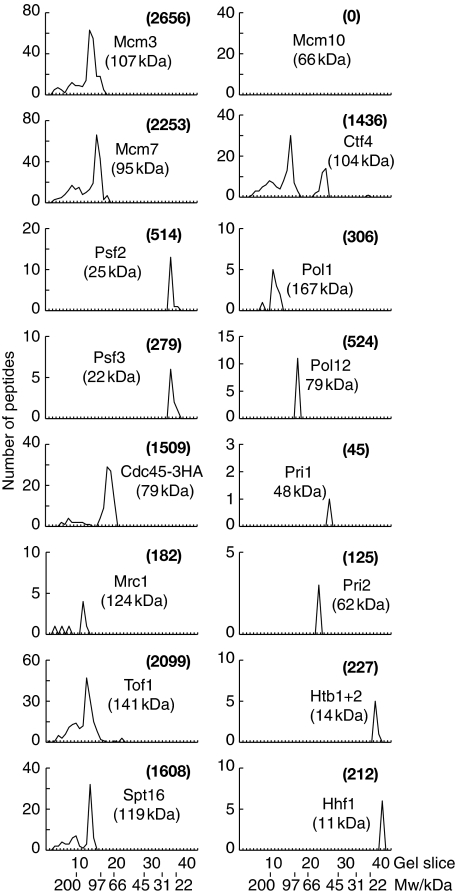
We used budding yeast cells in which the Sld5 subunit of GINS was fused to the TAP tag, and in which the Mcm4 subunit of the MCM2-7 helicase ended with five copies of the FLAG epitope. We grew an 8L asynchronous culture of TAP-SLD5 MCM4-5FLAG cells, together with a culture of MCM4-5FLAG control cells in which the Sld5 subunit of GINS was untagged. Cell extracts were generated in the presence of 50 mM potassium acetate and chromosomal DNA was digested to completion with DNase I. We then incubated the extracts with IgG Sepharose to isolate GINS from the TAP-SLD5 strain but not from the control, before purifying MCM2-7 from the resultant material to yield RPCs. The purified samples from the two strains were separated in an SDS–PAGE gel, and each lane was cut subsequently into 40 slices. Mass spectrometry was then used to identify all the factors that were only present in the sample purified from the TAP-SLD5 MCM4-5FLAG strain. As expected, the purified sample contained all of the previously identified RPC components with the exception of Mcm10 (Figure 1; Supplementary Figure 1). Strikingly, however, the purified RPC material contained four additional proteins that were not detected in our previous experiments using cell extracts containing 300 or 700 mM potassium acetate. These factors comprised the four subunits of DNA polymerase α (Figure 1), indicating that a fraction of Pol α exists in a common complex with MCM2-7 and GINS at DNA replication forks. These findings imply that one role of the RPC is to connect the MCM helicase to DNA polymerase α.
Figure 1.
The RPC associates with DNA polymerase α. A cell extract of TAP-SLD5 MCM4-5FLAG (YAG236-2) was generated in the presence of 50 mM potassium acetate and the RPC was purified as described in Materials and methods. The final material was separated in a 4–12% SDS–PAGE gradient gel and the lane was then cut into 40 slices. The graphs illustrate the total number of peptides that were identified by mass spectrometry in each slice of the gel, for a representative selection of the identified proteins. More details are shown in Supplementary Figure 1. The numbers in brackets indicate the combined Mascot score for each protein, corresponding to the sum of the unique peptide scores.
Ctf4 is required to couple DNA polymerase α to the RPC
To begin to explore the significance of the association of the RPC with DNA polymerase α, we wanted to determine which RPC subunits are responsible for the interaction. Work with yeast has shown that Mcm10, Ctf4 and FACT are all able to associate individually with Pol α (Miles and Formosa, 1992b; Wittmeyer and Formosa, 1997; Ricke and Bielinsky, 2004), and the orthologues of Mcm10 and Ctf4 in human cells and Xenopus laevis have also been found to interact with DNA polymerase α (Chattopadhyay and Bielinsky, 2007; Zhu et al, 2007). However, the contribution of these factors to the stable association at forks of DNA polymerase α with MCM2-7 helicase has not been addressed earlier. It is interesting to note that Pol α was found to co-purify with the RPC in our experiments under conditions in which Mcm10 is displaced from the RPC. This indicates that other factors contribute to the interaction between MCM2-7 and Pol α at DNA replication forks.
We found earlier that both Mcm10 and FACT co-purify with the MCM2-7 helicase during G1-phase and S-phase, whereas Ctf4 only interacts with MCM2-7 during S-phase as part of the RPC (Gambus et al, 2006). To determine when MCM2-7 associates with DNA polymerase α, we synchronised cells in G1-phase by treatment with mating pheromone, and then washed cells into fresh medium lacking mating pheromone so that they could enter S-phase (Figure 2A). At each time point, we generated cell extracts and digested the chromosomal DNA, before isolating the Mcm4 protein by immunoprecipitation and analysing the associated proteins by immunoblotting. In contrast to Mcm10 and FACT (Gambus et al, 2006), DNA polymerase α did not associate with the MCM2-7 helicase during G1-phase. Instead, Pol α co-purified with MCM2-7 during S-phase with similar kinetics to Ctf4 and other RPC components (Figure 2B). The interaction is likely to be independent of DNA, as the chromosomal DNA was digested to undetectable levels during the course of the experiment (Figure 2C), and the interaction was not affected by addition of ethidium bromide to the cell extract (Figure 2D). We note that Pol α is also recruited to chromatin later than Mcm10 during chromosome replication in extracts of Xenopus eggs, with similar timing to And-1/Ctf4 (Zhu et al, 2007). Taken together, these findings suggested that Ctf4 might be important to maintain the link between MCM2-7 and DNA polymerase α at replication forks.
Figure 2.
MCM2-7 associates with DNA polymerase α only during S-phase when RPCs are present. (A) An asynchronous culture of MCM4-5FLAG POL1-6HA PRI1-9MYC (YFJD62) was arrested in G1-phase at 24°C with mating pheromone before release into S-phase for the indicated times. DNA content was measured by flow cytometry. (B) Cell extracts were generated from the same experiment and treated with benzonase to digest chromosomal DNA as described in Materials and methods, before centrifugation and isolation of Mcm4-5FLAG by immunoprecipitation. The indicated proteins were analysed by immunoblotting. (C) Digestion of chromosomal DNA was monitored in an analogous experiment to that described in (A). Most DNA was digested during the initial 30′ treatment with Benzonase before centrifugation (compare lanes 1 and 2). The remainder was removed by centrifugation at 16 000 g for 30′ (lane 3). Even without centrifugation, all chromosomal DNA was degraded to undetectable levels during the time taken for the complete immunoprecipitation procedure (lane 4). (D) The experiment in (A) was repeated and samples taken 20′ after release from G1-phase. Benzonase and ethidium bromide were added to the initial extract as indicated, before immunoprecipitation of Mcm4.
