Abstract
An important property of NEMO, the core element of the IKK complex involved in NF-κB activation, resides in its ability to specifically recognize poly-ubiquitin chains. A small domain called NOA/UBAN has been suggested to be responsible for this property. We recently demonstrated that the C-terminal Zinc Finger (ZF) of NEMO is also able to bind ubiquitin. We show here by ZF swapping and mutagenesis that this represents its only function. While neither NOA nor ZF shows any preference for K63-linked chains, we demonstrate that together they form a bipartite high-affinity K63-specific ubiquitin-binding domain. A similar domain can be found in two other proteins, Optineurin and ABIN2, and can be freely exchanged with that of NEMO without interfering with its activity. This suggests that the main function of the C-terminal half of NEMO is to specifically bind K63-linked poly-ubiquitin chains. We also demonstrate that the recently described binding of NEMO to linear poly-ubiquitin chains is dependent on the NOA alone and does not require the presence of the ZF.
Keywords: K63-linked poly-ubiquitin chains, linear poly-ubiquitin chains, NF-κB/IKK/NEMO, ubiquitin-binding domain
Introduction
The NF-κB family of transcription factors plays a critical role in the immune and inflammatory responses, cell survival and cell proliferation. The NF-κB proteins are sequestered in the cytoplasm through physical interaction with inhibitors of the IκB family. In response to a number of extracellular signals, a cytoplasmic kinase complex, IKK, becomes activated and phosphorylates the IκB's, leading to their degradation through the ubiquitin–proteasome pathway (Häcker and Karin, 2006; Scheidereit, 2006; Hayden and Ghosh, 2008). NF-κB dimers then translocate to the nucleus and activate their target genes. The cytoplasmic IKK complex is composed of two kinase subunits, IKKα and IKKβ, and a regulatory subunit, NEMO/IKKγ. An alternative NF-κB activation pathway only relies on IKKα dimers and does not seem to require IKKβ or NEMO. Although devoid of catalytic activity, NEMO is absolutely required for the canonical NF-κB activation pathway (Yamaoka et al, 1998). Its N-terminus is involved in the interaction with the kinase subunits, whereas the C-terminal half of the molecule is involved in signal recognition. Until recently, few reports were available regarding the structure of NEMO. This protein is predicted to be essentially made of a series of coiled-coils (CC1 (aa 100–196), CC2 (aa 241–286) and LZ (aa 295–336; unless otherwise stated, coordinates are from murine NEMO), and of a C-terminal Zinc-Finger domain (ZF) of unknown function (aa 387–412). The co-crystal of peptides derived from NEMO (aa 44–111) and the C-terminal end of IKKβ indicates that the N-terminus of NEMO forms an elongated parallel two-helix bundle (Rushe et al, 2008). Co-crystallization of the central 150–272 region with the viral KSHV-derived vFLIP NF-κB transactivator indicates that this region forms a parallel intermolecular coiled-coil (Bagnéris et al, 2008). We recently determined the structure of the C-terminal ZF and showed that the integrity of its structure was required for NEMO activity (Cordier et al, 2008).
Few years after its cloning it was reported that mutations affecting the gene encoding NEMO, which is located on the X chromosome, caused severe human pathologies, classified as ID (immunodeficiency), EDA-ID (anhidrotic ectodermal dysplasia with immunodeficiency) and IP (incontinentia pigmenti, the most severe affection, which is embryonic lethal in affected males; Smahi et al, 2000; Courtois et al, 2001; Fusco et al, 2008; Hanson et al, 2008). These mutations have been found to be distributed throughout the entire molecule, and have been demonstrated to more or less severely affect the NF-κB response. Some of these mutations affect certain pathways more than others, in agreement with the hypothesis that NEMO serves as an integrating platform for a series of NF-κB activating signals.
