Abstract
The transcriptional response to damaging agents is of fundamental significance for understanding mechanisms responsible for cell survival and genome maintenance. However, how damage signals are transmitted to the transcriptional apparatus is poorly understood. Here we identify two new regulators of the UV response transcriptome: Snf1, a nutrient-sensing kinase, and Rad23, a nucleotide excision repair factor with no previously known function in transcriptional control. Over half of all UV-responsive genes are dependent on Snf1 or Rad23 for proper regulation. After irradiation, Snf1 targets the Mig3 repressor, a new effector of the UV response. Snf1 and Rad23 are both required for the displacement of Mig3 from the UV-activated HUG1 promoter, and Rad23's activity is functionally linked to the proteasome 19S regulatory particle. Our data reveal overlapping functions for Snf1 and Rad23 in UV-responsive transcriptional regulation and provide mechanistic insight into the action of these factors at a UV-activated promoter. These results also highlight how diverse environmental stimuli are processed by a limited repertoire of signalling molecules to result in tailored patterns of gene expression.
Keywords: AMPK, Mig3, Rad23, Snf1, UV response
Introduction
Cells are subjected to stress-inducing stimuli from the external environment, which vary in chemical complexity, duration, and dose. The proper response to cellular stress is essential for survival, and defects in stress response pathways are the underlying cause of many human disorders including cancer and diabetes (Luo et al, 2005; Vousden and Lane, 2007). In the face of great diversity in the external environment, cells respond with a more limited repertoire of signalling components. The response to stress requires the integration of multiple signals from the external and internal environment to generate a complex response tailored specifically to the physiological state of the cell. This involves coordination of signalling pathways and cellular processes to properly regulate downstream responses such as changes in gene expression. Thus, the cell reacts to diverse stimuli using a well-defined set of factors that ultimately control fundamental cellular processes such as metabolism, growth, and proliferation.
Although many of the basic signalling pathways and effectors involved in response to individual types of environmental stress such as nutrient starvation and DNA damage have been well studied, only recently have connections between these different pathways been established. For example, there is now evidence that AMP-activated protein kinase (AMPK) and p53 are involved in the response to both DNA damage and nutrient starvation (Feng et al, 2005; Jones et al, 2005; Kim et al, 2008). Studies in yeast show that DNA damage-induced cell cycle arrest is regulated by cAMP-dependent protein kinase (PKA), which functions in many of the same metabolic response pathways as the AMPK homologue, Snf1 (Searle et al, 2004). These findings suggest extensive crosstalk between nutrient-sensing and DNA damage response pathways that extends to all eukaryotes. A full understanding of these connections is imperative, as key players in these processes are emerging as important drug targets for cancer, diabetes, and cardiovascular disease (Hardie, 2007).
Snf1 kinase, the yeast homologue of the drug target AMPK, is an evolutionarily conserved sensor of cellular energy status and master regulator of metabolism (Hardie et al, 1998). The activation of Snf1 occurs when cells are grown in low glucose or non-optimal carbon sources and leads to transcriptional regulation of genes involved in glycolysis, glucose transport, and alternative carbon source utilization, among other processes. Snf1 acts on gene expression primarily by regulating target transcription factors such as the Mig1 repressor (Treitel et al, 1998; Papamichos-Chronakis et al, 2004). Snf1 is also involved in responses to nitrogen limitation, heat shock, salt, and genotoxic stress (Sanz, 2003; Dubacq et al, 2004; Hong and Carlson, 2007). Specifically, Snf1 is important for cellular resistance to the genotoxic agent, hydroxyurea (Dubacq et al, 2004). Although the putative transcription factor, Mig3, has been implicated as a target (Dubacq et al, 2004), functions of Snf1 and Mig3 in response to genotoxic stress remain unclear.
On the basis of these observations, we sought to explore the role of Snf1 in the DNA damage response in yeast. Remarkably, half of the UV-induced transcriptional response is dependent on Snf1. This function of Snf1 has remained elusive because it is phenotypically redundant with Rad23, a proteasome-associated nucleotide excision repair (NER) factor. Our data show that this redundancy is based on a previously undiscovered function of Rad23 in transcription regulation. We offer a molecular mechanism for the cooperative action by Snf1 and Rad23 through regulation of Mig3 occupancy at a UV-activated promoter. Additional evidence suggests that Rad23 regulates transcription through association with the proteasome 19S regulatory particle (RP). These findings not only reveal novel functions for Snf1 and Rad23, but also provide a molecular framework for understanding how kinase signalling and the 19S RP are integrated in transcriptional control.
Results
Snf1 kinase regulates UV-induced gene expression
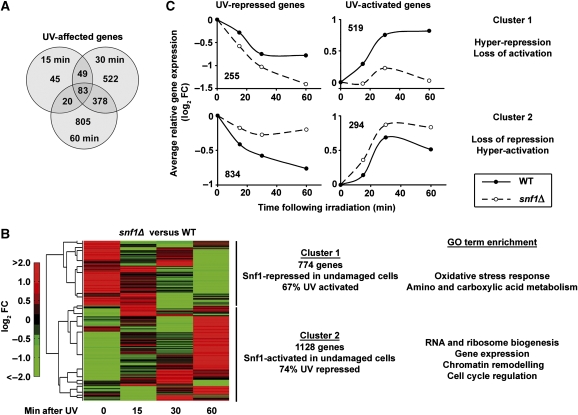
Genome-wide expression analysis was used to test the hypothesis that Snf1 regulates gene expression in response to DNA damage. We carried out microarray analysis on WT and snf1Δ cells before and at multiple time points after UV irradiation (see Materials and methods section). A total of 1903 genes were significantly affected at 15, 30 or 60 min after irradiation of WT cells (Figure 1A and Supplementary Table S1), with no bias for either gene activation or repression. Gene sets for all three time points were enriched for factors important to arrest cell growth, promote intermediary metabolism, and deal with DNA damage (Supplementary Table S2). These results are consistent with and extend previous studies regarding DNA damaging agents (Gasch et al, 2001; Birrell et al, 2002). In addition, many genes involved in metabolism and cell growth were regulated by Snf1 in the absence of DNA damage (Supplementary Table S3), showing that Snf1 has unanticipated effects on nutrient-related genes even in glucose-fed cells.
Figure 1.
Snf1 regulates UV-induced transcription. (A) Genes affected by UV irradiation in WT cells. Microarray analysis was carried out in duplicate, and genes with a significant change (false discovery rate <0.05) at the indicated time point after irradiation with 100 J/m2 of UV light were counted. (B) Two-dimensional hierarchical clustering of log-transformed fold change (FC) in gene expression between snf1Δ and WT cells at the indicated time point after irradiation. All 1903 UV-responsive genes were clustered. (C) Average UV response of all genes in Cluster 1 or 2 from panel B separated by UV activation or repression. Values are the average log-transformed FC of all genes in each category when comparing WT or snf1Δ cells after damage with the congenic undamaged cells.
Two-dimensional hierarchical clustering of UV-responsive genes showed a striking relationship between Snf1 and the UV response (Figure 1B). Most UV-responsive genes were reciprocally regulated by Snf1 in undamaged versus damaged cells. Cluster 1 comprised 774 genes repressed by Snf1 in undamaged cells, which conversely required Snf1 for activation in response to UV irradiation. The 1128 genes in Cluster 2 showed the opposite pattern; Snf1 was required to activate these genes in undamaged cells and repress them following UV exposure (Figure 1B). This binary regulatory pattern indicates that Snf1 is required for proper regulation of UV-responsive genes in both the undamaged and damaged states.
This conclusion is supported by the results in Figure 1C, which show the average UV response of all UV-regulated genes separated by cluster. Most UV-repressed genes showed, on average, a loss of repression in snf1Δ cells (lower left panel). Similarly, the majority of UV-induced genes showed a substantial loss of activation in this mutant (upper right panel). A small proportion of UV-responsive genes had an amplified UV response (Figure 1C, upper left and lower right panels). Strikingly, the gene regulatory patterns make intuitive biological sense: Cluster 1 genes are involved in processes, such as stress response and intermediary metabolism, which are kept in check by Snf1 in the absence of DNA damage and rely on Snf1 for UV activation (Figure 1B and C). Cluster 2 genes are enriched in factors involved in cell cycle regulation, ribosome biogenesis, and transcription whose expression in unirradiated cells and repression after DNA damage is Snf1 dependent (Figure 1B and C).