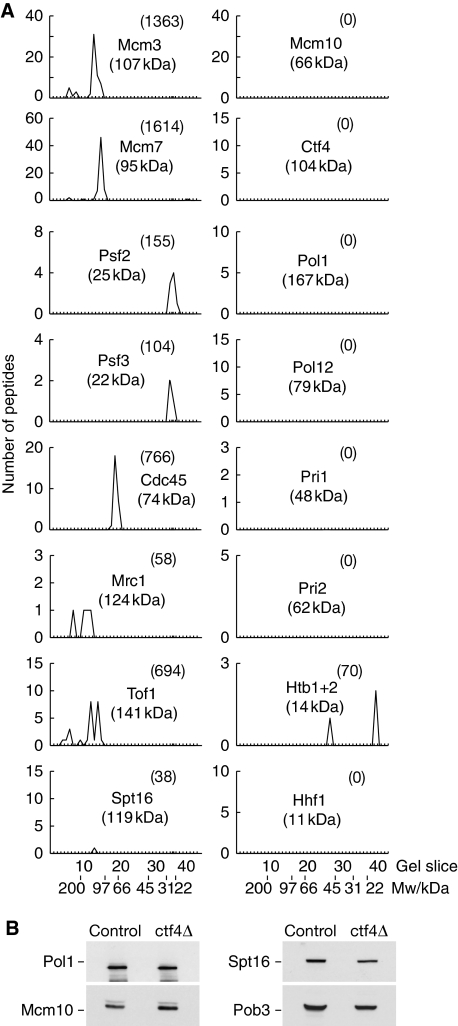
To test this idea more directly, we purified RPCs from extracts of ctf4Δ cells in the presence of 50 mM potassium acetate and then analysed the resulting material as above. As shown in Figure 3A and Supplementary Figure 2, Pol α did not co-purify with RPCs in the absence of Ctf4. In addition, however, the Spt16 and Pob3 subunits of the FACT complex were largely but not completely displaced from purified RPCs in the absence of Ctf4 (Figure 3A; Supplementary Figure 2). It was possible, therefore, that FACT also helps to maintain the link between MCM2-7 and Pol α, but our preliminary findings indicate that this is not the case (Magdalena Foltman and Karim Labib, unpublished data). Moreover, the stability of Pol α, Mcm10 and FACT in yeast cell extracts is independent of Ctf4 (Figure 3B). As Ctf4 is known to bind directly to Pol α (Miles and Formosa, 1992b), and both factors associate with MCM2-7 with similar kinetics (Figure 2; Gambus et al, 2006), these data indicate that Ctf4 has a direct role in preserving the link between PoI α and the RPC.
Figure 3.
Ctf4 is required for stable association of the RPC with DNA polymerase α. (A) RPC material was purified from ctf4Δ TAP-SLD5 MCM4-5FLAG (YAG374-2) and analysed as described above for Figure 1. The graphs illustrate a representative selection of the identified proteins, and the full data set is shown in Supplementary Figure 2. (B) The levels of Pol1, Mcm10, Spt16 and Pob3 were monitored by immunoblotting in extracts of control and ctf4Δ cells.
Ctf4 binds directly to the GINS component of the RPC
To understand how Ctf4 contributes to the link between Pol α and the MCM2-7 helicase within the RPC, it was necessary to determine how Ctf4 itself associates with the RPC. The interaction of most components of the RPC with either GINS or MCM2-7 only occurs during S-phase and is dependent on prior loading of the MCM2-7 helicase at origins of DNA replication. These interactions might require post-translational modifications that only occur during the initiation reaction at origins of DNA replication, or could involve multiple RPC components that are only brought together at origins when the whole complex is assembled. In contrast, Mcm10 and FACT co-purify with MCM2-7 (but not GINS) even from cells in G1-phase and these interactions are thought to be direct.
Ctf4 co-purifies with MCM2-7 only during S-phase (Figure 2; Gambus et al, 2006) but is unique amongst RPC subunits as a fraction of Ctf4 can associate with GINS throughout the cell cycle (Figure 4A). Consistent with this fact, Ctf4 still associates with GINS when RPC formation is blocked by inhibiting the prior loading of the MCM2-7 helicase at origins during G1-phase, in contrast to other RPC components such as MCM2-7 or Cdc45 (Figure 4B). These findings show that at least some fraction of GINS and Ctf4 form part of a common complex even away from the RPC. Moreover, we showed earlier that GINS is not only required to recruit Ctf4 to origins and to the MCM2-7 helicase, but is also needed subsequently to maintain the interaction of MCM2-7 with Ctf4 within the RPC (Gambus et al, 2006). Taken together, these data suggest that the association of Ctf4 with RPCs might involve direct interaction of Ctf4 with GINS.
Figure 4.
A fraction of Ctf4 can interact with GINS throughout the cell cycle and not just in RPCs. (A) Cultures of PSF2-TAP (YAG187-1) were grown at 24°C either asynchronously (Asyn.), or arrested in G1-phase with mating pheromone (G1), or released into S-phase from G1 in the presence of 0.2 M hydroxyurea for 60′ (S), or arrested in G2/M-phase with nocodazole (G2/M). Psf2-TAP was isolated by immunoprecipitation and the indicated proteins analysed by immunoblotting. (B) A cdc6Δ∷GAL-CDC6 TAP-SLD5 strain (YAG258-1) was grown at 24°C in rich medium containing galactose (YPGal), in parallel with a TAP-SLD5 control strain (YAG236-2), and cells were arrested in G2-M-phase with nocodazole. Expression of GAL-CDC6 was then repressed in fresh medium containing glucose (YPD) as well as nocodazole, before cells were released into fresh YPD medium to allow synchronization in the following G1-phase with mating pheromone. At this stage, the MCM helicase was assembled at origins into prereplicative complexes in the control strain (+ pre-RCs), but not in the GAL-CDC6 strain (− pre-RCs). Finally, cells were released into S-phase for 60′ in the presence of 0.2 M HU. TAP-Sld5 was isolated by immunoprecipitation and the indicated proteins analysed by immunoblotting.
To test whether GINS and Ctf4 do indeed bind each other directly, we determined whether these factors are able to interact in an extract of E. coli cells, in the absence of other eukaryotic proteins. We generated an E. coli strain that expressed the four GINS proteins as part of a common operon, and a second E. coli strain that expressed Ctf4. We then mixed the cells and generated a single cell extract containing GINS, Ctf4 and all the native E. coli proteins. We purified GINS from the extract by virtue of a GST tag on the Psf3 subunit of GINS, and found that GINS co-purified with Ctf4, in contrast to the 4000 native E. coli proteins (Figure 5A, E1). Ctf4 was then isolated from this material through a six-histidine tag at its amino terminus, and was again seen to co-purify specifically with the four GINS proteins (Figure 5A, E2). This shows that GINS and Ctf4 are able to interact directly to form a specific and stable complex.
Figure 5.
