Abstract
Mutations in RECQ4, a member of the RecQ family of DNA helicases, have been linked to the progeroid disease Rothmund–Thomson Syndrome. Attempts to understand the complex phenotypes observed in recq4-deficient cells suggest a potential involvement in DNA repair and replication, yet the molecular basis of the function of RECQ4 in these processes remains unknown. Here, we report the identification of a highly purified chromatin-bound RECQ4 complex from human cell extracts. We found that essential replisome factors MCM10, MCM2-7 helicase, CDC45 and GINS are the primary interaction partner proteins of human RECQ4. Importantly, complex formation and the association of RECQ4 with the replication origin are cell-cycle regulated. Furthermore, we show that MCM10 is essential for the integrity of the RECQ4–MCM replicative helicase complex. MCM10 interacts directly with RECQ4 and regulates its DNA unwinding activity, and that this interaction may be modulated by cyclin-dependent kinase phosphorylation. Thus, these studies show that RECQ4 is an integral component of the MCM replicative helicase complex participating in DNA replication in human cells.
Keywords: DNA replication, genome stability, RECQ, RECQ4, Rothmund–Thomson Syndrome
Introduction
The human RECQ family helicases have non-redundant roles in maintaining genome integrity, shown by the fact that mutations in three of the five RECQ homologues, BLM, WRN and RECQ4, have been implicated in distinct clinical diseases (Mohaghegh and Hickson, 2002; Singh et al, 2009). In particular, mutations in the RECQ4 gene have been associated with Rothmund–Thomson Syndrome (RTS), RAPADILINO and Baller–Gerold Syndrome. RTS patients exhibit various physical and mental developmental abnormalities and show signs of premature aging, such as early development of cataracts, hair loss and increased risk of osteosarcoma.
Most of the mutations associated with RTS lie within or after the conserved helicase domain located at the centre of the RECQ4 protein, and RTS symptoms were further recapitulated in mouse models containing deletion or nonsense mutations within the helicase domain (Kitao et al, 2002; Hoki et al, 2003; Mann et al, 2005). Cells derived from these recq4-deficient mice showed phenotypes ranging from aneuploidy to slow cell growth. The defect in cellular proliferation may be because of a cell-cycle block or problems with DNA damage repair. The latter possibility remains controversial, however, as mouse embryonic fibroblasts derived from the RECQ4 helicase-deficient mutants do not exhibit sensitivity to DNA damaging agents (Hoki et al, 2003). In addition, unlike other RECQ mutants, such as cells deficient in BLM and RECQ5 helicases, recq4-deficient mouse cells fail to show increased sister chromatid exchanges, chromosome breakages or fusions (Mann et al, 2005). These observations indicate that the primary function of RECQ4 is not a role in DNA damage repair. Nevertheless, cells from some, but not all RTS patients, are indeed sensitive to UV and ionizing radiation and show reduced DNA repair synthesis (Kitao et al, 2002; Singh et al, 2009).
An alternative explanation for the growth retardation and genome instability phenotypes of recq4 mutants is a defect in cell-cycle progression. Interestingly, sequence comparisons show that the N-terminus of Xenopus RECQ4 shares sequence homology with the yeast replication factor, Sld2 (Figure 1A; Sangrithi et al, 2005; Matsuno et al, 2006). Genetic studies show that yeast Sld2 interacts with Dpb11 to allow the onset of S phase (Kamimura et al, 1998; Tak et al, 2006), and this interaction is indeed mirrored between RECQ4 and the corresponding Dpb11 homologue in Xenopus, known as Cut5 (Matsuno et al, 2006). The potential of RECQ4 as the Sld2 homologue in Xenopus was further shown by the fact that DNA replication initiation is compromised in RECQ4-depleted Xenopus extracts (Sangrithi et al, 2005; Matsuno et al, 2006). It is possible that RECQ4 is the Sld2 homologue in vertebrates, in which case it would be expected to be essential for cell growth. Consistent with this prediction, deletion of the N-terminus of RECQ4 results in embryonic lethality in mice (Ichikawa et al, 2002). However, unlike Xenopus RECQ4, in which the sequence homology with Sld2 spans the entire 453a.a. region of Sld2, only the first 70a.a. of mammalian RECQ4 show detectable sequence homology with yeast Sld2 (Figure 1A). Additionally, the proposed Cut5 interaction domain in Xenopus RECQ4 is missing in the mammalian homologues. Presently, there is little mechanistic insight available to define the biochemical properties of the Sld2 protein and suggest a precise role for Sld2 in DNA replication initiation in yeast (Tanaka et al, 2007a). This very limited sequence homology, restricted to a small region of the Sld2 N-terminus, makes it difficult to sufficiently conclude whether RECQ4 functions as a true Sld2 homologue in DNA replication in mammalian cells.
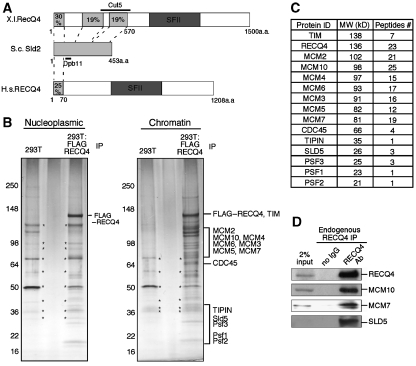
Figure 1.
Purification and identification of RECQ4 complex from human cells. (A) Schematic diagrams of Xenopus laevis (X.l.) RECQ4, Saccharomyces cervisaie (S.c.) Sld2 and human RECQ4 protein structures. The conserved superfamily II (SFII) helicase domains of RECQ4 are shown in dark grey. Regions of Xenopus and human RECQ4 that share sequence homology with S.c. Sld2 are shown in light grey and aligned with S.c. Sld2 protein with dashed lines. Sequence identity of each of the region to S.c. Sld2 is shown in %. Amino acids of S.c. Sld2 involved in Dpb11 interaction and the region that covers X.l. RECQ4–Cut5 interaction are indicated with black lines. (B) Visualization of the M2-agarose chromatography eluates of nucleoplasmic (left) and chromatin fractions (right) from control (293T) and FLAG–RECQ4-expressing cells on 4–15% gradient SDS–PAGE followed by silver staining. The identified polypeptides from the Mass Spec analysis were indicated on the right of the SDS–PAGE, where the proteins were expected based on their molecular weight. Protein bands that were also found in 293T immunoprecipitation (IP) were indicated with asterisks. (C) List of the polypeptides identified by mass spectrometric analysis (left), their corresponding molecular weight (centre) and number of peptides detected (right). (D) Co-immunoprecipitation of MCM10, MCM7 and SLD5 with the endogenous RECQ4 from normal 293T cells using rabbit anti-RECQ4 antibody (SDI). Immunopurified complex were analysed on SDS–PAGE followed by western blotting using the corresponding antibodies.