Snf1 is transiently activated following UV irradiation
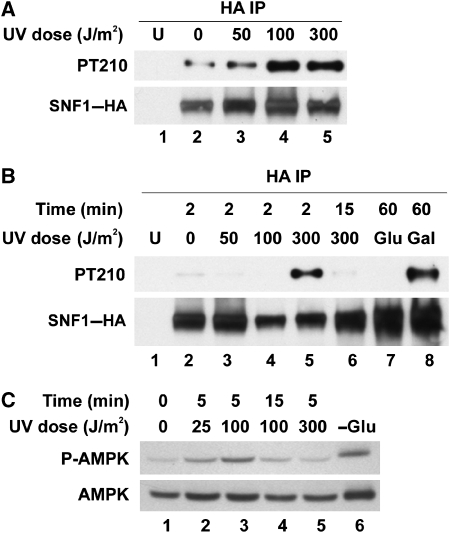
To determine whether Snf1 is activated after UV irradiation, a phospho-specific antibody was used to detect the phosphorylation of Snf1 T210, a hallmark of the activated kinase (McCartney and Schmidt, 2001). UV-induced activation of Snf1 was observed at 100 and 300 J/m2 UV light, though activation at the lower dose was not detectable in all experiments (Figure 2A and B). In proportion to the total level of Snf1, levels of activation in response to 300J/m2 UV irradiation were somewhat higher than those in cells switched to galactose for 1 h (Figure 2B). Activation occurred rapidly after irradiation and was undetectable after 15 min (Figure 2B). Similar to Snf1 in yeast, human AMPK is activated by the phosphorylation of a conserved threonine residue (Stein et al, 2000). Transient activation of AMPK by UV light was also observed in human cells (Figure 2C), indicating that this response is conserved.
Figure 2.
Transient activation of Snf1 following UV irradiation. (A and B) Phosphorylation of Snf1 Thr 210 after UV irradiation. HA-tagged Snf1 was immunoprecipitated from whole cell extracts and western blots of the precipitated material were probed for Snf1–HA or the phosphorylated form of Snf1 (PT210). Samples were taken 2 min after irradiation (A) or at the indicated times (B). Cells switched to either fresh glucose- or galactose-containing medium for 1 h were controls for Snf1 activation (B, lanes 7 and 8). U, untagged control. (C) Activation of human AMPK in response to UV irradiation. HEK-293T cells were irradiated and collected at the indicated time points. Western blotting was carried out for total AMPK and the phospho-T172 form of the enzyme. Growth for 4 h in the absence of glucose was used as a positive control for AMPK activation (lane 6).
Snf1 kinase cooperates with Rad23 in the UV response
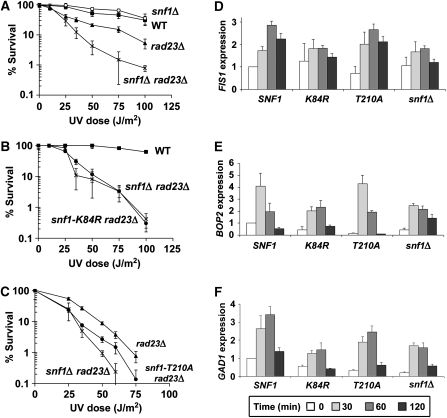
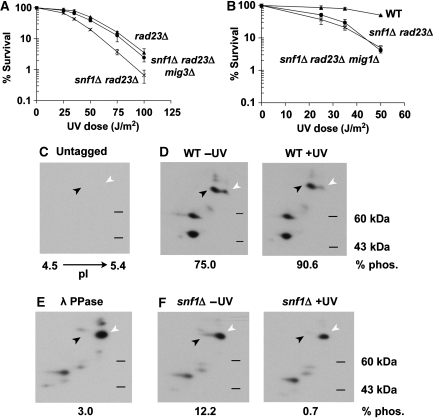
Although Snf1 is required for proper expression of a large proportion of UV-responsive genes, snf1Δ cells were not UV sensitive (Figure 3A). We reasoned that the function of Snf1 in UV resistance may be redundant with, and thus masked by, another factor involved in the UV response, a hypothesis consistent with previous observations highlighting the redundancy in the NER machinery (Ramsey et al, 2004). We turned to the DNA damage recognition protein, Rad23, because it shows synthetic UV sensitivity with other factors, including Elc1 (Ramsey et al, 2004), a protein which physically associates with a regulatory subunit of the Snf1 kinase complex (Jackson et al, 2000; Ramsey et al, 2004). Although yeast cells lacking RAD23 were UV sensitive, snf1Δ rad23Δ double mutants were 10-fold more sensitive to irradiation than rad23Δ cells (Figure 3A). This genetic interaction was specific to the UV response, as deletion of RAD23 did not exacerbate other phenotypes of snf1Δ cells (Supplementary Figure S1). This suggests that Snf1 contributes specifically to UV resistance in a manner redundant with Rad23 function.
Figure 3.
Snf1 kinase activity is important for UV resistance of rad23Δ cells and induction of Snf1-dependent UV responsive genes. (A) UV sensitivity of indicated yeast strains. Percent survival signifies the number of colonies on agar plates after irradiation relative to unirradiated controls. Experiments were done in triplicate and error bars represent the standard deviation (B and C) UV sensitivity of cells harbouring a kinase-dead snf1 point mutant, snf1-K84R, (B), or an activation-deficient snf1 mutant, snf1-T210A, (C). Results were obtained as in (A). (D–F) Expression of three UV-induced, Snf1-dependent genes in snf1Δ cells harbouring plasmid-borne WT SNF1, the kinase-dead snf1-K84R allele, the activation-deficient snf1-T210A allele or vector only control. Expression levels were determined by quantitative RT–PCR using primer sets specific to the FIS1 ORF (D), BOP2 ORF (E), or GAD1 ORF (F). RNA levels were normalized to ACT1 expression and were averaged from three independent experiments. Error bars represent the standard deviation.
We also verified genetically that kinase activity was important for Snf1-mediated UV resistance. The snf1-K84R allele encodes a substitution in the ATP binding domain that completely abrogates kinase activity (Celenza and Carlson, 1989). UV sensitivity of the snf1-K84R mutant in the rad23Δ background was identical to that of snf1Δ (Figure 3B), suggesting a requirement of Snf1 kinase activity for UV resistance. Different results were observed with the activation-defective snf1-T210A mutant. snf1-T210A rad23Δ cells had an intermediate phenotype compared with rad23Δ and snf1Δ rad23Δ cells (Figure 3C). This suggests that activation of Snf1 contributes to, but is not essential for the UV resistance conferred by Snf1, and that basal kinase activity contributes to UV survival as well. To further explore the function of Snf1 activation in the UV response, snf4Δ cells were analyzed. The Snf4 regulatory subunit is required for full activation of kinase activity under low glucose conditions but is not required for basal kinase activity (Celenza and Carlson, 1989; Leech et al, 2003). The deletion of SNF4 did not lead to the same synthetic UV phenotype as deletion of the catalytic subunit SNF1 (Supplementary Figure S2), supporting the conclusion that basal kinase activity is largely responsible for Snf1-mediated UV resistance. These observations are consistent with previous results regarding the function of Snf1 basal kinase activity in resistance to hydroxyurea (Dubacq et al, 2004). In contrast, a rad23Δ strain harbouring deletions of the three redundant kinases responsible for Snf1 T210 phosphorylation had a phenotype similar to snf1Δ rad23Δ cells (Supplementary Figure S2), suggesting the possibility that the three upstream kinases have other downstream targets important for UV resistance.