GINS and Ctf4 interact directly to form a stable complex. (A) Recombinant GINS was purified from an E. coli cell extract that also contained recombinant Ctf4 as described in Materials and methods, by virtue of a GST tag on the Psf3 subunit of GINS (E1). The purified material was separated in an SDS–PAGE gel that was stained with Coomassie blue. The band marked by an asterisk (*) was identified by mass spectrometry and corresponds to free GST that might have been derived as a degradation product from GST-Psf3. Ctf4 was then isolated from the purified GINS sample using a six-histidine tag at its amino terminus, and the figure shows the flow through of this purification step (FT), the washes (W1–W3), and the final eluate that represents purified GINS–Ctf4 complex (E2). The identity of the various bands was confirmed by mass spectrometry. (B) A similar experiment was performed using an E. coli extract containing recombinant Sld5–Psf2 subcomplex of GINS (with the GST tag on Sld5), together with 6His-Ctf4. (C) Summary of the interaction of truncated forms of recombinant Ctf4 with GINS in an E. coli extract, in an assay analogous to that described above. (D) An extract of E. coli was generated that contained recombinant Ctf4-ΔNT with 6His at its amino terminus, and recombinant GINS with the Streptag III epitope at the amino terminus of Psf3 as well as a truncated form of Psf1 (Psf1-ΔCT, amino acids 1–164) to improve visualisation of the four GINS proteins in the subsequent gel. StreptagIII-Psf3 was isolated from the extract and 6His-Ctf4ΔNT was then purified from the resultant material. The purified complex of GINS and Ctf4ΔNT was applied to a gel filtration column, and migrated with a retention volume of 50 ml (distinct from the void volume of 46 ml). The peak fractions were combined and concentrated before separation in an SDS–PAGE gel that was then stained with Coomassie blue. (E) Comparison of the migration through gel filtration columns of purified recombinant versions of GINS-Ctf4ΔNT, GINS and Ctf4ΔNT.
To investigate which subunits of GINS might be required to interact with Ctf4, we generated recombinant subcomplexes comprising Sld5–Psf1–Psf2 and Sld5–Psf2 and found that both could still interact directly with Ctf4 in extracts of E. coli cells (Figure 5B, and F van Deursen unpublished data). This indicates that Ctf4 interacts with either or both of the Sld5 and Psf2 subunits of the GINS complex. Unfortunately, Sld5 and Psf2 are not soluble on their own, and so a more complete understanding of the interaction between Ctf4 and Sld5–Psf2 awaits future structural studies of these factors.
Direct binding of Ctf4 to GINS and the amino terminus of Pol1 does not require the WD40 domain of Ctf4
We then investigated which domains of the Ctf4 protein are required for its interaction with GINS. The tertiary structure of Ctf4 has not been determined, but BLAST searches indicate that the protein begins with a WD40 domain, which is thought to mediate protein–protein interactions (Figure 5C). Orthologues of Ctf4 have a similar predicted structure although Ctf4 in higher eukaryotes has an additional HMG-like domain at the carboxy terminus.
We found that the WD40 domain is dispensable for the interaction of Ctf4 with GINS, which instead is mediated by the remainder of the protein (Figure 5C; Supplementary Figure 3). To illustrate this fact, we generated an extract of E. coli cells containing GINS and Ctf4-ΔNT, purified the GINS–Ctf4ΔNT complex in an analogous manner to that described above, and ran the purified material through a gel filtration column. This generated a single peak of protein that comprised the recombinant GINS–Ctf4ΔNT complex (Figure 5D), and that migrated more quickly than either GINS or Ctf4-ΔNT (Figure 5E). Note that the Ctf4 protein (and Ctf4-ΔNT) behaves as a multimer, and this is true of the endogenous protein in yeast extracts as well the purified recombinant protein (A Gambus and K Labib, unpublished data).
One of the original studies to identify Ctf4 showed that it could be isolated from a yeast extract using an affinity column bearing the Pol1 catalytic subunit of DNA polymerase α (Miles and Formosa, 1992b), although the nature of the interaction between Ctf4 and DNA polymerase α is not well understood. A later study combined Ctf4 in a two-hybrid assay with the first 550 amino acids of Pol1 that comprise a unique amino terminal region, before the exonuclease and polymerase domains, and found that they could interact (Zhou and Wang, 2004). We used libraries expressing random fragments of yeast proteins fused to the Gal4 transcriptional activation domain to screen through the two-hybrid assay for domains that interact with either the WD40 domain of Ctf4 (amino acids 1–383), or with the remainder of the protein (amino acids 461–927; Ctf4-ΔNT). The screen involving Ctf4-ΔNT identified 12 overlapping fragments from Pol1 that share a small region within the unique amino terminal domain of the Pol1 protein (Figure 6A). The same screen also identified three interacting clones representing the full-length Psf2 subunit of GINS, together with other factors that will be described elsewhere. In contrast, the WD40 domain was not found to interact with subunits of GINS or DNA polymerase α, but instead we isolated seven fragments from the Mms22 protein (Supplementary Figure 4), which forms part of an E3 ubiquitin ligase that is thought to regulate the progression of DNA replication forks (Zaidi et al, 2008). These findings indicate that Ctf4-ΔNT interacts with both GINS and with the amino terminus of Pol1, and suggest that the WD40 domain of Ctf4 has a different role that remains to be explored in the future.
Figure 6.
Direct interaction of Ctf4 with the amino terminus of Pol1 is independent of the WD40 domain of Ctf4. (A) Summary of fragments of Pol1 that were isolated in a two-hybrid screen with amino acids 461–927 of Ctf4 (Ctf4-ΔNT) as bait. (B) An extract of E. coli was generated that contained the indicated recombinant versions of Ctf4, each with 6His at the amino terminus, together with Pol1NT-Myc (left panel). The 6His-Ctf4 fragments were isolated from the extract (right panel) and the presence of Pol1NT-Myc and Ctf4 determined by immunoblotting.
We then studied the interaction between recombinant versions of Pol1-NT (amino acids 1–602) and Ctf4, using a similar approach to that described above for GINS. We generated extracts of E. coli cells that contained Pol1-NT and either full-length Ctf4, Ctf4-NT (the WD40 domain), or Ctf4-ΔNT. We isolated the various versions of Ctf4 through a six-histidine tag at the amino terminus of each protein, and used immunoblotting to determine whether they interacted with Pol1-NT. As shown in Figure 6B, Pol1-NT was able to interact with full-length Ctf4 and with Ctf4-ΔNT, but did not interact with the isolated WD40 domain itself. Taken together with the data described above, these findings indicate that Ctf4 binds GINS and the amino terminus of Pol1 through the region of the protein after the WD40 domain, whereas the WD40 domain probably has a separate function.