To understand the unique function of each of the RECQ helicases in the DNA metabolism and their relationships with different clinical diseases, recent efforts have focused on elucidating specific protein–protein interactions that provide insight into the cellular processes of a particular RECQ helicase (for review see Liu and West, 2008). In this work, we show that human RECQ4 is an integral part of the DNA replisome. Importantly, our data provide novel molecular insights into the regulation of human RECQ4 and its communication with the DNA replication machinery that is distinct from our current understanding on the proposed homologues in lower organisms.
Results
Purification and identification of the human RECQ4 complex
To facilitate purification of the RECQ4 complex, a 293T cell line stably expressing FLAG-tagged RECQ4 was established. The exogenous FLAG–RECQ4 expression was approximately two-fold of the endogenous RECQ4 protein in 293T cells. The nucleoplasmic fraction was prepared from these cells and chromatin-bound proteins were solublized by digesting the chromatin pellet with benzonase to remove nucleic acids as described earlier (Aygun et al, 2008). FLAG–RECQ4 complex was then purified by M2-agarose affinity chromatography and subjected to mass spectrometric analysis to identify co-purifying polypeptides. Although very few proteins are associated with nucleoplasmic RECQ4, SDS–PAGE showed several prominent bands for the RECQ4 complex purified from the soluble chromatin fraction (Figure 1B). After excluding the contaminants commonly found during FLAG immunopurification (Supplementary Figure 1; Guo et al, 2009), mass spectrometric analysis showed that the most abundant co-purified polypeptides in the chromatin-bound RECQ4 complex were MCM10, followed by MCM2-7 helicase, CDC45 and the GINS complex, containing SLD5, PSF1, PSF2 and PSF3 (Figure 1B and C). All of these factors are known to interact together to form the MCM replicative helicase complex important for DNA replication initiation and progression (for review see Sclafani and Holzen, 2007). In addition, within the RECQ4 complex, we identified the human homologue of yeast Tof1-Csm3, the TIMELESS (TIM)/TIPIN heterodimer, which is a known MCM2-7 helicase interacting protein important for fork progression and replication stress response (Errico et al, 2007; Gotter et al, 2007). The interaction between RECQ4 and the MCM replicative helicase complex was further confirmed by the co-immunoprecipitation of MCM10, MCM7 and SLD5 with endogenous RECQ4 protein from 293T cells without ectopic expression using a rabbit anti-RECQ4 antibody (Figure 1D).
Surprisingly, in contrast to the studies with Xenopus RECQ4, we failed to detect the presence of the human Dpb11/Cut5 homologue, TOPBP1, in the purified RECQ4 complex by both mass spectrometry analysis and western blotting. Moreover, though Xenopus RECQ4 has been shown to co-immunopurify with Cut5, other replication factors such as MCM2-7 helicase and CDC45 were reported not to associate with Xenopus RECQ4 (Matsuno et al, 2006). Our identification of the MCM proteins as the primary RECQ4 interacting partners on human chromatin provides the first evidence of a phyical link between human RECQ4 and the DNA replication machinery that is distinct from the Xenopus homologue.
Cell cycle regulated RECQ4–MCM complex formation
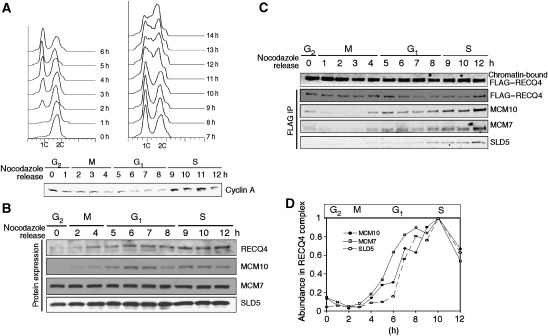
During our purification, we observed that the interaction of RECQ4 with MCM replicative complex only exists in actively dividing cells, but not in quiescent cells, leading us to hypothesize that RECQ4–MCM complex formation is cell-cycle regulated. Consistent with this proposal, complex formation among DNA replication factors is known to be highly regulated by post-translational modification or by cell cycle-dependent protein expression. Indeed, we found that both endogenous RECQ4 and MCM10 are predominantly expressed during G1 and S phases, in contrast to the constitutive expression of MCM2-7 helicase (represented by MCM7 expression) and GINS (represented by SLD5 expression; Figure 2B). To examine the dynamics of the RECQ4 complex during G1 and S phases, in which all the major components of the complex are present, 293T cells stably expressing FLAG–RECQ4 were synchronized in G2/M phase by serum starvation followed by nocodazole treatment to arrest the cells. At different time points after nocodazole release, cells were collected and their stage in the cell cycle was monitored by flow cytometry (Figure 2A, upper) and Cyclin A expression, which is required for S-phase progression (Figure 2A, lower; Yam et al, 2002). We established that the mitotic phase takes place between 1–4 h after nocodazole release, followed by G1 phase (5–8 h) and S phase (9–14+ h). Unlike endogenous RECQ4, FLAG–RECQ4 was constitutively expressed in these cells and could be detected in the chromatin-bound fraction at all cell-cycle stages (Figure 2C, top panel). When FLAG–RECQ4 was immunoprecipitated from the soluble chromatin-bound fraction at each time point, we found that MCM10 indeed associates with RECQ4 only during G1 and S phases because of the cell cycle regulated MCM10 expression. The kinetics of the association of MCM7 with RECQ4 was similar to that of MCM10, whereas SLD5 was only enriched significantly in the RECQ4–MCM complex starting at the G1/S-phase transition. The latter interaction is consistent with observations made in yeast that GINS initiates its interaction with MCM2-7 helicase at the G1/S-phase transition (for review see Sclafani and Holzen, 2007).