Next, we assayed UV-induced gene expression of Snf1-dependent genes in cells expressing WT SNF1 or one of the two snf1 point mutants, snf1-K84R or snf1-T210A. For all three genes tested, induction in the snf1-K84R mutant was equivalent to that seen in snf1Δ cells (Figure 3D–F), thus demonstrating the important role of Snf1 catalytic activity for full UV induction of these genes. Expression levels in the snf1-T210A mutant, however, more closely resembled those of WT cells. For FIS1 and BOP2, snf1-T210A cells were indistinguishable from WT (Figure 3D and E), whereas GAD1 induction levels in snf1-T210A cells were intermediate between those of WT and snf1Δ cells (Figure 3F). This correlates well with genetic data described above and altogether suggests that although basal kinase activity is critical for regulation of Snf1-dependent UV-responsive genes, Snf1 activation is not obligatory, but instead affects the expression of a subset of these genes.
Rad23 regulates transcription of Snf1-dependent and UV-responsive genes
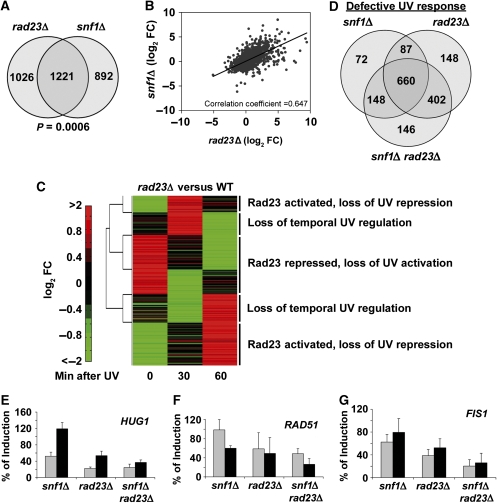
The broad role for Snf1 in UV-induced gene expression (Figure 1) suggested that Rad23 possesses a previously undiscovered function in gene regulation. Microarray analysis showed that surprisingly, a large number of genes were affected by deletion of RAD23 in the absence of DNA damage (Figure 4A). Interestingly, there was significant overlap and positive correlation between snf1Δ and rad23Δ data sets, indicating that these factors regulate many of the same genes in undamaged cells (Figure 4A and B and Supplementary Table S4). This was also reflected by the fact that the mutant data sets were enriched for genes in very similar functional classes (Supplementary Table S3).
Figure 4.
Rad23 regulation of Snf1-dependent, UV-responsive genes. (A) Venn diagram showing the number of genes significantly affected in snf1Δ and rad23Δ cells compared with WT cells in the absence of irradiation. (B) Correlation of snf1Δ and rad23Δ microarray data sets. For genes with a significant change in either snf1Δ or rad23Δ cells compared with WT, log-transformed FC in gene expression in snf1Δ cells was plotted against the log-transformed FC in rad23Δ cells and the correlation coefficient was calculated. (C) Two-dimensional hierarchical clustering performed on rad23Δ data as in Figure 1B. Five distinct clusters are indicated. (D) Venn diagram showing overlap in genes with a significantly different expression level after DNA damage compared with WT cells. (E–G) Induction level of HUG1, RAD51, and FIS1 in snf1Δ, rad23Δ and snf1Δ rad23Δ cells compared with WT. The graphs show relative RNA levels obtained by quantification of northern blots of total RNA isolated from the indicated strains at 1 h (grey bars) or 2 h (black bars) after irradiation with 100 J/m2 UV light. In each case, the expression level of the indicated gene at the indicated time in WT cells was defined as 100%. Labelled probes were specific for the HUG1 ORF (E), the RAD51 ORF (F), or the FIS1 ORF (G) and expression levels were normalized to ACT1 expression. For quantification of RNA levels over a more extended time course, see Supplementary Figure S5. Values are the average of three independent experiments and error bars represent standard deviation.
To determine whether Rad23 regulates UV-responsive transcription, expression of UV-responsive genes was compared with the WT controls. Overall, loss of Rad23 caused a global mis-regulation of the UV response transcriptome (Figure 4C). The rad23Δ-induced changes in UV-responsive gene expression showed a similar clustering pattern as the Snf1-regulated gene set: genes that were Rad23-activated or repressed in undamaged cells subsequently lost UV-induced repression or activation, respectively (Figure 4C and Supplementary Figure S3). Two additional clusters included genes that were not regulated by Rad23 in the absence of damage but showed temporal changes in UV-induced gene expression (Figure 4C and Supplementary Figure S3).
Consistent with the broad roles of Rad23 and Snf1 in the UV-mediated transcriptional response, there was extensive overlap in the UV-responsive genes regulated by these two factors (Figure 4D). The Venn diagram shows genes with significantly different expression after DNA damage in mutant cells compared with WT. These mis-regulated genes represent 50, 68 and 71% of all UV-responsive genes for snf1Δ, rad23Δ and snf1Δ rad23Δ cells, respectively. A total of 1663 UV-responsive genes, or 87% of the cellular UV response, were affected by deletion of SNF1, RAD23, or both. This gene set includes about 40% of the general environmental stress response genes identified by Gasch et al (2000) (Supplementary Figure S4). These results not only show that both Snf1 and Rad23 function in UV-induced transcriptional regulation, but that these functions are largely overlapping and redundant, a relationship which probably contributes to the increased UV sensitivity of snf1Δ rad23Δ cells. In addition, the data show that the global interplay between Snf1 and Rad23 is complex, implying that there are probably several molecular mechanisms by which these factors cooperate to control the UV response. Below, we explore the mechanism of action of Snf1 and Rad23 at a specific UV-responsive promoter, but it is important to bear in mind that the genome-scale analyses indicate that multiple regulatory effects are likely at work.
Relationship of Snf1 and Rad23 to DNA damage responses
A number of genes were cooperatively regulated by Snf1 and Rad23 in response to UV irradiation. A loss of induction of these genes probably contributes to the observed synthetic phenotype. FIS1, HUG1, and RAD51 are examples of such genes whose expression was studied in more depth (Figure 4E–G and Supplementary Figure S5). In WT cells, significant induction of these genes was observed at 1–2 h after irradiation. For each gene, the greatest diminution in UV-induced expression was observed in the snf1Δ rad23Δ strain (Figure 4E–G). The Rad23-dependent and Snf1-dependent regulation of repair factors such as RAD51 suggested the idea that a transcriptional defect in snf1Δ rad23Δ cells may compromise DNA repair efficiency. However, analysis of DNA repair rates in WT and mutant cells indicated that Snf1 does not significantly contribute to DNA repair (Supplementary Figure S6).
Another possibility, based on the mis-regulation of factors involved in cell cycle arrest such as HUG1 (Basrai et al, 1999), was that improper transcriptional regulation leads to cell cycle arrest defects in snf1Δ rad23Δ cells. To address this question, we tested the UV damage-induced arrest and recovery of α-factor-arrested or nocodazole-synchronized cells by flow cytometry. This revealed a significant delay in recovery from the G2/M arrest in snf1Δ rad23Δ cells compared with either single deletion alone, and this may well contribute to their increased UV sensitivity (Supplementary Figures S7 and S15). Given the roles of Snf1 and Rad23 in HUG1 expression (Figure 4E), and evidence implicating HUG1 in the MEC1 damage response pathway (Basrai et al, 1999), we tested the possibility that a defect in HUG1 expression was responsible for reduced cell survival in response to irradiation. However, high-copy HUG1 did not rescue the UV phenotype of snf1Δ rad23Δ cells (Supplementary Figure S8). Taken together, the results suggest that the synthetic UV phenotype observed in snf1Δ rad23Δ cells is due to the additive effect of gene expression changes that influence cell cycle arrest pathways and possibly other DNA damage response mechanisms.
Mig3 is a downstream target of the Snf1 UV response pathway
During metabolic stress, Snf1 controls transcription by targeting transcriptional repressors and activators, as well as histone H3 (Lo et al, 2001; Papamichos-Chronakis et al, 2004). For example, phosphorylation of H3-Ser 10 by Snf1 is important for the induction of the INO1 gene in the absence of inositol (Lo et al, 2001). Genetic analysis ruled out H3-Ser 10 as a substrate involved in Snf1-mediated UV resistance (Supplementary Figure S9), suggesting instead that transcriptional activators or repressors are involved.