While poly-ubiquitination using Lys48 (K48)-linked chains had long been known to be involved in the NF-κB cascade, in particular through the proteasome-mediated degradation of the IκB inhibitors, recent studies have shown that poly-ubiquitination of signalling proteins through K63-linked poly-ubiquitin chains plays an important role in the activation of the NF-κB cascade (Chen, 2005; Israel, 2006). Contrary to K48 chains, K63-linked poly-ubiquitin chains represent protein–protein interaction motifs that allow inducible recruitment of ubiquitinated proteins or their interacting partners to specific subcellular localizations. These chains are recognized by ubiquitin-binding domains (UBD), which have been described in a large number of proteins (Hicke et al, 2005; Hurley et al, 2006). Recently it was demonstrated that NEMO specifically binds K63-linked poly-ubiquitin chains, and that it also becomes poly-ubiquitinated following NF-κB activation (Ea et al, 2006; Sebban et al, 2006; Wu et al, 2006). In particular, it has been suggested that following TNF treatment, the adaptor molecule RIP1 is recruited to the TNF receptor, becomes poly-ubiquitinated by K63-linked chains, and recruits the IKK complex in a NEMO-dependent manner. Simultaneous recruitment of another kinase complex, the TAK1/TAB1/TAB2 complex, to identical or separate poly-ubiquitin chains through the UBD of TAB2 (or TAB3, a related molecule) has been postulated to allow phosphorylation and activation of the IKK kinase subunits by TAK1, an upstream kinase for the IKK complex. Alternatively poly-ubiquitinated NEMO may directly recruit the TAK1/TAB complex.
Within the minimum region of NEMO required to bind K63-linked chains (aa 259–325; Ea et al, 2006; Wu et al, 2006), a small conserved ±30 amino-acid region embedded in the LZ domain, and called UBAN or NUB, was found to be conserved in five proteins: NEMO, Optineurin and ABIN-1,2,3 (Sebban et al, 2006; Wagner et al, 2008). We made the same observation and called this small domain NOA (Nemo Optineurin Abin). Whereas the function of ABINs is currently poorly understood, it has been shown that Optineurin can compete for binding of NEMO to poly-ubiquitinated RIP1, and therefore behaves as a negative regulator of TNF-mediated NF-κB activation (Zhu et al, 2007). On the other hand, ABIN molecules have been originally cloned as interactors of A20, a K63-specific de-ubiquitinase, which negatively regulates the NF-κB pathway, and proposed to act as NF-κB inhibitors whose activity is dependent on their ability to bind K63 chains. It is, however, unclear whether ABINs act as competitors, similar to Optineurin, or whether they recruit other negative regulators to the IKK complex.
Recently we demonstrated that the C-terminal ZF of NEMO represents a second UBD (Cordier et al, 2009). We also observed that the hydrophobic side of the ZF α-helix interacts, like a majority of the known UBDs, with the hydrophobic region of ubiquitin centred around Ile44, and that mutations that affect the ability of the ZF to bind ubiquitin (such as M415S in human NEMO) also inhibit TNF-mediated NF-κB response. In this report we have replaced the ZF of NEMO by a series of ubiquitin-binding ZFs derived from four proteins, Optineurin, ABIN2, WRNIP1 and Rabex5, and observed that in the four cases this allows to complement the loss of NF-κB activation resulting from deletion of the NEMO ZF. Mutagenesis of the critical ubiquitin-interacting residues in these ubiquitin-binding ZFs allowed us to demonstrate that the main, if not unique, function of the C-terminal ZF of NEMO is to bind ubiquitin.
More recently we have been able to determine the structure of a fragment of murine NEMO (aa 255–344) encompassing the CC2-LZ region (and therefore the NOA domain) and this allowed us through NMR analysis and mutagenesis to build a model of ubiquitin bound to this domain (Grubisha et al, 2009). In this model, the CC2-LZ region also forms an elongated parallel intermolecular coiled-coil. The NOA domain (aa 298–330) contacts ubiquitin through specific residues that include D311 (which contacts H68 of ubiquitin; the D311N mutation gives rise to EDA-ID in male patients; Doffinger et al, 2001) and F312 (which contacts L8 of ubiquitin).