Figure 2.
Cell cycle-dependent RECQ4–MCM complex formation. (A) (Upper) flow cytometry of 293T:FLAG–RECQ4 cells at different time points after release from serum starvation and nocodazole double synchronization. Unreplicated cells are identified with DNA content 1C, whereas 2C DNA content represents replicated cells. (Lower) whole-cell extracts of the synchronized cells were blotted with Cyclin A antibody to confirm S and G2 phases. (B) Cell cycle-dependent protein expressions of the endogenous RECQ4, MCM10, MCM7 and Sld5 by western blotting analysis. (C) Western analysis of the chromatin-bound (top panel) and immunopurified FLAG–RECQ4 (2nd–4th panels from top) using M2-agarose from cells synchronized at different time points after nocodazole release. The purified chromatin-bound complexes were analysed with the indicated antibodies to detect the presence of RECQ4, MCM10, MCM7 and SLD5. (D) The abundance of MCM10, MCM7 and SLD5 relative to RECQ4 protein concentration in the purified complex from different time points is quantified based on the western blotting signals shown in part (C).
Cell cycle-dependent association of RECQ4 with origins of replication
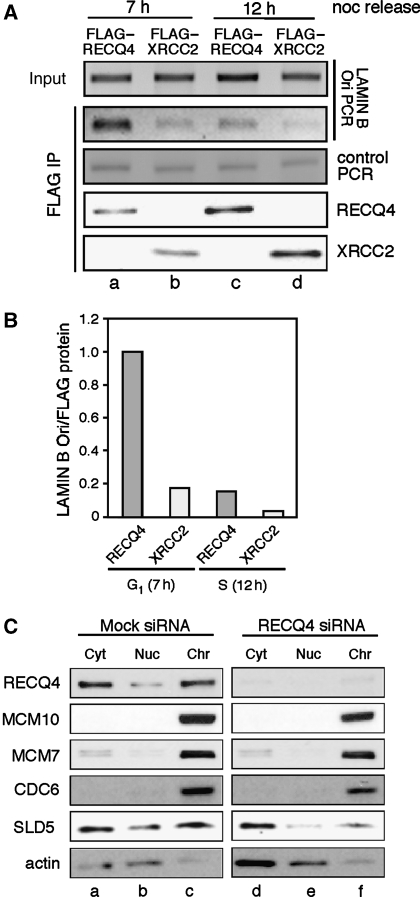
Given that RECQ4 associates with MCM2-7 helicase during G1 phase and that MCM2-7 helicase is part of pre-RC complex (for review see Sclafani and Holzen, 2007), we further established that RECQ4 is recruited to the origins of replication before replication initiation. To do this, we analysed the association of FLAG–RECQ4 with a 225 bp LAMIN B sequence containing one of the most well-characterized mammalian replication origins (Todorovic et al, 1999). Extracts containing an equal amount of DNA input from G1 (7 h post-nocodazole release) and S-phase cells (12 h post-nocodazole release; Figure 3A, top panel) were incubated with M2-agarose to isolate FLAG-tagged proteins at each time point. The 225 bp fragment from the M2-bound protein–DNA complex was amplified by PCR showing that the LAMIN B Ori sequence was five-fold enriched in the purified RECQ4–DNA complex from G1 cells compared to those from S phase (Figure 3A, lanes a and c, and Figure 3B). In contrast, this cell cycle-dependent enrichment at the LAMIN B Ori was not seen in a 309 bp-control PCR at the p53 gene locus (Figure 3A, 3rd panel). In addition, a control chromatin-bound protein, FLAG–XRCC2, failed to exhibit similar cell cycle-dependent LAMIN B Ori association (Figure 3A, lanes b and d, and Figure 3B). Our observations indicate that RECQ4 is recruited to the origin of replication before replication initiation.
Figure 3.
Replication origin association by RECQ4. (A) ChIP analysis of FLAG–RECQ4 association with LAMIN B origin at 7 h (G1) and 12 h (S) after nocodazole release, compared with cells expressing FLAG–XRCC2 as control. (Top panel) 225 bp PCR products flanking LAMIN B origin from extract before immunoprecipitation were analysed on 2% agarose gel. (2nd Panel from top) 225 bp PCR products after FLAG purification using M2-agarose. (3rd Panel from top) control PCR products from p53 gene locus after FLAG purification. Western blot analysis of the purified FLAG–RECQ4 (4th panel from top) and FLAG–XRCC2 (bottom panel) using rabbit anti-FLAG antibody. (B) Quantification of the PCR products relative to FLAG protein concentration bound to M2-agarose beads shown in (A). (C) Effects of the depletion of RECQ4 on protein stability and chromatin association of CDC6, MCM7, MCM10 and SLD5. Equal concentrations of protein extracts from cytosol, nucleoplasmic and chromatin fractions were loaded onto the SDS–PAGE and analysed by western blotting.
The recruitment of RECQ4 to the origin of replication led us to ask whether RECQ4 has a function in pre-RC establishment on human chromatin. For this, RECQ4 siRNA knockdown 293T cells were separated into cytosolic, nucleoplasmic and soluble chromatin-bound fractions (Figure 3C). The lysates were probed with antibodies to detect RECQ4, MCM10, MCM7, SLD5 and CDC6. Consistent with earlier observation (Yin et al, 2004), RECQ4 was found in both the cytoplasm and the nucleus (Figure 3C, top panel, lanes a–c). In contrast, MCM10, MCM7 and CDC6 were enriched in the chromatin fraction. When RECQ4 was downregulated, neither the protein stability nor the chromatin association of CDC6, MCM10 and MCM7 was affected (Figure 3C, lanes d–f), suggesting that RECQ4 is downstream of MCM10 and MCM2-7 in pre-RC establishment. On the other hand, the amount of chromatin-bound SLD5 was significantly reduced in the RECQ4 knockdown cells (Figure 3C, 5th panel from top, comparing lanes c and f). This observation not only is consistent with our cell-cycle analysis indicating that RECQ4 associates with MCM2-7 helicase before GINS during G1 (Figure 2C), but also suggests that RECQ4 is required for the recruitment of GINS to the human chromatin and its interaction with MCM2-7 helicase.