Cells lacking both Ctf4 and Mrc1 experience chronic checkpoint activation during S-phase and cannot complete the cell cycle
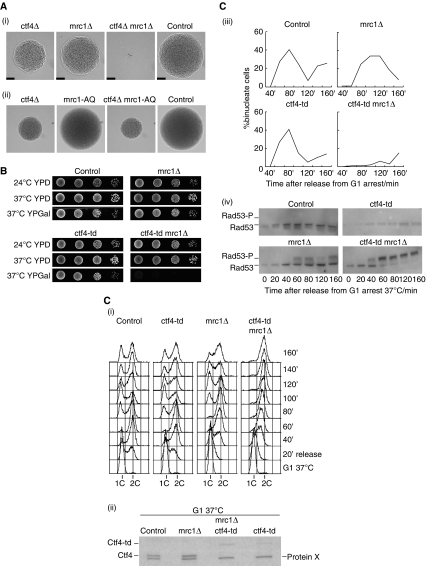
The data described above show that the Ctf4 component of the RPC has an important function in maintaining a stable link between DNA polymerase α and the MCM2-7 helicase during chromosome replication. Budding yeast cells can survive without this stable connection, however, and both early and late origins are activated in the absence of Ctf4 (Supplementary Figure 5). Nevertheless, ctf4Δ cells have a major delay in cell cycle progression, a high rate of genome instability, and are defective in establishing cohesion between sister chromatids (Kouprina et al, 1992; Miles and Formosa, 1992a; Hanna et al, 2001). Cells can also survive in the laboratory without the RPC components Mrc1, Top1, Tof1 and Csm3, but like Ctf4 these proteins are important for preserving genome integrity during chromosome replication. Importantly, budding yeast cells lacking any two of the above factors are viable with one exception—simultaneous loss of Mrc1 and Ctf4 is lethal (Warren et al, 2004) and germinating mrc1Δ ctf4Δ spores usually die in the first few cell cycles (Figure 7A (i)). It seems that the inviability of cells lacking both Mrc1 and Ctf4 is unlikely to be due to the role of Mrc1 in checkpoint activation, as the mrc1-AQ allele that is defective in checkpoint activation but otherwise competent for replication (Osborn and Elledge, 2003) is viable in combination with ctf4Δ (Figure 7A (ii); Supplementary Figure 6).
Figure 7.
Cells lacking both Ctf4 and Mrc1 undergo chronic checkpoint activation during chromosome replication and cannot complete the cell cycle. (A) (i) A ctf4Δ strain was crossed to mrc1Δ and the subsequent diploid was sporulated and subjected to tetrad analysis. Cells were grown at 30°C for 24 h and then photographed, and the images show a complete tetrad that illustrates the growth of control and mutant cells. The scale-bar corresponds to 50 μm. (ii) Similar analysis of a cross of ctf4Δ to mrc1-AQ. (B) Serial dilutions of the indicated strains were placed on the indicated media (YPD, yeast extract, peptone, dextrose; YPGal, yeast extract, peptone, galactose) and cells were grown for 48 h before the images were taken. (C) Asynchronous cultures of control (YKL200), mrc1Δ (YBH81), ctf4-td (YAG138-1) and ctf4-td mrc1Δ (YAG156-1) were grown at 24°C in YPRaffinose medium, and then synchronized in G1-phase with mating pheromone. Cells were transferred to YPGalactose medium for 35 min at 24°C to induce expression of the Ubr1 E3 ubiquitin ligase, and then shifted to 37°C for 1 h to inactivate Ctf4-td. Cells were transferred subsequently to fresh medium lacking mating pheromone to allow them to proceed with the cell cycle, and samples were taken at the indicated times and processed for flow cytometry (i), immunoblotting to monitor Ctf4—Protein X indicates a non-specific band in the immunoblot (ii), fluorescence microscopy to analyse nuclear division (iii), or used to generate protein extracts to monitor phosphorylation of the Rad53 checkpoint kinase. Rad53-P indicates the hyperphosphorylated form of Rad53 (iv).
Previously, it was not possible to characterise why cells die in the absence of Mrc1 and Ctf4. To circumvent this problem, we made a strain in which the Ctf4 protein is fused at its amino terminus to the heat-inducible degron (Dohmen et al, 1994; Labib et al, 2000), allowing us to inactivate Ctf4 in a conditional manner. Control cells, mrc1Δ cells, and the ctf4-td strain (td, temperature sensitive degron) were all able to grow at both 24 and 37°C in the presence or absence of the Ubr1 E3 ubiquitin ligase that induces ubiquitylation and degradation of the heat-inducible degron (Figure 7B). In contrast, the mrc1Δ ctf4-td strain was able to grow at 24 or 37°C in the absence of Ubr1, but was specifically unable to grow at 37°C in the presence of Ubr1 (Figure 7B). This indicated that we could use the mrc1Δ ctf4-td strain to analyse how cells die after removal of both Mrc1 and Ctf4.
We grew asynchronous cultures of control, mrc1Δ, ctf4-td and mrc1Δ ctf4-td cells at 24°C before synchronising cells in G1-phase with mating pheromone (Figure 7C (i)). After inducing expression of Ubr1, the cells were incubated at 37°C to inactivate Ctf4-td (Figure 7C (ii)), before washing into fresh medium lacking mating pheromone to allow cells to enter S-phase. Both control cells and ctf4-td cells replicated their chromosomes in the subsequent 40–60 min (Figure 7C (i)), and the peak of binucleate cells was observed after 80 min (Figure 7C (iii)). As predicted by earlier work (Alcasabas et al, 2001), chromosome replication began on schedule in the mrc1Δ strain but was completed more slowly, with all cells reaching a 2C DNA content by 60–80′ (Figure 7C (i)). Previous work showed that defective replication in the mrc1Δ strain is associated with transient hyperphosphorylation and thus activation of the Rad53 checkpoint kinase, in a manner dependent on the Rad9 protein that normally mediates activation of the DNA damage checkpoint (Alcasabas et al, 2001). We observed that a fraction of Rad53 became hyperphosphorylated between 60′ and 80′ as mrc1Δ cells completed replication (Figure 7C (iv), mrc1Δ), before subsequently dividing their nuclei (Figure 7C (iii)). This effect was not seen in control cells or in the ctf4-td strain (perhaps reflecting incomplete inactivation of Ctf4 in the latter case).
Chromosome replication in the mrc1Δ ctf4-td strain proceeded very slowly (Figure 7C (i)). Crucially, hyperphosphorylation of Rad53 was detected by 40′ and thus earlier than in the mrc1Δ strain (Figure 7C (iv), mrc1Δ ctf4-td). By 60′ almost all of the Rad53 protein was in the hyperphosphorylated form, and this remained the case until the end of the experiment (Figure 7C (iv), mrc1Δ ctf4-td). Consistent with this fact, most cells retained a single undivided nucleus throughout the experiment (Figure 7C (iii)). These findings indicate that cells lacking both Mrc1 and Ctf4 experience severe problems during chromosome replication that lead to chronic activation of the Rad53 checkpoint kinase. It thus appears that cells can survive if they lose either the Ctf4-dependent link between MCM and Pol α, or lose Mrc1 that might link RPCs to DNA polymerase epsilon, but cells are not able to survive chromosome replication in the absence of both of these regulatory components.