Mig3 emerged as a good candidate to mediate the Snf1 UV response because it is phosphorylated by Snf1 in vitro and has been implicated as a target of Snf1 in response to hydroxyurea (Dubacq et al, 2004). Mig3 is homologous to Mig1, a downstream target of Snf1 during glucose de-repression, but does not repress glucose responsive genes (Lutfiyya et al, 1998). Under low glucose conditions, Snf1 phosphorylates Mig1, resulting in activation of glucose-repressed genes (Treitel et al, 1998). If Snf1 targets Mig3 in a similar manner after irradiation, we reasoned that deletion of MIG3 would suppress the snf1Δ-dependent defect in the UV response. Indeed, deletion of MIG3 completely suppressed the synthetic UV sensitivity in snf1Δ rad23Δ cells (Figure 5A). Suppression was not observed with deletion of MIG1 (Figure 5B), and conversely, mig1Δ was able to suppress the general growth defect of snf1Δ rad23Δ cells, but mig3Δ could not (Supplementary Figure S10). This demonstrates the specificity of these two downstream targets of Snf1 and supports a model in which Snf1 negatively regulates Mig3 after DNA damage to modulate the expression of UV-responsive genes.
Figure 5.
Mig3 is a downstream target of Snf1. (A and B) Effect of mig3Δ and mig1Δ on the UV sensitivity of snf1Δ rad23Δ cells. UV survival curves were obtained as in Figure 3A. (C–F) Two-dimensional gel electrophoresis and western blotting of whole cell extracts from untagged (C) or Mig3-myc containing (D-F) cells. Samples were taken from WT (D) or snf1Δ cells (F) before and 5 min after UV irradiation, as indicated. Myc-tagged Mig3 migrates with an apparent molecular weight of 70 kDa on a standard one-dimensional denaturing protein gel. Two major species of Mig3-myc consistent with this molecular weight were detectable, and are indicated by the black (phosphorylated) and white (unphosphorylated) arrows. Loss of phosphorylation is evident in lambda phosphatase-treated WT sample (E).
To verify that Mig3 phosphorylation is regulated by Snf1 kinase in vivo, Mig3 isoforms were visualized by two-dimensional gel electrophoresis. Both phosphorylated and unphosphorylated forms of Mig3 were detected in WT extracts from undamaged cells (Figure 5D and Supplementary Figure S11). The identities of the phosphorylated and unphosphorylated species were confirmed by control experiments using untagged or phosphatase-treated extracts (Figure 5C and E). Mig3 phosphorylation was increased in response to UV treatment, and phosphorylation was dependent on Snf1 (Figure 5D and F, and Supplementary Figure S11). The combined genetic and biochemical results described thus far establish the existence of a UV-induced, Snf1-mediated signal transduction pathway critical for UV-responsive transcription with Mig3 as an important downstream target.
Direct regulation of UV-responsive genes by Mig3 is modulated by Snf1 and Rad23
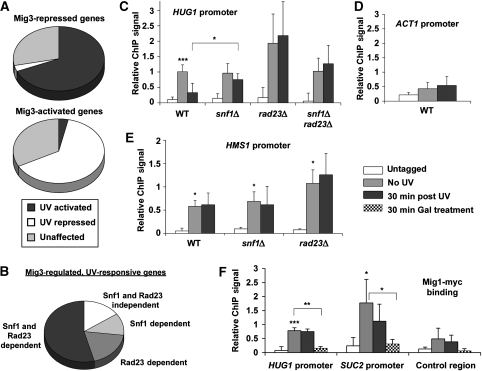
To define the Mig3-regulated UV-responsive gene set, microarray expression analysis was carried out on mig3Δ cells. The expression levels of 245 genes were affected by deletion of MIG3 (99 repressed and 146 activated), and importantly, the majority were UV-responsive genes. Strikingly, 64% of Mig3-activated genes were UV repressed, and 69% of Mig3-repressed genes were activated by UV irradiation (Figure 6A). This suggests that Mig3 repressed UV-responsive genes in the absence of DNA damage, and loss of MIG3 precociously activated the UV response. In addition, the majority of UV-responsive, Mig3-regulated genes were dependent on Snf1, Rad23, or both for a proper UV response (Figure 6B). These results support the idea that Mig3 functions downstream in a transcriptional network including Snf1 and Rad23. Moreover, these results suggest that Mig3 is inactivated by Snf1 after irradiation, and this inactivation regulates Mig3 target genes. Although the combined genetic and microarray results show an important function for Mig3 in the UV response, the relatively small number of genes regulated by Mig3 in comparison with Snf1 and Rad23 implicate other downstream effectors. Thus, the UV response seems to share similarities with the response to nutrient limitation in targeting several transcriptional regulators to result in an effective and coordinated response (Schuller, 2003).
Figure 6.
Mig3 directly regulates UV-responsive, Snf1- and Rad23-regulated genes. (A) UV dependence of Mig3-regulated genes. Comparison of microarray data sets for UV-irradiated WT cells with the unirradiated mig3Δ data set shows the proportion of Mig3-regulated genes that were significantly affected by UV irradiation. (B) Snf1- and Rad23-dependence of the UV response of Mig3-regulated genes. Comparison of the UV response in snf1Δ, rad23Δ, and snf1Δ rad23Δ cells with WT for Mig3-regulated, UV-responsive genes indicates that most were Snf1- or Rad23-dependent. (C–E) Mig3 binding to the promoters of Mig3-regulated genes. ChIP and RT–PCR were performed for Myc-tagged Mig3 using primers specific for the HUG1 promoter (C), ACT1 promoter (D), and HMS1 promoter (E). Relative ChIP signal was obtained by subtracting mock IP samples and normalizing to the total input chromatin for each sample. ChIP signal was calculated for cells containing untagged Mig3 as a negative control and cells harbouring Myc-tagged Mig3 before irradiation and 30 min after irradiation. Values are the average of three independent experiments and error bars represent standard deviation. Statistical significance was measured using a two-tailed Student's t-test with paired variables (***P<0.01, **P<0.03, *P<0.05; compared to untagged unless otherwise noted). Although there was significant variability associated with the triplicate ChIP results obtained using rad23Δ cells, the failure of Mig3 to vacate the HUG1 promoter in rad23Δ cells after irradiation (panel C) was highly reproducible (average ratio of ChIP signal for irradiated/unirradiated cells was 1.13±0.16). Thus, the error bars reflect the day-to-day variability in the Mig3 occupancy versus untagged control rather than variation in the Mig3 occupancy when comparing samples with and without UV treatment prepared in parallel. (F) Mig1 binding to Mig3- and Mig1-regulated promoters. ChIP and RT–PCR were carried out for Myc-tagged Mig1 using primers specific for the HUG1 promoter, SUC2 promoter, and an intergenic region on chromosome V. ChIP signal was obtained as in panels (C–E).
Chromatin immunoprecipitation (ChIP) experiments were carried out to test whether Mig3 was associated with the promoters of Mig3-regulated, UV-responsive genes. HUG1 showed the largest UV induction across the genome, as well as the largest repression by Mig3, so it was an ideal candidate for ChIP. A 10-fold enrichment in ChIP signal was seen at the HUG1 promoter for Mig3-myc, whereas no such localization was observed at the ACT1 promoter (Figure 6C and D). In addition, Mig3 was released from the HUG1 promoter after UV irradiation, and this release was Snf1 dependent (Figure 6C). Most interestingly, the release of Mig3 from the HUG1 promoter was also dependent on Rad23 (Figure 6C). The displacement of Mig3 from the promoter preceded the accumulation of HUG1 mRNA, which peaked at 60 min (Figure 4E and Supplementary Figure S5), as expected if the events were mechanistically coupled. The requirement for both Snf1 and Rad23 for the release of Mig3 from the HUG1 promoter offers a molecular explanation for the contributions of these two factors to UV-dependent HUG1 expression.