MCM10 mediates RECQ4 association with MCM2-7 helicase/GINS complex
Surprisingly, even though MCM7 and SLD5 are present throughout the cell cycle (Figure 2B, 3rd and 4th panels from top), their interactions with constitutively expressed FLAG–RECQ4 were still limited to G1 and S phases, when MCM10 was expressed (Figure 2C and D). To determine whether the RECQ4 interactions with MCM2-7 helicase and GINS require the presence of MCM10, we analysed FLAG–RECQ4 complex in MCM10 siRNA knockdown cells (Figure 4A). In the absence of MCM10, neither the level of RECQ4 nor that of MCM7 in the chromatin-bound fraction was affected. The latter was in agreement with the earlier report that MCM10 is not required for the chromatin binding of MCM2-7 helicase (Wohlschlegel et al, 2002). On the other hand, similar to the RECQ4 knockdown cells (Figure 3C), the amount of chromatin-bound SLD5 was noticably reduced in the MCM10 knockdown cells, consistent with a function of MCM10 upstream of GINS and CDC45 during DNA replication initiation (Figure 4A, lane b, bottom panel; Wohlschlegel et al, 2002). Nevertheless, in the absence of MCM10, RECQ4 failed to co-purify with both MCM7 and SLD5 (Figure 4A, comparing lanes c and d). This result indicates that MCM10 is required for the formation and the integrity of the RECQ4–MCM replicative complex.
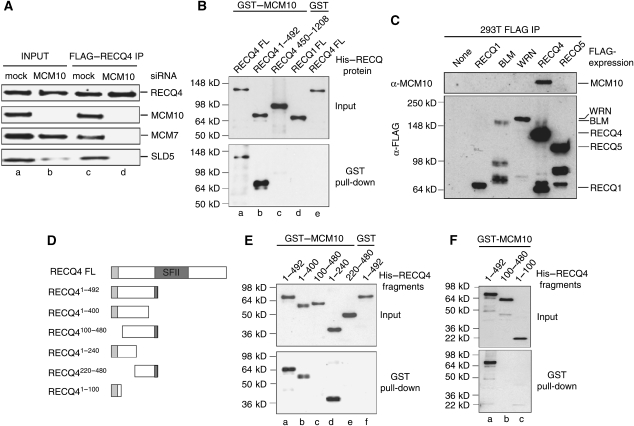
Figure 4.
Direct interaction of RECQ4 and MCM10. (A) Western blot analysis of the chromatin input (a, b) and immunopurified FLAG–RECQ4 complex (c, d) from mock and MCM10 siRNA knockdown cells using the indicated antibodies. For the input lanes, equal amounts of total chromatin-bound proteins from each samples were loaded onto the gel. (B) GST or GST–MCM10 proteins attached to glutathione resins were incubated with His-tagged full-length RECQ4 and RECQ4 fragments. His-tagged RECQ1 was used as control. After washing, the beads were boiled, and bound proteins were analysed by SDS–PAGE followed by western blotting using anti-His antibodies. (Upper panel) 30% of total input of each His-tagged proteins. (Bottom panel) His-tagged proteins after pull-down. (C) MCM10 specifically interacts with RECQ4 but not with other RECQ family helicases. FLAG–RECQ complexes were immunopurified from 293T cells using M2-agarose and analysed for the presence of MCM10 by western blotting using rabbit anti-MCM10 antibody. (D) Schematic diagram of the full-length RECQ4 and RECQ4 fragments. The conserved SFII helicase domain is shown in dark grey, whereas the Sld2-like domain is shown in light grey. (E) GST pull-down experiment as described in (B), except different His–RECQ4 N-terminal fragments were used. (F) GST pull-down experiment as described in (B) to test MCM10 interaction with the RECQ41−100 fragment.
Direct interaction between RECQ4 and MCM10
As MCM10 has a crucial role in the complex formation of RECQ4 with the DNA replication factors, we next determined whether RECQ4 directly interacts with MCM10 using purified recombinant proteins. For this, we overexpressed and purified full-length RECQ4 and various RECQ4 fragments as N-terminal His-tagged and C-terminal FLAG-tagged (Xu and Liu, 2009). We also cloned and purified MCM10 as GST-tagged to homogeneity. We found that GST–MCM10, but not GST alone, bound to glutathione resin, efficiently pulled down full-length RECQ4 (Figure 4B, lanes a and e). No interactions were detected between another RECQ helicase, His–RECQ1 and GST–MCM10 under the same conditions (Figure 4B, lane d).
Given that humans have five RECQ helicases, we next wished to determine the specificity of the MCM10 interaction among different RECQ proteins in vivo. Earlier, we have described that a stable protein complex consisted of RECQ5 and RNA Polymerase II (Aygun et al, 2008). We have also purified FLAG-tagged BLM and WRN complexes from 293T cells. In agreement with earlier reports, mass spectrometry analysis showed that our BLM complex also consisted of Topoisomerase IIIα, RMI1/2, RPA and FANCM, whereas WRN primarily associated with the Ku heterodimer and DNA-PKcs (for review see Liu and West, 2008). Mass spectrometry analysis failed to detect the presence of MCM10 in any of the RECQ complexes except RECQ4. The unique interaction of MCM10 with RECQ4 was further confirmed by western blotting of the purified RECQ complexes (Figure 4C).
Interestingly, using different RECQ4 fragments, the interaction domain was mapped to the N-terminus of RECQ4 (Figure 4B, lane b). When smaller fragments containing sequences derived from the N-terminus were generated and analysed (Figure 4D), we found that all RECQ4 fragments containing the first 99a.a. with the Sld2-like sequence showed a positive interaction with MCM10, suggesting that this region is crucial for the MCM10 interaction (Figure 4E, lanes a, b and d). However, the Sld2-like domain alone is not sufficient for the interaction, as shown by the negative interaction using a RECQ4 fragment containing only the first 100a.a. (Figure 4F, lane c). These data together indicate that MCM10 interacts with the first 200a.a. of RECQ4 containing the Sld2-like sequence.