The ability of Ctf4 to bind GINS and DNA polymerase α is critically important in cells lacking Mrc1
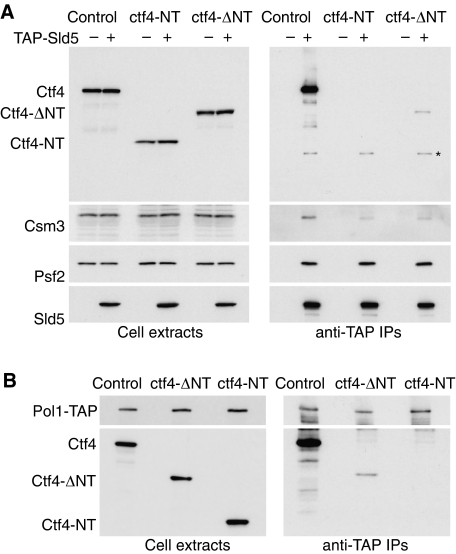
To explore whether the lethal problems experienced during S-phase in the absence of Mrc1 and Ctf4 are due to loss of the ability of Ctf4 to bind GINS and Pol1, and thus couple Pol α to MCM helicase, we made yeast strains expressing truncated forms of Ctf4 from the endogenous CTF4 promoter (Figures 8 and 9). We modified the CTF4 locus so that cells would either express the WD40 domain alone (Ctf4-NT), or the remainder of the protein (Ctf4-ΔNT). Figure 8 shows that the truncated forms of Ctf4 were expressed to a similar level to the full-length protein, and as expected we found that Ctf4-ΔNT but not Ctf4-NT could interact with both GINS and Pol1 (note that these interactions are less efficient than seen with recombinant proteins, perhaps due to problems in subcellular localisation of the truncated Ctf4 proteins, or to cell cycle defects in the mutant strains, or to competition with other factors that bind Pol1 and GINS).
Figure 8.
The WD40 domain of Ctf4 is not essential for interaction of Ctf4 with GINS and DNA polymerase α in vivo. (A) Cell extracts were generated from asynchronous cultures of the indicated strains, and TAP-Sld5 isolated by immunoprecipitation with IgG-Sepharose, before detection of the indicated proteins by immunoblotting. The asterisk in the Ctf4 immunoblot of the immunoprecipitates indicates TAP-Sld5, which binds in a non-specific manner to antibodies by virtue of the TAP tag. (B) A similar experiment was performed with asynchronous cultures of the indicated strains expressing Pol1-TAP.
Figure 9.
The ability of Ctf4 to bind GINS and DNA polymerase α is uniquely important in cells lacking Mrc1. (A) Spores of the indicated genotypes were identified by tetrad analysis and grown for 24 h at 30°C before imaging. The scale-bar corresponds to 50 μm. (B) Asynchronous cultures of the indicated strains were grown at 30°C and images were taken by phase contrast microscopy (left panels)—the scale-bar corresponds to 5 μm. Samples were also processed for flow cytometry (right panels). (C) (i) Cells were grown on the indicated medium for 48 h at 30°C before the images were taken. (ii) Strains lacking Ctf4 or with the indicated truncated versions of Ctf4 were crossed to a ctf18Δ strain and subjected to tetrad analysis. Images of the indicated double mutants are shown after 24 h growth at 30°C. (iii) Strains with the same range of ctf4 mutations were crossed to an mrc1Δ strain and processed as above.
We then compared the phenotypes of the ctf4Δ, ctf4-ΔNT and ctf4-NT strains. Cells expressing the truncated forms of Ctf4 were viable (Figure 9A), but showed a similar delay in progression through the cell cycle as ctf4Δ cells, with an accumulation of cells with a large bud and a 2C DNA content (Figure 9B). Both ctf4-ΔNT and ctf4-NT behaved similarly to ctf4Δ cells when treated with drugs like camptothecin that cause problems during chromosome replication (Figure 9C (i); we used the minimal dose that blocked growth of ctf4Δ cells). Moreover, ctf4Δ, ctf4-ΔNT and ctf4-NT all caused lethality in the first couple of cell cycles in the absence of the Ctf18 protein that forms an alternative form of the RFC complex that loads PCNA at replication forks (Figure 9C (ii)). Strikingly, however, only ctf4-NT that lacks the ability to bind GINS or DNA polymerase α was synthetic lethal with mrc1Δ (Figure 9C (iii); Supplementary Figure 6). In contrast, removal of the WD40 domain did not cause lethality in combination with mrc1Δ (Figure 9C (iii), ctf4-ΔNT mrc1Δ; Supplementary Figure 6). This indicates that the ability of Ctf4 to couple DNA polymerase α to MCM helicase through the GINS complex is particularly crucial in cells that lack Mrc1.
Discussion
Our data show for the first time that MCM2-7, GINS and DNA polymerase α all form part of a common complex at DNA replication forks. The demonstration that DNA polymerase α associates with the RPC thus represents an important step towards characterising the eukaryotic replisome. In the E. coli replisome, physical links between DNA helicase, DNA polymerase and primase regulate fork progression and allow unwinding to be coordinated with DNA synthesis. Our findings indicate that an important feature of the eukaryotic replisome will be that RPC components link the MCM helicase to other factors, such as DNA polymerase α that acts on the lagging strand template (Figure 10). Others have shown that Mrc1 is a prime candidate for a factor that couples MCM to DNA polymerase epsilon on the leading strand template (Lou et al, 2008), although this remains to be shown directly. Further characterisation of the eukaryotic replisome is likely to require new approaches and a better understanding of the interactions between the many eukaryotic DNA replication factors.
Figure 10.
Ctf4 couples DNA polymerase α to the RPC. Ctf4 binds directly to DNA polymerase α and to GINS in budding yeast, and is required to link DNA polymerase α to the RPC at replication forks. Mrc1 is a strong candidate for a factor that might couple DNA polymerase epsilon to the RPC, although this remains to be shown directly. The eukaryotic replisome will contain other components in addition to those depicted, but the interaction of such factors with the RPC and with DNA polymerases remains to be characterized in the future.
It is interesting that viruses of eukaryotic cells such as SV40 and HPV evolved a simplified form of eukaryotic DNA replication that uses a viral DNA helicase, perhaps to escape the more complex regulation of eukaryotic DNA replication forks. The use of SV40 T-antigen and HPV E1 protein might allow the viruses to dispense with many RPC components, but it is striking that both DNA helicases interact with Topoisomerase I and also with DNA polymerase α (Smale and Tjian, 1986; Park et al, 1994; Simmons et al, 1996; Clower et al, 2006). The latter interaction is analogous to the link between the MCM2-7 helicase and DNA polymerase α, but the situation at eukaryotic forks is clearly more complicated as the interaction is mediated by components of the RPC, probably to allow greater regulation of chromosome replication. Our data indicate that a complex of GINS and Ctf4 has an important function in coupling MCM2-7 helicase to DNA polymerase α in budding yeast. GINS, Ctf4 and Pol α all interact with MCM2-7 with similar kinetics, the interaction of Pol α with the RPC requires Ctf4, and the interaction of Ctf4 with MCM2-7 requires GINS. It is not clear at present how GINS interacts with MCM2-7 but this seems likely to be a direct interaction, analogous to the situation in the archaeons Sulfolobus solfataricus and Pyrococcus furiosus (Marinsek et al, 2006; Yoshimochi et al, 2008). Archaea lack a direct equivalent to DNA polymerase α, but it is interesting to note that Sulfolobus GINS has been found to interact with Primase (Marinsek et al, 2006), suggesting that archaeal GINS might also serve to link MCM to Primase on the lagging strand template.