Our studies, as well as other reports, have shown that mutations in NOA or ZF that affect binding to ubiquitin also interfere with activation of IKK and NF-κB. We, thus, determined which of these two domains was actually responsible for high-affinity and specific binding to K63-linked poly-ubiquitin chains. Surprisingly we observed that neither NOA nor ZF showed any specificity for this type of chains, and that their affinity for poly-ubiquitin chains was relatively low. However, a 170-aa fragment of NEMO that contains the two UBDs (encompassing the CC2-LZ-ZF region) exhibits high affinity and high specificity for K63 poly-ubiquitin chains. Interestingly, this bipartite UBD (that we called NOAZ) turned out to be conserved in two other proteins, Optineurin and ABIN2. Replacing the NOAZ of NEMO by that of Optineurin or ABIN2 allowed the chimaeric molecule to fully complement NEMO-deficient cells, strongly suggesting that the essential, if not unique, function of the C-terminal half of NEMO is to specifically bind K63 poly-ubiquitin chains.
As NEMO has been recently described to bind linear poly-ubiquitin chains with high affinity, we studied the determinants of binding to K63-linked versus linear chains by affinity measurements and mutagenesis, and observed that binding to linear chains was only dependent on the NOA domain and did not require the presence of the C-terminal ZF. In addition, we observed that the affinity of full-length NEMO for K63-linked chains was stronger than for linear chains.
Results
The ubiquitin-binding ability of the NEMO ZF is critical for its activity
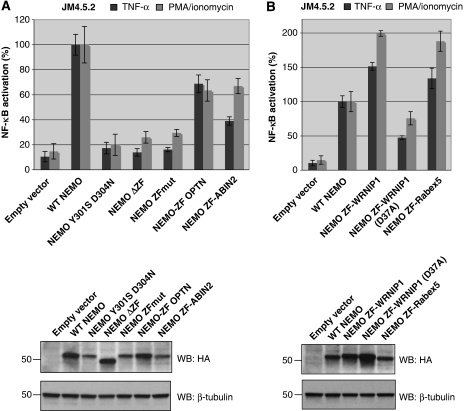
The recent demonstration that the C-terminal ZF of NEMO binds ubiquitin led us to verify whether this was an important function of this region. To answer this question, we replaced this ZF by ubiquitin-binding ZFs derived from other proteins. We first used the C-terminal ZF of Optineurin that shows strong homology with NEMO. As we already showed in a previous report (Figure 3A in reference Schwamborn et al, 2000), replacing the ZF of NEMO by that of Optineurin fully complements the NF-κB response to lipopolysacharide (LPS) in NEMO-deficient 1.3E2 cells. We confirmed that this was also the case for the NF-κB response to TNF and PMA/ionomycin in JM4.5.2 NEMO-deficient T cells (Figure 2A; NEMO-ZF OPTN), when compared with the activity of a NEMO mutant either lacking the ZF (NEMO ΔZF) or mutated on the last two zinc-coordinating amino acids (H406A/C410A) (NEMO ZFmut), which both show an ∼80% reduction in activity. A double mutation of the NOA domain that abolishes binding to ubiquitin (NEMO Y301S D304N identical to human Y308S/D311N: the D311N mutation is associated with the EDA-ID pathology and is impaired in its ability to bind K63-linked poly-ubiquitin chains (Wu et al, 2006), whereas the Y308S mutation has been shown to interfere with binding to K63-linked poly-ubiquitin chains and with IKK activation in a reconstituted in vitro system (Ea et al, 2006)) is shown for comparison.