MCM10 regulates RECQ4 helicase activity
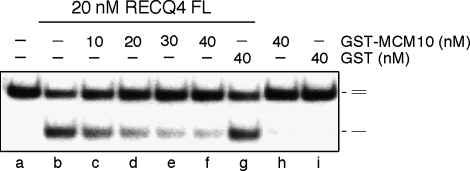
Earlier, we showed that RECQ4 is an active DNA helicase capable of unwinding DNA structures including splayed arms, bubbles and blunt-end duplex DNA (Xu and Liu, 2009). We further showed that the first 99a.a. containing the Sld2-like domain is required for the efficient DNA unwinding activity by promoting protein–DNA interaction. As this Sld2-like domain is also involved in the interaction with MCM10, we next tested the effect of MCM10 interaction on RECQ4 helicase activity. Interestingly, the addition of GST–MCM10 to the RECQ4 reaction efficiently inhibits RECQ4 helicase activity (Figure 5, lanes c–f). Under the same condition, GST showed no effect on the RECQ4 helicase reaction (Figure 5, lane g). The inhibition may be because of the competition for DNA substrate binding by MCM10; however, this is unlikely because the reaction contains excess of unlabelled ssDNA relative to protein concentration (Xu and Liu, 2009). Most likely, the inhibition is due to the inability of MCM10-associated RECQ4 to directly bind to DNA substrate. Our result suggests that RECQ4 is subject to regulation through its direct interaction with MCM10.
Figure 5.
MCM10 inhibits RECQ4 helicase activity. Helicase activity of the recombinant RECQ4 proteins on 32P-labelled duplex oligos in the presence of increasing amounts of purified MCM10 proteins. 32P-labelled single-stranded DNA products were visualized by autoradiography following neutral PAGE.
Regulation of RECQ4–MCM10 interaction through cyclin-dependent kinase phosphorylation sites
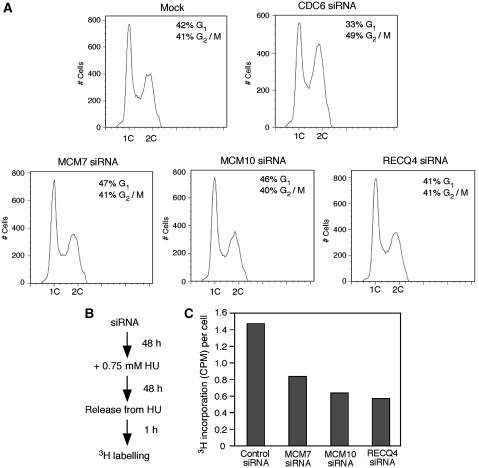
In mammalian cells, excess of MCM2-7 molecules are found on chromatin, and the majority of these molecules are kept inactive during an unperturbed S-phase progression (Ge et al, 2007; Ibarra et al, 2008). Only a limited number of MCM2-7 helicases are needed for origin activation and replication progression. Most of the MCM2-7 helicases associate with dormant origins that serve as backup origins for re-starting DNA synthesis when cells are challenged with DNA replication stress that either stalls or slows down normal replication forks. Consistent with the dormant origin model, we found that MCM7, MCM10 and RECQ4 siRNA knockdown cells showed little defect in cell cycle progression compared with the mock-controlled cells (Figure 6A). In contrast, depletion of CDC6, a protein important for pre-RC establishment, resulted in cell cycle arrest at G2/M phase (Figure 6A).
Figure 6.
RECQ4–MCM complex is required for replication-restart after hydroxyurea treatment. (A) Flow cytometry of 293T cells 48 h after transfecting with mock, CDC6, MCM7, MCM10 and RECQ4 siRNA. (B) Outline of 3H-thymidine labelling of the siRNA knockdown cells after hydroxyurea treatment. (C) DNA synthesis of 293T siRNA knockdown cells after hydroxyurea treatment was monitored by [3H]-thymidine labelling. Count per minute (CPM) was divided by the actual number of cells of each of the siRNA knockdowns to obtain CPM per cell (y-axis).
We next examined the ability of RECQ4 and MCM knockdown cells to restart DNA synthesis after DNA replication stress. For this, we treated the knockdown cells with hydroxyurea for 48 h (Figure 6B). After the cells were released from hydroxyurea, we measured the rate of DNA synthesis by in vivo 3H-thymidine labelling. We found that MCM7, MCM10 and RECQ4 siRNA knockdown cells all showed much reduced efficiency in DNA synthesis. This observation suggests that similar to the MCM proteins, RECQ4 is involved in replication fork restart and/or initiation of dormant origins for the efficient recovery from DNA replication stress (Figure 6C).
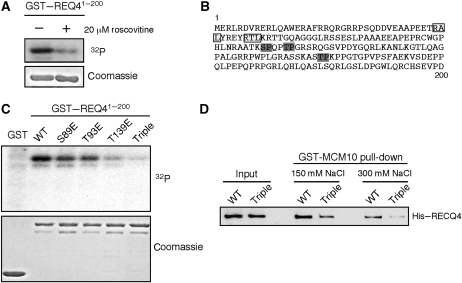
In yeast, both MCM2-7 helicase and Sld2 are activated by hyperphosphorylation to engage in DNA replication (Masumoto et al, 2002; Sheu and Stillman, 2006; Zegerman and Diffley, 2007; Tanaka et al, 2007a, 2007b). Human RECQ4 may also be subjected to cell cycle control by phosphorylation. Indeed, we found that a purified recombinant GST–RECQ41−200 fragment containing the Sld2-like sequence was phosphorylated in vitro by human cell extracts (Figure 7A, left lane). The kinase activity on the GST–RECQ41−200 was significantly reduced by the addition of roscovitine, a cyclin-dependent kinase (CDK) inhibitor, to the extracts (Figure 7A, right lane). RECQ41−200 contains three S/T-P CDK target sites (S89, T93, T139; Figure 7B, dark grey boxes) and two CDK-binding motifs (RxL; Figure 7B, light grey boxes). Phospho-mimicking mutations of any of the CDK target S/T sites to glutamate (E) reduce the amount of 32P-label on the GST–RECQ41−200 fragment (Figure 7C). The reduction is most profound in the fragment containing mutations at all three CDK target sites (e.g. triple mutant, Figure 7C), suggesting that all the three CDK target sites are potentially phosphorylated in vivo. Interestingly, even though the RECQ4 triple-glutamate mutant is proficient in DNA unwinding (Supplementary Figure 2), the mutations significantly weaken the interaction of RECQ4 with MCM10 in a salt-dependent manner compared with WT RECQ4 (Figure 7D). Overall, our data suggest that though MCM10 negatively regulates RECQ4 helicase activity through direct protein–protein interaction, CDK phosphorylation at the Sld2-like domain of RECQ4 may serve as a switch to activate RECQ4 helicase activity during replication by allowing a transient dissociation of RECQ4 from MCM10.