Ctf4 is not essential for viability in budding yeast but is a key determinant of genome stability. Fission yeast cells lacking the Mcl1 protein that is orthologous to Ctf4 are unable to grow at high temperatures but are viable at 25 or 27°C (Williams and McIntosh, 2002; Mamnun et al, 2006), whereas depletion of And-1/Ctf4 from an extract of Xenopus eggs has been reported to reduce the rate of replication to 40% of the normal value (Zhu et al, 2007). Presumably, the interaction of DNA polymerase α with other factors such as Mcm10, FACT or other replisome components allows DNA synthesis to be completed in the absence of Ctf4, at least in yeasts. Nevertheless, budding yeast cells lacking Ctf4 form spontaneous subnuclear foci of the recombination factor Rad52 during S-phase and have greatly reduced viability in the absence of Rad52, indicating defective regulation of DNA replication forks (Kouprina et al, 1992; Alvaro et al, 2007).
A recent study found that inactivation of the Psf1 subunit of GINS in fission yeast did not block recruitment of DNA polymerase α to chromatin (Pai et al, 2009). It remains to be determined whether GINS interacts with Mcl1 in fission yeast, and whether this putative interaction is lost after inactivation of Psf1, but we note that budding yeast Psf1 is dispensable for the interaction of GINS with Ctf4 (Figure 5).
Our data indicate that the stable link between MCM2-7 and DNA polymerase α is particularly important in cells lacking Mrc1 that binds DNA polymerase epsilon. Chromosome replication is very slow in cells lacking both Ctf4 and Mrc1, and this is associated with chronic activation of the DNA damage checkpoint and with cell cycle arrest. The ability of Ctf4 to bind GINS and DNA polymerase α is required for viability in the absence of Mrc1, and these findings indicate that Ctf4's ability to couple MCM2-7 helicase to DNA polymerase α is crucial for the preservation of genome stability, together with the role played by the Mrc1 protein.
In contrast, the WD40 domain of Ctf4 is not required for interaction with GINS or DNA polymerase α and is dispensable in cells lacking Mrc1. Cells lacking the WD40 domain of Ctf4 do share other phenotypes with cells that completely lack Ctf4, and our findings indicate that the WD40 domain of Ctf4 has a distinct role that still remains to be characterised. In this regard, it is interesting that our two-hybrid screen identified the Mms22 protein as a potential partner for the WD40 domain. Recent work indicates that Mms22 associates with Mms1 and Rtt101 to form an E3 Ubiquitin ligase complex that is thought to act at certain kinds of stalled DNA replication fork to preserve genome stability (Duro et al, 2008; Zaidi et al, 2008). It seems clear that there is still much to be learnt in the future regarding the complex mechanisms by which eukaryotic cells preserve genome stability during chromosome replication.
Materials and methods
Yeast strains
The strains used in this study are listed in Table I. Cells were grown as described earlier (Gambus et al, 2006).
Table 1.
Strains used in this study (all based on W303-1a)
| Strain | Genotype |
|---|---|
| W303-1a | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 |
| YAG112-a | CTF4-9MYC (K.l.TRP1) pep4Δ∷URA3 ADE2 |
| YAG137-1 | ctf4-td (hphNT) GAL-UBR1 (HIS3) |
| YAG138-1 | ctf4-td (kanMX) GAL-UBR1 (HIS3) |
| YAG156-1 | ctf4-td (kanMX) GAL-UBR1 (HIS3) mrc1Δ∷hphNT |
| YAG161-1 | POL1-TAP (kanMX) CTF4-9MYC (K.l.TRP1) pep4Δ∷URA3 ADE2 |
| YAG172-1 | ctf4ΔNT-9MYC (K.l.TRP; kanMX) |
| YAG184 | MATa/MATα CTF4/ctf4ΔNT-9MYC (kanMX; K.l.TRP1) MRC1/mrc1Δ∷hphNT |
| YAG187-1 | PSF2-TAP (kanMX) MRC1-18MYC (K.l.TRP1) pep4Δ∷URA3 CDC45-3HA (K.l.TRP1) ADE2 |
| YAG190-1 | 9MYC-ctf4NT (K.l.TRP1; kanMX) |
| YAG192 | MATa/MATα CTF4/9MYC-ctf4NT (K.l.TRP1; kanMX) MRC1/mrc1Δ∷hphNT |
| YAG205-1 | POL1-TAP (kanMX) ctf4ΔNT-9MYC (K.l.TRP1; kanMX) pep4Δ∷URA3 ADE2 |
| YAG205-4 | ctf4ΔNT-9MYC (K.l.TRP1; kanMX) pep4Δ∷URA3 ADE2 |
| YAG206-2 | POL1-TAP (kanMX) 9MYC-ctf4NT (K.l.TRP1; kanMX) pep4Δ∷URA3 ADE2 |
| YAG207-3 | 9MYC-ctf4NT (K.l.TRP1; kanMX) pep4Δ∷URA3 ADE2 |
| YAG236-2 | TAP-SLD5 (kanMX) MCM4-5FLAG (hphNT) CDC45-3HA (TRP1) CSM3-18MYC (K.l.TRP1) pep4Δ∷URA3 ADE2 |
| YAG237-7 | MCM4-5FLAG (hphNT) CDC45-3HA (TRP1) CSM3-18MYC (K.l.TRP1) pep4Δ∷URA3 ADE2 |
| YAG249 | MATa/MATα CTF4/ctf4Δ∷kanMX MRC1/mrc1AQ-13MYC (his5+) |
| YAG258-1 | TAP-SLD5 (kanMX) MCM4-5FLAG (hphNT) CDC45-3HA (TRP1) CSM3-18MYC (K.l.TRP1) cdc6Δ∷GAL-CDC6 (LEU2) pep4Δ∷URA3 ADE2 |
| YAG374-2 | TAP-SLD5 (kanMX) pep4Δ∷URA3 MCM4-5FLAG (hphNT) ctf4Δ∷kanMX ADE2 |
| YAG398-1 | TAP-SLD5 (kanMX) 9MYC-ctf4NT (K.l.TRP1; kanMX) pep4Δ∷URA3 ADE2 |
| YAG399-2 | TAP-SLD5 (kanMX) ctf4ΔNT-9MYC (K.l.TRP1; kanMX) pep4Δ∷URA3 ADE2 |
| YASD429 | TAP-SLD5 (kanMX) CTF4-9MYC (K.l.TRP1) pep4Δ∷URA3 ADE2 |
| YBH81 | mrc1Δ∷hphNT |
| YDP14 | MATa/MATα CTF4/ctf4Δ∷kanMX CTF18/ctf18Δ∷hpHNT |
| YDP16 | MATa/MATα CTF18/ctf18Δ∷hpHNT CTF4/ctf4ΔNT-9MYC (K.l.TRP; kanMX) |
| YDP17 | MATa/MATα CTF18/ctf18Δ∷hpHNT CTF4/9MYC-ctf4NT (K.l.TRP1; kanMX) |
| YFJD62 | MCM4-5FLAG (hphNT) POL1-6HA (K.l.TRP1) PRI1-9MYC (K.l.TRP1) pep4Δ∷URA3 ADE2 |
| YKL200 | GAL-HA-UBR1 (HIS3) |
Purification and immunoprecipitation of proteins from yeast cell extracts
The RPC was isolated as described earlier in full detail (Gambus et al, 2006), except that we used 50 mM potassium acetate in all the buffers. Briefly, cell pellets were resuspended in a small volume of buffer before freezing in liquid nitrogen. Frozen cells were ground in the presence of liquid nitrogen using a Retsch RM100 mortar grinder, DNase I was used to digest chromosomal DNA in the resultant extract, before centrifugation and isolation of TAP-Sld5 on IgG-Sepharose. Mcm4-5FLAG was purified from the resultant material using M2 anti-FLAG agarose, and the final sample was precipitated with TCA before separation in a 4–12% NuPAGE gel (Invitrogen). The lane was cut into 40 bands and the protein content of each band determined by mass spectrometry after digestion with trypsin.