By comparison, HMS1 was one of the most UV-repressed and Mig3-activated genes. As with the HUG1 promoter, Mig3-myc specifically localized to the HMS1 promoter. Unlike the HUG1 promoter, Mig3 remained bound to the HMS1 promoter after irradiation and its binding was not affected by the deletion of either SNF1 or RAD23 (Figure 6E). This suggests different mechanisms of Mig3 regulation at repressed and activated promoters. Eight other Mig3-regulated genes were examined but did not show significant Mig3 binding (data qnot shown), suggesting that Mig3 exerts indirect effects on transcription as well. In fact, HMS1 encodes a basic helix-loop-helix factor similar to members of the Myc family of transcription factors (Robinson and Lopes, 2000).
If activated Snf1 can target several transcriptional regulators; how is the functional specificity achieved in response to different environmental stimuli? To address this question, we compared the promoter localization of Mig1 and Mig3 in irradiated or galactose-grown cells. The DNA binding specificities of Mig1 and Mig3 were reported to be similar in vitro (Lutfiyya et al, 1998). Accordingly, Mig1 was readily detectable at the Mig3-regulated HUG1 and HMS1 promoters, as well as Mig1-regulated promoters (Figure 6F and Supplementary Figure S12). In addition, expression analysis showed that Mig1 also represses HUG1 expression, although not to the extent achieved by Mig3 (Supplementary Table S5). Similarly, Mig3 binding was detectable at Mig1-regulated SUC2 and GAL1 promoters, but in these cases Mig3 did not regulate these promoters (Supplementary Figure S12 and Supplementary Table S5). Importantly, however, release of Mig3 after UV irradiation was specific to the UV-regulated HUG1 promoter and did not occur at SUC2 or GAL1 (Figure 6C and Supplementary Figure S12). Moreover, phosphorylation of Mig1 and release from promoter DNA occurred only when cells were grown in galactose and not following UV irradiation (Figure 6F and Supplementary Figure S12). These results indicate that although Mig1 and Mig3 bind promoters promiscuously in vivo, each responds specifically to a particular environmental stimulus registered by Snf1.
Rad23–proteasome interaction is required for transcriptional regulation
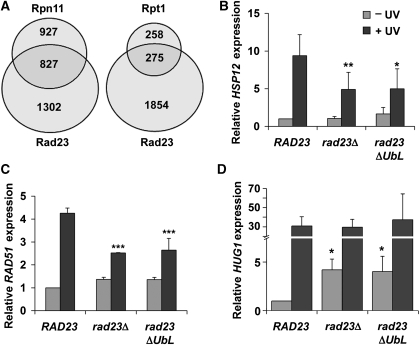
A major finding of this study is the novel role for Rad23 in the regulation of transcription. In addition to participating directly in the NER reaction, Rad23 has been identified as a regulator of ubiquitylated substrate turnover, which occurs through direct interaction with the proteasome (Ortolan et al, 2000; Chen and Madura, 2002). As the ubiquitin–proteasome pathway is now understood to have direct and global functions in transcriptional regulation (Collins and Tansey, 2006), we hypothesized that Rad23 affects transcription through its interaction with the proteasome. To test this idea, we compared previously reported expression data for proteasome mutants (Auld et al, 2006) with our rad23Δ data set. There was significant overlap between Rad23-regulated genes and those regulated by Rpn11, a 19S RP lid subunit, and Rpt1, a 19S RP base subunit (Figure 7A). Statistical analyses confirmed that the data sets are enriched in Rad23/Rpn11-co-regulated genes (P=0.0003) and Rad23/Rpt1-co-regulated genes (P=0.0004). A comparison of the rad23Δ data set with data from the mutant 20S core particle subunit Pre1, however, showed no enrichment of co-regulated genes (P=0.2447). In addition, there was a positive correlation between Rad23-mediated effects and Rpn11- and Rpt1-mediated effects that was not seen for Pre1 (see Supplementary Table S4 for full analysis). This shows that Rad23 regulates many of the same genes in the same way as the 19S RP. The simplest interpretation is that Rad23 acts in conjunction with the 19S RP to regulate transcription at UV-responsive promoters.
Figure 7.
Rad23–proteasome interaction is important for transcriptional regulation. (A) Comparison of rad23Δ data set to existing proteasome mutant data sets. Venn diagrams show overlap between lists of significantly affected genes from our rad23Δ data set (from undamaged cells) and two different proteasome 19S RP mutant data sets (Auld et al, 2006). (B–D) Expression levels of HSP12, RAD51, and HUG1 in rad23Δ cells harbouring plasmid-borne RAD23, rad23ΔUbL, or empty vector. Quantification of northern blots of total RNA isolated from the indicated strains before and 1 h after irradiation is shown. Labelled probes were specific for the HSP12 ORF (B), RAD51 ORF (C), or HUG1 ORF (D), and levels were normalized to ACT1 expression. Cells were grown in appropriate synthetic drop-out medium to maintain plasmids. It should be noted that expression levels and induction kinetics varied from those in YPD (Supplementary Figure S5). Although this dependence on growth medium is not understood in detail, this variation is consistent with the idea that these genes are regulated by factors that also control gene expression in response to nutritional stimuli. Values plotted are an average of three independent experiments. Error bars represent standard deviation. Statistical significance was measured using a two-tailed Student's t-test with paired variables (***P<0.01, **P<0.03, *P<0.05; compared to WT).
To further test this hypothesis, we used a mutant form of Rad23 that lacks the N-terminal ubiquitin-like (UbL) domain required for proteasome binding (Schauber et al, 1998). Previous results implicate the Rad23 UbL domain in UV resistance (Ortolan et al, 2004), although how it participates in the process was unexplained. We compared the Rad23-dependent, UV-induced transcriptional response of a number of genes in WT, rad23Δ and rad23ΔUbL cells. For all genes tested, the expression profile was identical in rad23Δ and rad23ΔUbL cells (Figure 7B–D, data not shown). For HSP12 and RAD51, UV induction in both rad23Δ and rad23ΔUbL cells was about half of that in WT cells, whereas HUG1 expression was de-repressed in undamaged rad23Δ and rad23ΔUbL cells (Figure 7B–D). We conclude that interaction with the proteasome is critical for proper Rad23-mediated transcriptional regulation.
Discussion
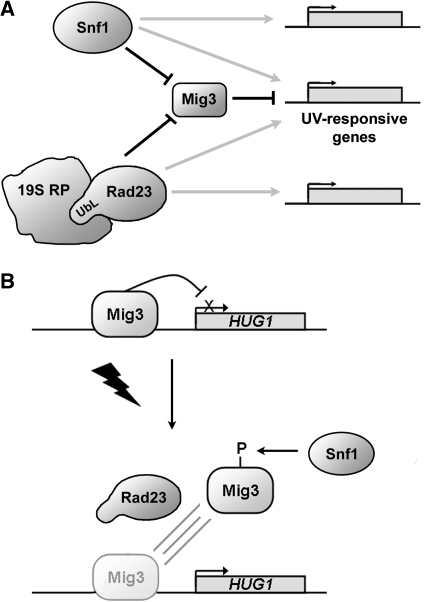
The results presented here reveal a novel function for the nutrient-sensing kinase, Snf1, and its downstream target, Mig3, in the transcriptional response to UV irradiation. This is distinct from the function of Snf1 in nutrient sensing and shows how components of signal transduction pathways are co-opted for different purposes in response to diverse stimuli. In addition, we show that the Rad23 ‘repair factor' plays a general role in gene regulation in undamaged and damaged cells. This represents a completely novel function for Rad23 as well. The mechanistic link between Rad23- and 19S RP-mediated transcriptional regulation provides new insight into how functional specificity of the 19S RP is achieved. It is striking that although the DNA damage response has been extensively investigated, the combined actions of Snf1 and Rad23 reported here contribute to a majority of the UV-induced transcriptional response. The expression data show a complex picture in which overlapping pathways involving Snf1, Rad23, and the 19S RP coordinate the regulation of gene expression (Figure 8A).