Figure 7.
Regulation of RECQ4–MCM10 interaction by CDK phosphorylation. (A) Phosphorylation of GST–RECQ41−200 by 293T extracts. Glutathione beads with GST–RECQ41−200 fragment were incubated with asynchronized 293T extracts without (left) or with (right) roscovitine in the presence of γ32P-ATP. After washing, 32P-labelled products were detected by SDS–PAGE followed by Coomassie blue staining and autoradiography. (B) Sequence of the first 200a.a. of RECQ4 containing the Sld2 domain. The light grey boxes indicate the cyclin recognition motif (RxL); the dark grey boxes indicate the potential CDK target sites. (C) Phosphorylation of GST–RECQ41−200 WT or glutamate mutants by 293T extracts as described in (A). (D) GST–MCM10 pull-down of full-length WT RECQ4 or glutamate substituted triple mutant at S89, T93 and T139. Inputs and bound fractions were analysed on SDS–PAGE followed by western blotting using anti-His antibody.
Discussion
DNA replication is essential for cell proliferation. Improper DNA replication may compromise chromosome integrity, leading to apoptosis or cell transformation. Studies in mice indicate that the N-terminal domain of RECQ4 is crucial for embryonic development and cell survival, whereas the conserved helicase domain is required for preventing premature aging and cancer predisposition (Ichikawa et al, 2002; Hoki et al, 2003; Mann et al, 2005). In this study, we report that RECQ4 is a part of the replication initiation complex and forms a stable association with essential DNA replication factors, MCM10, MCM2-7 helicase, CDC45 and GINS in a cell cycle-dependent manner. After replication initiation, RECQ4 may also be a part of the replication progression complex along with the replication progression factors, TIM/TIPIN heterodimer. Among these replication factors, we showed that MCM10 directly interacts with RECQ4 and regulates its DNA helicase activity. This interaction is specific to RECQ4 but not other RECQ helicases, suggesting a unique role of RECQ4 in DNA replication. Importantly, we further showed that MCM10 is required for the interactions of RECQ4 with MCM2-7 helicase and GINS. As MCM10 is known to directly interact with MCM2-7 (Homesley et al, 2000), it is possible that RECQ4 indirectly associates with MCM2-7 and GINS through MCM10. Alternatively, we cannot rule out the possibility that a direct interaction between RECQ4 and MCM2-7 helicase requires the MCM10-dependent post-translational modification events (Lee et al, 2003).
Our identification of a cell cycle regulated human RECQ4–MCM replicative helicase complex indicates that the primary role of RECQ4 is in DNA replication, and allows us to suggest a model for the recruitment and activation of RECQ4 in DNA replication. After cell division, RECQ4 establishes a stable interaction with MCM10 and MCM2-7 helicase and is recruited to the origins of replication at G1 phase before replication initiation. Evidence in the Xenopus study suggests that RECQ4 helicase activity is required for efficient origin melting (Sangrithi et al, 2005); therefore, we postulate that RECQ4 activity requires tight regulation by MCM10 to prevent unlicensed replication initiation. Similar to MCM2-7 helicase, RECQ4 may be activated temporarily for the limited number of origin firings during the G1/S transition and S phases, which could be achieved through CDK phosphorylations that release RECQ4 from MCM10, allowing RECQ4 to directly interact with DNA at origins and unwind DNA. Supporting this model, we found that the RECQ4 fragment containing the first 200a.a. was phosphorylated in vitro in a CDK-dependent manner. Importantly, phospho-mimicking mutations at the CDK target sites within this region of RECQ4 greatly reduce its affinity to MCM10 at physiological salt concentrations (e.g. 150 mM). The interesting question that remains to be addressed is why do two DNA helicases (e.g. MCM2-7 and RECQ4) associate with each other during S phase? One possibility is that RECQ4 helicase activity is required to facilitate the opening of duplex DNA at the origin by the MCM2-7 helicase to allow the subsequent loading of RPA and DNA polymerase onto chromatin (Sangrithi et al, 2005).
The data presented here indicate that human RECQ4 is an integral component of the MCM replicative helicase complex during DNA replication. The implication that RECQ4 acts in DNA replication initiation has been suggested from studies of Xenopus extracts, based on its sequence similarity to the yeast replication initiation factor, Sld2 (Sangrithi et al, 2005; Matsuno et al, 2006). However, doubts that this function is evolutionally conserved in mammals were raised. First, not only there is less than 20% sequence identity found between the N-termini of human and Xenopus RECQ4, but also only the first 70a.a of the human RECQ4 protein sequence can be aligned with a small fragment of the yeast Sld2 protein. Second, no interaction between human RECQ4 and TOPBP1 was found in normal cycling cells in our study. Our findings suggest that though the participation of RECQ4 in DNA replication is evolutionally conserved in vertebrates, the molecular basis of the role of RECQ4 in DNA replication may likely have diverged among different organisms.
Although yeast Mcm10 and Sld2 are both important factors for DNA replication initiation, no reports have suggested a direct interaction between Mcm10 and Sld2 in lower eukaryotes. Nonetheless, we cannot exclude the possibility that an association between Sld2 and Mcm10 may also exist but is yet to be shown in lower organisms. Indeed, in humans, MCM10 interacts with RECQ4 through its Sld2-like region. The failure to detect an interaction between RECQ4 and MCM complex in Xenopus egg extracts may be because of the fact that this interaction primarily forms on chromatin rather than in chromatin-free extracts. In addition, though cumulative genetic studies so far demonstrate the requirement of Sld2 for DNA replication initiation, it is not clear how Sld2 communicates with pre-RC in licensing replication (Tanaka et al, 2007a). Our data presented here not only show the complex interaction between human RECQ4 and DNA replication machinery but may also provide an important clue to how Sld2 in yeast potentially interacts with the replication complex containing MCM2-7 helicase, GINS and Cdc45 during replication initiation.