We noticed that the C-terminal end of ABIN2 (one of the proteins that also contain a NOA-related domain; Sebban et al, 2006; Wagner et al, 2008) also forms a putative ZF (see Figure 1) that exhibits conservation with NEMO and Optineurin of some critical residues involved in either the structure of the ZF or its ability to contact ubiquitin, such as P398 (in human NEMO), or D406 (whose mutation leads to the EDA-ID pathology). We confirmed that a peptide encompassing this domain also binds ubiquitin in a Zn-dependent manner, with an affinity of 1.3 μM for K63-linked poly-ubiquitin3–7 (Table I). The affinity of the NEMO ZF under the same conditions was 16 μM. We, therefore, replaced the ZF of NEMO by that of ABIN2, and observed that this chimaeric molecule could restore, at least partially, the NF-κB response to TNF and PMA/ionomycin in NEMO-deficient T cells (Figure 2A, NEMO ZF-ABIN2). The ZFs of NEMO, Optineurin and ABIN2 form a subfamily of ubiquitin-binding ZFs. We, thus, decided to test an unrelated ubiquitin-binding ZF. We chose WRNIP1 (Werner helicase-interacting protein 1; Bish and Myers, 2007; Bish et al, 2008), a DNA repair protein containing an N-terminal ubiquitin binding ZF of the UBZ family (Bienko et al, 2005; Hurley et al, 2006). This UBD contacts the classical Ile44-centred hydrophobic surface of ubiquitin and does not seem to show specificity for a given poly-ubiquitin chain linkage (Crosetto et al, 2008). Replacement of the NEMO ZF by that of WRNIP1 (aa 15–44) fully complemented NEMO-deficient cells for the NF-κB response to TNF and PMA/ionomycin (Figure 2B, NEMO ZF-WRNIP1). The level of NF-κB activation was even 1.5- to 2-fold higher than with WT NEMO, despite an identical level of expression. We then introduced a mutation of the WRNIP1 ZF that interferes with its ability to bind ubiquitin (D37A; Bish and Myers, 2007) but not with its structure (Bomar et al, 2007). This mutation reduced by 60–70% the ability of the chimaeric molecule to complement NEMO-deficient JM4.5.2 cells (Figure 2B, NEMO ZF-WRNIP1(D37A)).
Figure 1.
Alignment of the C-terminal region (LZ-linker-ZF for NEMO) of human NEMO, Optineurin (Optn) and ABIN2. The NOA which is part of the LZ is indicated. Amino acids conserved in two proteins are indicated in green, and in red when conserved in the three proteins. The last amino acid indicated for each protein is the actual C-terminus.
Table 1.
Affinity of NEMO and ABIN2 ZFs for different types of poly-ubiquitin chains
| KD (chain concentration, μM) | |
|---|---|
| NEMO ZF | |
| K63-Tri-Ub | 17±2a |
| K63-Poly-Ub3–7 | 16±2 |
| K48-Tri-Ub | 18±0.5 |
| Linear tetra-Ub | 27±3 |
| ABIN2 ZF | |
| K63-Tri-Ub | 4±0.7 |
| K63-Poly-Ub3–7 | 1.3±0.2 |
| K48-Tri-Ub | 3±0.3 |
| ZF, zinc finger; Ub, ubiquitin. | |
| aThe affinity of N-terminally fluorescein-coupled peptides for poly-ubiquitin chains was measured as described under section Materials and methods. The affinity for monoubiquitin was too weak to be measured by this technique in the presence of 150 mM KCl. However, we previously calculated a value of 253±15 μM using an F395W mutant (to allow fluorescence measurement) of the human NEMO ZF in a buffer without salt (Cordier et al, 2009). | |
Figure 2.
NF-κB activity elicited by ZF-swap NEMO derivatives in NEMO-deficient T cells. NF-κB activation by TNF-α or PMA/ionomycin was measured in NEMO-deficient JM4.5.2 T cells following transfection of various NEMO derivatives (panels A and B represent separate series of experiments) together with a NF-κB-dependent firefly luciferase reporter construct and a control Renilla luciferase-expressing plasmid. The results shown were obtained from an experiment done in triplicate (see section Materials and methods). 100% corresponds to the ratio between stimulated and non-stimulated cells following transfection of WT NEMO; it corresponds to 9.6- and 6.8-fold for TNF-α and PMA/ionomycin, respectively. Each condition was repeated at least three times and gave similar results. The expression level of the different constructs was determined in a typical experiment by western blot analysis using an anti-HA antibody (shown below the graphs).