Figure 8.
Model for transcriptional regulation by Snf1 and Rad23. (A) Schematic summarizing the function of Snf1 and Rad23 in UV-responsive gene regulation. The diagram highlights the myriad effects on overlapping and distinct gene sets by Snf1 and Rad23 evident from the microarray data (grey arrows). The model also shows the specific effects on the transcription factor Mig3 (in black). The results in this study implicate Mig3 as a repressor of the UV-response, which is inhibited by Snf1 and Rad23 after irradiation. The results presented here also document a function of the linkage (both physical and functional) between Rad23 and the 19S RP in the UV response. (B) The UV-induced, Mig3-repressed HUG1 promoter provides a mechanistic example of Snf1- and Rad23-mediated gene regulation. HUG1 is repressed by Mig3 in the absence of damage. After irradiation, Mig3 is released from the promoter through a process dependent on both Rad23 and Snf1-mediated phosphorylation.
Transcriptional regulation by Snf1 and Rad23
It is clear from these data that Snf1 and Rad23, in conjunction with the 19S RP, regulate both overlapping and distinct sets of UV-regulated genes before and after DNA damage (Figures 1, 4 and 7A). In undamaged cells, these factors ensure appropriate expression of many genes, including UV-responsive genes involved in cellular growth and proliferation (Figure 1 and 4, and Supplementary Table S3). In response to DNA damage, the major trend is that the functions of Snf1 and Rad23 are reversed. Genes repressed by Snf1 and Rad23 require these factors for damage-mediated induction, whereas activated genes rely on Snf1 and Rad23 for repression following damage (Figures 1 and 4). Snf1 and Rad23 probably achieve these myriad transcriptional effects through regulation of multiple downstream effectors.
Here we have focused on the identification and characterization of the downstream target Mig3 and its action at the HUG1 promoter, the most highly UV-activated promoter in yeast. Genetic and biochemical data support a model in which Mig3 represses the UV response and, upon irradiation, is deactivated in an Snf1- and Rad23-dependent manner, leading to the induction of the UV response (Figure 8A). Multiple observations support this model. Mig3 suppressed the synthetic UV phenotype of snf1Δ rad23Δ cells (Figure 5). In addition, Snf1 was activated after irradiation coincident with increased phosphorylation of Mig3, which was Snf1 dependent (Figures 2, 5 and Supplementary Figure S11). The expression data are consistent with this model as well: Mig3-regulated genes were conversely regulated by UV treatment (Figure 6A) and the majority of Mig3-regulated, UV-responsive genes were dependent on Snf1 and/or Rad23 for proper UV induction (Figure 6B).
At the HUG1 promoter, Mig3 was bound in the absence of DNA damage and repressed transcription (Figures 6C and 8B). The expression level of HUG1 was increased about 20-fold in mig3Δ compared with WT cells (Supplementary Table S5). Following irradiation, transient activation of Snf1 (Figures 2A and B) was correlated with increased phosphorylation of Mig3 (Figure 5C–F and Supplementary Figure S11). Within 30 min, Mig3 was released from the promoter and this release was dependent on both Snf1 and Rad23 (Figure 6C). We hypothesize that the release of promoter-associated Mig3, in turn, allows activator binding and subsequent activation. This conjecture is supported by the fact that timely induction of HUG1 required both Snf1 and Rad23. These factors also have some redundant effects because although the induction was slow in snf1Δ and reduced in rad23Δ cells, induction levels were further reduced in snf1Δ rad23Δ double mutants within the 2 h time course examined (Figure 4E).
Selectivity in the Snf1 response: Mig1 versus Mig3
Mig1 and Mig3 are homologous transcriptional repressors with highly related DNA binding domains, yet they have distinct functional specificities in vivo (Lutfiyya et al, 1998; Treitel et al, 1998; Figures 5, 6 and Supplementary Figure S12). Genetic results presented here show that there is a fairly clean delineation in the response of Mig1 or Mig3 to a particular stimulus (Figure 5A, B and Supplementary Figure S10). In addition, Mig1 is phosphorylated and vacates promoters in response to glucose limitation but not irradiation and Mig3 vacates only UV-responsive promoters after irradiation (Figure 6C, F and Supplementary Figure S12). However, despite the lack of evidence for a functional role of Mig3 in the starvation response, it is modified and degraded in response to glucose deprivation (Supplementary Figure S12; Dubacq et al, 2004) and there is an overlap in the DNA binding site preferences of Mig1 and Mig3 (Figure 6C–F and Supplementary Figure S12; Lutfiyya et al, 1998). These results imply that Mig1 and Mig3 are necessary but insufficient for dictating the observed transcriptional regulatory patterns, and that other co-activators/co-repressors probably delimit how and where Mig1 and Mig3 act.
Along these lines, we note that Snf1- and Rad23-dependent release of Mig3 from the HUG1 promoter would allow the binding of UV-induced activators and/or facilitate the remodelling of promoter chromatin to an active state. Mig1 was still detectable by ChIP after damage at the HUG1 promoter and can repress HUG1, however, to a lesser extent than Mig3. This supports the idea that the HUG1 promoter is more likely bound by Mig3 and bound only transiently or less effectively by Mig1. Thus, loss of HUG1 promoter-bound Mig3 is probably important for activator binding, whereas Mig1 is less effective for repression and does not interfere with subsequent activation steps. In contrast, at the Mig3-activated HMS1 promoter, Mig3 occupancy was not affected by UV irradiation. This suggests a different mechanism in which phosphorylation of Mig3 changes its function at the promoter, perhaps by altering interactions with other co-activators or co-repressors. Such a mechanism is analogous to the function of Mig1 at the GAL1 promoter, at which Mig1 remains bound during activation and phosphorylation regulates its interaction with the Cyc8–Tup1 co-repressor (Papamichos-Chronakis et al, 2004).
Implications of genetic data and expression analysis
Although Snf1 is transiently phosphorylated after UV irradiation, genetic data questioned the importance of this activation for UV survival. Loss of full Snf1 activation did not lead to the same phenotype as snf1Δ or loss of catalytic activity (Figure 3 and Supplementary Figure S2). This is consistent with expression data suggesting that although basal kinase activity is important, full activation is not as important for proper regulation of UV-induced, Snf1-dependent genes. These observations, along with microarray results from unirradiated snf1Δ cells, suggest a previously unappreciated role for Snf1 in conditions in which it is not fully activated.
The loss of Mig3 fully suppressed the synthetic UV sensitivity of snf1Δ rad23Δ cells (Figure 5A), arguing that the de-regulation of Mig3 target genes is responsible for the enhanced UV-sensitive phenotype. However, many genes regulated by Snf1 and Rad23 were not Mig3 dependent, consistent with the idea that Snf1 and Rad23 function through other downstream targets as well. It is also likely that the mig3Δ microarray data presented here do not capture the full impact of Mig3 on the UV response. As inhibition of Mig3 function after DNA damage is unlikely to be the only event required for the UV-induced transcriptional response, deletion of MIG3 is unlikely to fully mimic the UV-regulated state at all Mig3 target genes. Snf1 targets several transcriptional factors in response to glucose limitation (Schuller, 2003), and given the complexity in promoter organization and regulatory interactions, there are undoubtedly multiple mechanisms by which Snf1 and Rad23 transduce their effects. It remains to be seen if other known Snf1 substrates also participate in the UV response.
Mig3 directly regulates the transcription of two of the most UV-responsive genes in the yeast genome, one of which, HMS1, encodes a transcription factor. This implies that the phenotype seen in snf1Δ rad23Δ cells is due to the mis-regulation of a few key genes or that regulation of transcription factors led to myriad indirect effects. In addition, the cell cycle data suggest that the synthetic phenotype of snf1Δ rad23Δ cells is due at least in part to the inability of these cells to recover properly from UV damage-induced cell cycle arrest (Supplementary Figure S7). The defect in HUG1 activation in these cells could be related to this observation; however high-copy HUG1 did not rescue the UV sensitivity (Supplementary Figure S8). We therefore favour the interpretation that a combination of transcriptional effects in snf1Δ rad23Δ cells led to delayed recovery from damage-induced arrest, thereby contributing to increased UV sensitivity.