Immunoprecipitation experiments and immunoblots were performed as described earlier in detail (Gambus et al, 2006), except that we added 800 units of Benzonase (71206-3; Merck Biosciences) to the initial extract to digest chromosomal DNA, instead of using DNase I. After 30′ digestion at 4°C, the extract was spun at 16 000 g for 30′, and then at 100 000 g for 60′. The final supernatant did not contain any detectable chromosomal DNA and was used subsequently for immunoprecipitation. For the experiment in Figure 7C (iv), we detected Rad53 using a goat polyclonal antibody at a dilution of 1:1000 (Santa Cruz sc-6749).
Expression and purification of recombinant proteins in E. coli
We generated plasmids based on the ‘pET' series (Novagen) for expression of GINS or Ctf4 in E. coli. The plasmids are summarized in Table II and sequence details are available on request. The four GINS proteins (or the Sld5–Psf2 subcomplex) were expressed together from a single plasmid, and separate plasmids were then used to express derivatives of Ctf4 or Pol1-NT. To examine the interaction between GINS and Ctf4, or between Ctf4 and the amino terminus of Pol1, we grew one culture of E. coli cells expressing Ctf4 and a second culture expressing either GINS or Pol1-NT. After inducing expression of the recombinant proteins at 37°C, we mixed the cultures and generated a single pellet that was stored at −20°C. Subsequently this was used to generate a cell extract from which we purified GINS-Ctf4 or Ctf4-Pol1NT using Glutathione Sepharose (17-0756-01, GE Healthcare), Ni-NTA Agarose (30230, Qiagen), or Strep-Tactin Superflow (2-1206-025, IBA GmbH). Gel filtration was performed with Akta Explorer or Pharmacia SMART systems, using Superdex 200 or Superose 6 columns.
Table 2.
Plasmids used to express recombinant proteins in E. coli
| Plasmid | Expressed proteins |
|---|---|
| pFJD12 | GST-Psf3; Sld5; Psf1; Psf2 |
| pFJD7 | 6His-Ctf4 |
| pFJD24 | GST-Sld5; Psf2 |
| pFJD25 | GST-Sld5; Psf1; Psf2 |
| pFJD8 | 6His-Ctf4NT |
| pFJD9 | 6His-Ctf4ΔNT |
| pKL653 | StreptagIII-Psf3; Sld5; Psf1ΔCT; Psf2 |
| pKL710 | Pol1NT-Myc |
Flow cytometry
Samples were fixed with 70% ethanol and processed for flow cytometry as described earlier (Kanemaki et al, 2003), before analysis with a Becton Dickinson FACScan machine and CellQuest software.
Microscopy
Pictures of cells and colonies on agar plates were taken with a Nikon CoolPix 995 camera attached to a Nikon Eclipse E400 microscope. Phase contrast images of cells grown in liquid culture were taken using a Zeiss Axiovert 200 M microscope with a Cool Snap HQ camera (Photometrics) as described earlier (Sanchez-Diaz et al, 2008). For the experiment in Figure 7C (ii), we determined the proportion of binucleate cells by examining samples that had been processed for flow cytometry and were thus stained with propidium iodide after digestion of RNA. We examined 100 cells for each sample.
Two-hybrid analysis
The two-hybrid screens involving Ctf4 1–383 and Ctf4 461–927 were performed by the company Hybrigenics, using a library of random fragments of yeast genomic DNA.