Materials and methods
Cell culture and siRNA
Human 293T cells were cultured in DMEM medium supplemented with 10% v/v foetal bovine serum (FBS) and streptomycin/penicillin (100 U/ml). For synchronization, cells were cultured in DMEM without FBS for 22 h and released from serum starvation by the addition of FBS to a final concentration of 10%. After 2 h incubation at 37°C supplemented with 5% CO2, nocodazole was added to the medium to a final concentration of 50 ng/ml. After 16 h of incubation, cells were released by washing 2 × with complete DMEM medium. FACS analysis was carried out using a standard propidium iodide method. siRNA knockdown cells were generated using SMARTpool siRNAs (Dharmacon) for RECQ4, MCM10 and MCM7. CDC6 siRNA was generated as described (Mailand and Diffley, 2005). siRNAs were transfected into 293T cells using RNAiMax transfection reagent (Invitrogen) according to the manufacturer's protocol. For hydroxyurea treatment, 48 h after siRNA knockdown, 293T cells were incubated with 0.75 mM hydroxyurea for 48 h. Cells were released from hydroxyurea treatment by washing twice with pre-warm medium and further incubated for 1 h in complete medium before the addition of 5 uCi/ml [3H]thymidine (76 Ci/mmole, Perkin Elmer) for 2 h. Cells were processed as described (Liu et al, 1999).
Cell fractionation, immunoprecipitation and ChIP
293T parental cell line and 293T cells stably expressing FLAG-tagged RECQ constructs were fractionated to cytosolic, nucleoplasmic and benzonase-treated soluble chromatin-bound fractions performed as described earlier (Aygun et al, 2008) with the exception that MgCl2 and KCl were used in lysis and nuclease buffers. To immunopurify FLAG-tagged proteins, chromatin or nucleoplasmic extracts were incubated overnight with M2-agarose (Sigma) at 4°C. After binding of the protein complexes, beads were washed extensively with FLAG-A-binding buffer (10 mM HEPES pH7.9, 1.5 mM MgCl2, 0.3 M NaCl, 10 mM KCl, 0.2% Triton X-100, 10% glycerol). The purified FLAG-tagged protein complexes were eluted by using FLAG elution A buffer (10 mM HEPES 7.9, 0.2 M NaCl, 0.2 mM EDTA, 0.05% Triton-X, 0.3 mg/ml FLAG peptide, 10% glycerol), and subjected to western blotting by standard methods or mass spectrometric analysis at the Taplin Biological Mass Spectrometry Facility at Harvard University. For ChIP analyses, chromatin pellet was obtained as described (Aygun et al, 2008) but was re-suspended in chromatin buffer (20 mM HEPES pH7.9, 1.5 mM MgCl2, 150 mM KCl, 10% glycerol, protease and phosphatase inhibitors) followed by sonication. FLAG-tagged protein–DNA complex was immunopurified on M2-agarose, eluted with FLAG elution A buffer. After proteinase K digestion, the co-purified DNA was cleaned up by PCR purification kit (Qiagen) before PCR analysis using primers, 5′-CTGCAGCTGGGGCTGGCATG-3′ and 5′-GACATCCGCTTCATTAGGGCAG-3′ for LAMIN B Ori locus. For the control PCR, 5′CTGCCTCTTGCTTCTCTTTTCC-3′ and 5′GGTTTCTTCTTTGGCTGGG-3′ were used to amplify the p53 gene locus. For immunoprecipitation of endogenous RECQ4, rabbit anti-RECQ4 antibody (SDI) was conjugated to protein-A beads, incubated with 293T extracts and washed with FLAG-A-binding buffer to remove non-specific proteins. For western blotting, rabbit anti-MCM10 (Proteintech Group), rabbit anti-RECQ4 (4–11), goat anti-RECQ4 (Santa Cruz), rabbit anti-MCM7 (Abcam), rabbit anti-SLD5 (SDI) and rabbit anti-CDC6 (Santa Cruz) were used.
Proteins
Recombinant RECQ4 FL and fragments were prepared as described (Xu and Liu, 2009). GST–MCM10 and GST in pGEX-4T-1 vector (GE Healthcare) were expressed in Rosetta (DE3) pLysS cells by induction with 0.1 mM isopropyl-β-D-thio-galactoside overnight at 16°C. Cell pellets were suspended in buffer H (1 × PBS, pH7.4, 10% glycerol, 0.1 mM EDTA, 0.5% Triton X-100), plus 1 × protease inhibitor cocktail (Roche) and lysed by sonication. The supernatant after centrifugation was incubated with glutathione agarose beads (Piercenet) overnight at 4°C. The beads were washed five times with buffer H and eluted with 10 mM glutathione. For GST–MCM10 containing a C-terminal 2xFLAG tag, the eluate from glutathione agarose beads was further purified with anti-FLAG M2 beads (Sigma) as descried for the RECQ4 proteins.
In vitro protein interactions and helicase assays
GST–MCM10/GST bound to the glutathione beads were first blocked with 5 mg/ml of BSA in binding buffer (40 mM Tris, pH 7.4, 10% glycerol, 0.2% Triton X-100, 0.1 mM EDTA) containing 300 mM NaCl (150 mM NaCl as indicated in Figure 7D), and incubated for 5 min at 4°C. His-tagged RECQ4 FL and fragments (40 nM) were then added to the bead mixture and incubated further for 1 h at 4°C. The bound proteins were washed extensively with binding buffer supplemented with NaCl, boiled in SDS sample buffer and analysed on SDS–PAGE followed by western blotting. RECQ4 helicase assays were performed as described (Xu and Liu, 2009).