Mechanisms for Rad23 function in transcription
A major finding of this study is that Rad23 has a previously unrecognized, global impact on transcription. These effects are not explained by a defect in DNA repair because relatively few genes were affected by the deletion of rad4Δ, which completely abrogates repair (Supplementary Figure S13). This argues against an important function for the Rad23–Rad4 complex (Guzder et al, 1998) in transcriptional control. Thus, the transcriptional effects in rad23Δ cells are not the indirect result of a failure in the repair of endogenous damage. Furthermore, we did not detected a function for Rad23 in regulating the UV-induced turnover of stalled RNA polymerase II (Supplementary Figure S14), again arguing for direct effects on transcription initiation.
On the basis of the extensive overlap in Rad23- and 19S RP-regulated gene sets and the requirement of the Rad23 UbL (Figure 7 and Supplementary Table S4), it is likely that transcriptional regulation by Rad23 is mediated through its interaction with the 19S proteasomal cap (Figure 8). The lack of correlation between the Rad23- and Pre1-dependent gene sets points to a non-proteolytic function for this transcriptional control circuit. There is mounting evidence that the 19S RP and the APIS subcomplex are important for regulating transcriptional activation (Gonzalez et al, 2002; Lee et al, 2005; Collins and Tansey, 2006; Ferdous et al, 2007). The 19S RP can regulate transcription factor binding at promoters, and there is evidence that the APIS 19S RP subcomplex has a function in general transcription (Sun et al, 2002; Lee et al, 2005; Ferdous et al, 2007). The ubiquitylation of histones and transcription factors has an important role in dictating proteasome function at promoters (Ezhkova and Tansey, 2004; Ferdous et al, 2007). As a bridge between ubiquitylated substrates and the proteasome, Rad23 is an excellent candidate regulator of such events. The requirement for Rad23 for the displacement of Mig3 from the HUG1 promoter suggests a simple model in which gene activation involves 19S RP-catalysed removal of Mig3. In this scenario, Snf1-mediated phosphorylation of Mig3 may be required to mark Mig3 for Rad23–19S RP action. The broad requirement for Rad23 in transcriptional control (about one-third of the transcriptome) suggests that there are other transcriptional regulators whose promoter occupancy or activity is regulated by the Rad23–19S RP complex.
Materials and methods
Yeast strains and plasmids
Saccharomyces cerevisiae strains derived from YPH499 used in this study are described in Supplementary Table S6. Null alleles were generated by ORF deletion using PCR-amplified HIS3, NATMX, or KANMX cassettes with gene-specific primers for yeast transformation. Cassette integration was confirmed by PCR. Tagged genes were constructed as previously described (Ramsey et al, 2004). Supplementary Table S7 describes the plasmids used in this study. The snf1-K84R point mutant was obtained using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) and confirmed by sequencing. RAD23 and rad23ΔUbL-encoding plasmids were constructed by standard PCR and subcloning procedures. HA-tagged Snf1 and the snf1-T210A point mutant were generously provided by Dr Martin Schmidt.
UV survival assays
Cells were grown at 30°C in YPD or synthetic amino-acid drop-out medium to an optical density at 600 nm (OD600) of 1.0. Cells were plated on YPD or appropriate drop-out plates, irradiated in a Stratalinker 1800 (Stratagene) with the indicated dose of UV light and incubated in the dark at 30°C for 2–5 days. Percent survival was calculated from the number of surviving colonies relative to unirradiated plates.
Snf1 and AMPK activation assay
Antibody specific to Snf1 phosphorylated at Thr 210 was obtained from Dr Martin Schmidt (McCartney and Schmidt, 2001). Cells expressing HA-tagged Snf1 were grown in appropriate drop-out media to an OD600 of 1.0. Cells were collected by filtration on 0.65-μm nitrocellulose filters (Millipore) to avoid spurious activation of the kinase by centrifugation (Wilson et al, 1996). The filters were placed on YPD plates and irradiated with the indicated dose of UV light. Extracts were prepared by NaOH extraction as described by McCartney and Schmidt (2001).
Immunoprecipitation of HA-tagged Snf1 was carried out by incubating whole cell extracts with anti-HA affinity matrix (Roche) in RIPA buffer containing 5 mM sodium pyrophosphate, 50 mM sodium fluoride, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 2 μg/ml chymostatin, 3.5 μg/ml pepstatin, and 10 μg/ml aprotinin. Western blot analysis of HA-tagged protein was carried out as previously described (Ramsey et al, 2004). For phosphorylated Snf1, anti-PT210 antibody was used at a dilution of 1:1000 in TBST. Detection was completed using ECL Plus detection system (GE Healthcare) according to the manufacturer's instructions.
For AMPK activation, HEK-293T cells were grown to 80% confluence in DMEM containing 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin. As a positive control for AMPK activation, cells were grown for an additional 4 h in the same medium lacking glucose. For UV irradiation, medium was removed and cells were irradiated as described above. Medium was immediately replaced and cells were incubated for the indicated time. Cells were lysed in RIPA buffer containing protease inhibitor cocktail. Western blotting was done using primary antibodies against AMPK and P-T172-specific antibody (Cell Signaling Technology).
Two-dimensional gel electrophoresis
Cells were grown, collected by filtration and irradiated as described above. Cell pellets were lysed in osmotic lysis buffer (10 mM Tris (pH 7.4), 0.3% SDS) containing 5 mM sodium pyrophosphate, 50 mM sodium fluoride, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 2 μg/ml chymostatin, 3.5 μg/ml pepstatin, and 10 μg/ml aprotinin. For phosphatase treatment, phosphatase inhibitors were left out and extracts were treated with lambda phosphatase (New England Biolabs) according to the manufacturer's instructions. Two-dimensional gel electrophoresis and transfer to PVDF was carried out by Kendrick Laboratories (Madison, WI; see Supplementary data for details). Western blotting was carried out as previously described (Ramsey et al, 2004) using anti-myc (9E10) primary antibody.
Microarray
Cells were grown in YPD and left untreated or irradiated with 100 J/m2 UV light as described above. Samples were taken before and at 15, 30 and 60 min after irradiation. Total RNA was isolated using a hot acid/phenol method (Schmitt et al, 1990). Sample preparation, cRNA labelling, hybridization to Affymetrix Yeast Genome 2.0 expression arrays, and scanning were performed at the UVA Biomolecular Research Facility using the Affymetrix GeneChip System. The .cel files were quantile normalized and expression values estimated using GC-RMA (Gentleman, 2005). We applied a modified t-test using the limma package in Bioconductor to DNA damage time course and mutant samples versus control to identify differentially expressed genes (Gentleman, 2005). To arrive at the lists of genes for every comparison, we first corrected for multiple hypotheses testing by applying a false discovery rate (FDR) correction to the P-values and used a 5% FDR cutoff. Microarray data have been deposited with the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/): accession number GSE16799.
To identify genes with similar expression profiles across our time course, clustering analysis was carried out using the clustergram package from the Bioinformatics toolbox in MATLAB (Eisen et al, 1998; Bar-Joseph et al, 2001). This package was used to generate the heat maps shown in Figures 1 and 4. The Spearman rank correlation was used as a measure of gene expression profile similarity to generate the hierarchical tree, and the clustering was carried out on both genes and on strains (i.e. two-way clustering).
Expression analysis
Growth, treatment of cells and RNA isolation was done as described for microarray samples. Northern analysis was carried out as previously described (Dasgupta et al, 2005). Quantification of northern blots was accomplished using ImageQuant software (Molecular Dynamics). RT–PCR was carried out using the iScript Select cDNA Synthesis Kit (Bio-Rad) according to manufacturer's instructions. cDNA was quantified by RT–PCR using iQ SYBR Green Supermix (Bio-Rad) and the Bio-Rad MyiQ Single Color Real time PCR detection system.