Supplementary Material
Supplementary Information
Review Process File
Acknowledgments
We are very grateful to Cancer Research UK who funded this work, and thank the EMBO Young Investigator Programme for their support. AG is now supported by a Sir Henry Wellcome Postdoctoral Fellowship.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aguilera A, Gomez-Gonzalez B (2008) Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 9: 204–217 [DOI] [PubMed] [Google Scholar]
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ (2001) Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol 3: 958–965 [DOI] [PubMed] [Google Scholar]
- Alvaro D, Lisby M, Rothstein R (2007) Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet 3: e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio OM, Weinstein DM, Bell SP (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM complexes and Cdc45p during S phase. Cell 91: 59–69 [DOI] [PubMed] [Google Scholar]
- Aparicio T, Guillou E, Coloma J, Montoya G, Mendez J (2009) The human GINS complex associates with Cdc45 and MCM and is essential for DNA replication. Nucleic Acids Res 37: 2087–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Dutta A (2005) Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol 6: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M (2007) Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair (Amst) 6: 994–1003 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Bielinsky AK (2007) Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage during replication. Mol Biol Cell 18: 4085–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clower RV, Fisk JC, Melendy T (2006) Papillomavirus E1 protein binds to and stimulates human topoisomerase I. J Virol 80: 1584–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML (ed) (2006) DNA Replication and Human Disease. New York: CSHL Press [Google Scholar]
- Dohmen RJ, Wu P, Varshavsky A (1994) Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science 263: 1273–1276 [DOI] [PubMed] [Google Scholar]
- Duro E, Vaisica JA, Brown GW, Rouse J (2008) Budding yeast Mms22 and Mms1 regulate homologous recombination induced by replisome blockage. DNA Repair (Amst) 7: 811–818 [DOI] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K (2006) GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8: 358–366 [DOI] [PubMed] [Google Scholar]
- Hanna JS, Kroll ES, Lundblad V, Spencer FA (2001) Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol 21: 3144–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson B, Calzada A, Labib K (2007) Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol Biol Cell 18: 3894–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homesley L, Lei M, Kawasaki Y, Sawyer S, Christensen T, Tye BK (2000) Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev 14: 913–926 [PMC free article] [PubMed] [Google Scholar]
- Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K (2003) Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423: 720–725 [DOI] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Kouprina N, Kroll E, Bannikov V, Bliskovsky V, Gizatullin R, Kirillov A, Shestopalov B, Zakharyev V, Hieter P, Spencer F (1992) CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae [published erratum appears in Mol Cell Biol 1993 Nov;13(11):7202]. Mol Cell Biol 12: 5736–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Takase Y, Komori Y, Hashimoto Y, Arata T, Kamimura Y, Araki H, Takisawa H (2003) A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev 17: 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Burgers PM (2008) Dividing the workload at a eukaryotic replication fork. Trends Cell Biol 18: 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Tercero JA, Diffley JFX (2000) Uninterrupted MCM2-7 function required for DNA replication fork progression. Science 288: 1643–1647 [DOI] [PubMed] [Google Scholar]
- Lou H, Komata M, Katou Y, Guan Z, Reis C, Budd M, Shirahige K, Campbell J (2008) Mol Cell 32: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamnun YM, Katayama S, Toda T (2006) Fission yeast Mcl1 interacts with SCF(Pof3) and is required for centromere formation. Biochem Biophys Res Commun 350: 125–130 [DOI] [PubMed] [Google Scholar]
- Marinsek N, Barry ER, Makarova KS, Dionne I, Koonin EV, Bell SD (2006) GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep 7: 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles J, Formosa T (1992a) Evidence that POB1, a Saccharomyces cerevisiae protein that binds to DNA polymerase alpha, acts in DNA metabolism in vivo. Mol Cell Biol 12: 5724–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles J, Formosa T (1992b) Protein affinity chromatography with purified yeast DNA polymerase alpha detects proteins that bind to DNA polymerase. Proc Natl Acad Sci USA 89: 1276–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR (2006) Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA 103: 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelcheva MN, Roguev A, Dolapchiev LB, Shevchenko A, Taskov HB, Stewart AF, Stoynov SS (2005) Uncoupling of unwinding from DNA synthesis implies regulation of MCM helicase by Tof1/Mrc1/Csm3 checkpoint complex. J Mol Biol 347: 509–521 [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA (2008) Division of labor at the eukaryotic replication fork. Mol Cell 30: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn AJ, Elledge SJ (2003) Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev 17: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC (2006) Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21: 581–587 [DOI] [PubMed] [Google Scholar]
- Pacek M, Walter JC (2004) A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J 23: 3667–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CC, Garcia I, Wang SW, Cotterill S, Macneill SA, Kearsey SE (2009) GINS inactivation phenotypes reveal two pathways for chromatin association of replicative alpha and epsilon DNA polymerases in fission yeast. Mol Biol Cell 20: 1213–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park P, Copeland W, Yang L, Wang T, Botchan MR, Mohr IJ (1994) The cellular DNA polymerase alpha-primase is required for papillomavirus DNA replication and associates with the viral E1 helicase. Proc Natl Acad Sci USA 91: 8700–8704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz RT, O'Donnell M (2007) Replisome mechanics: insights into a twin DNA polymerase machine. Trends Microbiol 15: 156–164 [DOI] [PubMed] [Google Scholar]
- Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA (2007) Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science 317: 127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke RM, Bielinsky AK (2004) Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol Cell 16: 173–185 [DOI] [PubMed] [Google Scholar]
- Sanchez-Diaz A, Marchesi V, Murray S, Jones R, Pereira G, Edmondson R, Allen T, Labib K (2008) Inn1 couples contraction of the actomyosin ring to membrane ingression during cytokinesis in budding yeast. Nat Cell Biol 10: 395–406 [DOI] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM (2007) Cell cycle regulation of DNA replication. Annu Rev Genet 41: 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DT, Melendy T, Usher D, Stillman B (1996) Simian virus 40 large T antigen binds to topoisomerase I. Virology 222: 365–374 [DOI] [PubMed] [Google Scholar]
- Smale ST, Tjian R (1986) T-antigen-DNA polymerase alpha complex implicated in simian virus 40 DNA replication. Mol Cell Biol 6: 4077–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B (2008) DNA polymerases at the replication fork in eukaryotes. Mol Cell 30: 259–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyjka SJ, Viggiani CJ, Aparicio OM (2005) Mrc1 is required for normal progression of replication forks throughout chromatin in S. cerevisiae. Mol Cell 19: 691–697 [DOI] [PubMed] [Google Scholar]
- Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H (2003) GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev 17: 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JA, Labib K, Diffley JFX (2000) DNA synthesis at individual replication forks requires the essential initiation factor, Cdc45p. EMBO J 19: 2082–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourriere H, Pasero P (2007) Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst) 6: 900–913 [DOI] [PubMed] [Google Scholar]
- Tourriere H, Versini G, Cordon-Preciado V, Alabert C, Pasero P (2005) Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell 19: 699–706 [DOI] [PubMed] [Google Scholar]
- Warren CD, Eckley DM, Lee MS, Hanna JS, Hughes A, Peyser B, Jie C, Irizarry R, Spencer FA (2004) S-phase checkpoint genes safeguard high-fidelity sister chromatid cohesion. Mol Biol Cell 15: 1724–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DR, McIntosh JR (2002) mcl1+, the Schizosaccharomyces pombe homologue of CTF4, is important for chromosome replication, cohesion, and segregation. Eukaryot Cell 1: 758–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmeyer J, Formosa T (1997) The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol Cell Biol 17: 4178–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao NY, O'Donnell M (2008) Replisome dynamics and use of DNA trombone loops to bypass replication blocks. Mol Biosyst 4: 1075–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimochi T, Fujikane R, Kawanami M, Matsunaga F, Ishino Y (2008) The GINS complex from Pyrococcus furiosus stimulates the MCM helicase activity. J Biol Chem 283: 1601–1609 [DOI] [PubMed] [Google Scholar]
- Zaidi IW, Rabut G, Poveda A, Scheel H, Malmstrom J, Ulrich H, Hofmann K, Pasero P, Peter M, Luke B (2008) Rtt101 and Mms1 in budding yeast form a CUL4(DDB1)-like ubiquitin ligase that promotes replication through damaged DNA. EMBO Rep 9: 1034–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wang TS (2004) A coordinated temporal interplay of nucleosome reorganization factor, sister chromatin cohesion factor, and DNA polymerase alpha facilitates DNA replication. Mol Cell Biol 24: 9568–9579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Ukomadu C, Jha S, Senga T, Dhar SK, Wohlschlegel JA, Nutt LK, Kornbluth S, Dutta A (2007) Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev 21: 2288–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File