Kinase assays
293T cell extracts were prepared as described (Esashi et al, 2005). GST–RECQ41−200 WT and mutants bound to glutathione beads were phosphorylated in kinase buffer (10 mM HEPES pH 7.5, 50 mM NaCl, 10 mM MgCl2, 10 mM MnCl2, 5 μM ATP, 1 mM DTT, 2 uCi [γ-32P]ATP) by the addition of 293T cell extract. After 30 min incubation at 30°C, reactions were stopped by washing the glutathione beads extensively with T0G buffer (20 mM Tris pH8.0, 0.2% Triton X-100, 0.5 M NaCl, 3 mM β-mercaptoethanol, 10% glycerol) and boiled in SDS sample buffer for 5 min. Proteins were analysed on SDS–PAGE followed by Coomassie blue staining and autoradiography.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure Legends
Review Process File
Acknowledgments
This work was supported by a post-doctoral fellowship from Fonds de la Recherche en Sante du Quebec to PJR, and grants from American Federation for Aging Research and Yale Rudolph J Anderson Endowed Fellowship to YL. We thank Dr Stephen West for 293 cell-line expressing FLAG–XRCC2.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aygun O, Svejstrup J, Liu Y (2008) A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc Natl Acad Sci USA 105: 8580–8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico A, Costanzo V, Hunt T (2007) Tipin is required for stalled replication forks to resume DNA replication after removal of aphidicolin in Xenopus egg extracts. Proc Natl Acad Sci USA 104: 14929–14934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esashi F, Chris N, Gannon J, Liu Y, Hunt T, Jasin M, West SC (2005) CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature 434: 598–604 [DOI] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ (2007) Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev 21: 3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter AL, Suppa C, Emanuel BS (2007) Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors. J Mol Biol 366: 36–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Xu D, Wang W (2009) Identification and analysis of new proteins involved in the DNA damage response network of Fanconi anemia and Bloom syndrome. Methods 48: 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoki Y, Araki R, Fujimori A, Ohhata T, Koseki H, Fukumua R, Nakamura M, Takahashi H, Noda Y, Kito S, Abe M (2003) Growth retardation and skin abnormalities of the Recql4-deficient mouse. Hum Mol Genet 12: 2293–2299 [DOI] [PubMed] [Google Scholar]
- Homesley L, Lei M, Kawasaki Y, Sawyer S, Christensen T, Tye BK (2000) Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev 14: 913–926 [PMC free article] [PubMed] [Google Scholar]
- Ibarra A, Schwob E, Méndez J (2008) Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci USA 105: 8956–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa K, Noda T, Furuichi Y (2002) Preparation of the gene targeted knockout mice for human premature aging diseases, Werner syndrome, and Rothmund-Thomson syndrome caused by the mutation of DNA helicases. Nippon Yakurigaku Zasshi 119: 219–226 [DOI] [PubMed] [Google Scholar]
- Kamimura Y, Masumoto H, Sugino A, Araki H (1998) Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol Cell Biol 10: 6102–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitao S, Shimamoto A, Furuichi Y (2002) Molecular biology of Rothmund-Thomson Syndrome. In Chromosomal Instability and Aging, pp 223–244. New York, NY: Marcel Dekker, Inc. [Google Scholar]
- Lee JK, Seo YS, Hurwitz J (2003) The Cdc23 (Mcm10) protein is required for the phosphorylation of minichromosome maintenance complex by the Dfp1-Hsk1 kinase. Proc Natl Acad Sci USA 100: 2334–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li M, Lee EY, Maizels N (1999) Localization and dynamic relocalization of mammalian Rad52 during the cell cycle and in response to DNA damage. Curr Biol 9: 975–978 [DOI] [PubMed] [Google Scholar]
- Liu Y, West SC (2008) More complexity to the Bloom's syndrome complex. Genes Dev 22: 2737–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Diffley JF (2005) CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell 122: 915–926 [DOI] [PubMed] [Google Scholar]
- Mann MB, Hodges CA, Barnes E, Vogel H, Hassold TJ, Luo G (2005) Defective sister-chromatid cohesion, aneuploidy and cancer predisposition in a mouse model of type II Rothmund-Thomson syndrome. Hum Mol Genet 14: 813–825 [DOI] [PubMed] [Google Scholar]
- Masumoto H, Muramatsu S, Kamimura Y, Araki H (2002) S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 415: 651–655 [DOI] [PubMed] [Google Scholar]
- Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H (2006) The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol Cell Biol 26: 4843–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohaghegh P, Hickson ID (2002) Premature aging in RecQ helicase-deficient human syndromes. Int J Biochem Cell Biol 34: 1496–1501 [DOI] [PubMed] [Google Scholar]
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR (2005) Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell 121: 887–898 [DOI] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM (2007) Cell cycle regulation of DNA replication. Annu Rev Genet 41: 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B (2006) Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell 24: 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, Ahn B, Bohr VA (2009) Roles of RECQ helicases in recombination based DNA repair, genomic stability and aging. Biogerontology 10: 235–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Tak YS, Araki H (2007a) The role of CDK in the initiation step of DNA replication in eukaryotes. Cell Div 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H (2007b) CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332 [DOI] [PubMed] [Google Scholar]
- Tak YS, Tanaka Y, Endo S, Kamimura Y, Araki H (2006) A CDK-catalysed regulatory phosphorylation for formation of the DNA replication complex Sld2-Dpb11. EMBO J 25: 1987–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorovic V, Falaschi A, Giacca M (1999) Replication origins of mammalian chromosomes: the happy few. Front Biosci 4: D859–D868 [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dhar SK, Prokhorova TA, Dutta A, Walter JC (2002) Xenopus Mcm10 binds to origins of DNA replication after Mcm2-7 and stimulates origin binding of Cdc45. Mol Cell 9: 233–240 [DOI] [PubMed] [Google Scholar]
- Xu X, Liu Y (2009) Dual DNA unwinding activities of the Rothmund-Thomson Syndrome protein, RECQ4. EMBO J 28: 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam CH, Fung TK, Poon RY (2002) Cyclin A in cell cycle control and cancer. Cell Mol Life Sci 59: 1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Kwon YT, Varshavsky A, Wang W (2004) RECQL4, mutated in the Rothmund-Thomson and RAPADILINO syndromes, interacts with ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Hum Mol Genet 13: 2421–2430 [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF (2007) Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure Legends
Review Process File