Chromatin immunoprecipitation
Cells were grown in YPD and UV treated after filtration as described for Snf1 activation. ChIP was carried out using anti-myc (9E10) antibody as previously described (Dasgupta et al, 2005). Quantification of immunoprecipitated DNA was carried out by RT–PCR as described above for expression analysis.
Supplementary Material
Supplementary Information
Supplementary Data
Review Process File
Acknowledgments
We thank Marty Mayo and David Allison for providing HEK-293T cells. We are especially grateful to Dan Burke, Martin Schmidt and Joe Reese for reagents and discussions, and to Patrick Grant and members of the Auble lab for discussions and comments on the paper. This study was supported by an award from the March of Dimes Birth Defects Foundation and NIH grant GM55763 to DTA. SLW was supported by NIH/NCI Cancer Research Training Grant T32CA009109.
References
- Auld KL, Brown CR, Casolari JM, Komili S, Silver PA (2006) Genomic association of the proteasome demonstrates overlapping gene regulatory activity with transcription factor substrates. Mol Cell 21: 861–871 [DOI] [PubMed] [Google Scholar]
- Bar-Joseph Z, Gifford DK, Jaakkola TS (2001) Fast optimal leaf ordering for hierarchical clustering. Bioinformatics 17: S22–S29 [DOI] [PubMed] [Google Scholar]
- Basrai MA, Velculescu VE, Kinzler KW, Hieter P (1999) NORF5/HUG1 is a component of the MEC1-mediated checkpoint response to DNA damage and replication arrest in Saccharomyces cerevisiae. Mol Cell Biol 19: 7041–7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrell GW, Brown JA, Wu HI, Giaever G, Chu AM, Davis RW, Brown JM (2002) Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc Natl Acad Sci USA 99: 8778–8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Carlson M (1989) Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol Cell Biol 9: 5034–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Madura K (2002) Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol 22: 4902–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins GA, Tansey WP (2006) The proteasome: a utility tool for transcription? Curr Opin Genet Dev 16: 197–202 [DOI] [PubMed] [Google Scholar]
- Dasgupta A, Juedes SA, Sprouse RO, Auble DT (2005) Mot1-mediated control of transcription complex assembly and activity. EMBO J 24: 1717–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubacq C, Chevalier A, Mann C (2004) The protein kinase Snf1 is required for tolerance to the ribonucleotide reductase inhibitor hydroxyurea. Mol Cell Biol 24: 2560–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Tansey WP (2004) Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol Cell 13: 435–442 [DOI] [PubMed] [Google Scholar]
- Feng Z, Zhang H, Levine AJ, Jin S (2005) The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA 102: 8204–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous A, Sikder D, Gillette T, Nalley K, Kodadek T, Johnston SA (2007) The role of the proteasomal ATPases and activator monoubiquitylation in regulating Gal4 binding to promoters. Genes Dev 21: 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO (2001) Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell 12: 2987–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R (2005) Bioinformatics and computational biology solutions using R and Bioconductor. New York: Springer Science+Business Media [Google Scholar]
- Gonzalez F, Delahodde A, Kodadek T, Johnston SA (2002) Recruitment of a 19S proteasome subcomplex to an activated promoter. Science 296: 548–550 [DOI] [PubMed] [Google Scholar]
- Guzder SN, Sung P, Prakash L, Prakash S (1998) Affinity of yeast nucleotide excision repair factor 2, consisting of the Rad4 and Rad23 proteins, for ultraviolet damaged DNA. J Biol Chem 273: 31541–31546 [DOI] [PubMed] [Google Scholar]
- Hardie DG (2007) AMP-activated protein kinase as a drug target. Annu Rev Pharmacol Toxicol 47: 185–210 [DOI] [PubMed] [Google Scholar]
- Hardie DG, Carling D, Carlson M (1998) The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem 67: 821–855 [DOI] [PubMed] [Google Scholar]
- Hong SP, Carlson M (2007) Regulation of snf1 protein kinase in response to environmental stress. J Biol Chem 282: 16838–16845 [DOI] [PubMed] [Google Scholar]
- Jackson T, Kwon E, Chachulska AM, Hyman LE (2000) Novel roles for elongin C in yeast. Biochim Biophys Acta 1491: 161–176 [DOI] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB (2005) AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell 18: 283–293 [DOI] [PubMed] [Google Scholar]
- Kim HS, Hwang JT, Yun H, Chi SG, Lee SJ, Kang I, Yoon KS, Choe WJ, Kim SS, Ha J (2008) Inhibition of AMP-activated protein kinase sensitizes cancer cells to cisplatin-induced apoptosis via hyper-induction of p53. J Biol Chem 283: 3731–3742 [DOI] [PubMed] [Google Scholar]
- Lee D, Ezhkova E, Li B, Pattenden SG, Tansey WP, Workman JL (2005) The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell 123: 423–436 [DOI] [PubMed] [Google Scholar]
- Leech A, Nath N, McCartney RR, Schmidt MC (2003) Isolation of mutations in the catalytic domain of the snf1 kinase that render its activity independent of the snf4 subunit. Eukaryot Cell 2: 265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL (2001) Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293: 1142–1146 [DOI] [PubMed] [Google Scholar]
- Luo Z, Saha AK, Xiang X, Ruderman NB (2005) AMPK, the metabolic syndrome and cancer. Trends Pharmacol Sci 26: 69–76 [DOI] [PubMed] [Google Scholar]
- Lutfiyya LL, Iyer VR, DeRisi J, DeVit MJ, Brown PO, Johnston M (1998) Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics 150: 1377–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney RR, Schmidt MC (2001) Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J Biol Chem 276: 36460–36466 [DOI] [PubMed] [Google Scholar]
- Ortolan TG, Chen L, Tongaonkar P, Madura K (2004) Rad23 stabilizes Rad4 from degradation by the Ub/proteasome pathway. Nucleic Acids Res 32: 6490–6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortolan TG, Tongaonkar P, Lambertson D, Chen L, Schauber C, Madura K (2000) The DNA repair protein rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat Cell Biol 2: 601–608 [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Gligoris T, Tzamarias D (2004) The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tup1 co-repressor. EMBO Rep 5: 368–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KL, Smith JJ, Dasgupta A, Maqani N, Grant P, Auble DT (2004) The NEF4 complex regulates Rad4 levels and utilizes Snf2/Swi2-related ATPase activity for nucleotide excision repair. Mol Cell Biol 24: 6362–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson KA, Lopes JM (2000) SURVEY AND SUMMARY: Saccharomyces cerevisiae basic helix-loop-helix proteins regulate diverse biological processes. Nucleic Acids Res 28: 1499–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz P (2003) Snf1 protein kinase: a key player in the response to cellular stress in yeast. Biochem Soc Trans 31: 178–181 [DOI] [PubMed] [Google Scholar]
- Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K (1998) Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391: 715–718 [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18: 3091–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller HJ (2003) Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet 43: 139–160 [DOI] [PubMed] [Google Scholar]
- Searle JS, Schollaert KL, Wilkins BJ, Sanchez Y (2004) The DNA damage checkpoint and PKA pathways converge on APC substrates and Cdc20 to regulate mitotic progression. Nat Cell Biol 6: 138–145 [DOI] [PubMed] [Google Scholar]
- Stein SC, Woods A, Jones NA, Davison MD, Carling D (2000) The regulation of AMP-activated protein kinase by phosphorylation. Biochem J 345: 437–443 [PMC free article] [PubMed] [Google Scholar]
- Sun L, Johnston SA, Kodadek T (2002) Physical association of the APIS complex and general transcription factors. Biochem Biophys Res Commun 296: 991–999 [DOI] [PubMed] [Google Scholar]
- Treitel MA, Kuchin S, Carlson M (1998) Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol Cell Biol 18: 6273–6280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Lane DP (2007) p53 in health and disease. Nat Rev Mol Cell Biol 8: 275–283 [DOI] [PubMed] [Google Scholar]
- Wilson WA, Hawley SA, Hardie DG (1996) Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol 6: 1426–1434 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Data
Review Process File