All the ZFs we tested so far have been demonstrated to bind ubiquitin through the classical Ile44-centred hydrophobic patch (for NEMO see reference Cordier et al, 2009; for WRNIP1 see reference Crosetto et al, 2008; ABIN2, data not shown). We, therefore, decided to exchange the ZF of NEMO with an ubiquitin-binding ZF that contacts ubiquitin through a different surface. We selected the ZF of Rabex5, a guanine nucleotide exchange factor, which contains two UBDs (Lee et al, 2006; Penengo et al, 2006). This ZF interacts with ubiquitin through a surface centred on Asp58. We observed that similar to WRNIP1, the Rabex5 ZF can replace that of NEMO and give rise to NF-κB activation in response to TNF and PMA/ionomycin (Figure 2B, NEMO ZF-Rabex5), which is almost twofold that obtained with WT NEMO.
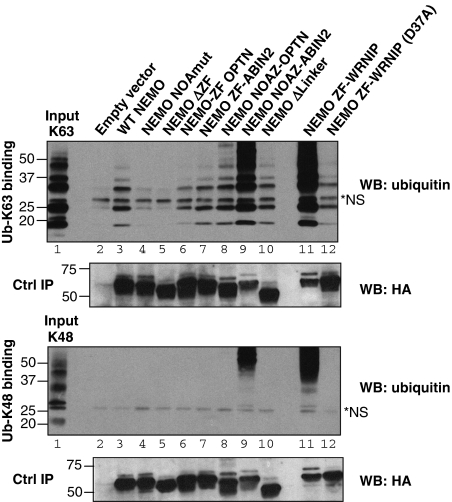
We then verified that the chimaeric molecules still exhibit specificity for K63-linked chains (Figure 3). Pull downs of K48 or K63-linked chains (length 3–7) showed that NEMO deleted of its ZF (lane 5), as well as the Y301S/D304N NOA mutant (lane 4), exhibited strongly reduced binding to K63-linked chains compared with WT NEMO (lane 3). None of these three constructs exhibited binding to K48-linked chains. These results were confirmed using WT or mutated recombinant peptides corresponding to the C-terminal region of NEMO (Supplementary Figure S1A).
Figure 3.
Pull-down activity of NEMO derivatives for K48- and K63-linked chains. HA-tagged NEMO derivatives were immunoprecipitated from transfected 293T cells and incubated with K48- or K63-linked poly-ubiquitin chains (3–7 in length). The interactions were set up as described under section Materials and methods; immunoprecipitated NEMO derivatives were incubated with poly-ubiquitin chains, washed and blotted for ubiquitin (WB: ubiquitin) and for HA-NEMO (WB: HA). K48 and K63 inputs correspond to 8% of the material used for the binding reactions. A non-specific band detected by the anti-ubiquitin antibody in the poly-ubiquitin preparations is shown by an asterisk. The dark background in lanes 9 and 11 is due to cellular ubiquitinated proteins co-precipitated with NEMO. Despite this background the NEMO-NOAZ ABIN2 and NEMO ZF-WRNIP1 maintain their specificity for K63-linked chains.
Similar to WT NEMO, the Optineurin and ABIN2 chimaeric molecules showed strong preference for K63-linked chains (Figure 3, lanes 6 and 7). This was also the case for the WRNIP1 (lane 11) and Rabex5 (not shown) chimaera. Interestingly the D37A mutation of the WRNIP1 ZF strongly interfered with its ability to bind K63-linked poly-ubiquitin chains (Figure 3, lane 12). The larger amount of K63 ladder pulled down by the WRNIP1 chimaera relative to WT NEMO (lane 11 versus 3) indicates it binds poly-ubiquitin chains with an affinity higher than WT NEMO. This was confirmed by the strong background (lanes 11 in the K63 and K48 panels), due to association with cellular ubiquitinated proteins during immunoprecipitation of the chimaeric molecule. This higher affinity may explain the stronger NF-κB activity generated by this construct in transfected JM4.5.2 cells.
Based on these experiments we can conclude that the essential, if not unique, function of the C-terminal ZF of NEMO is to bind ubiquitin.
Specificity for K63 poly-ubiquitin chains requires both the NOA and ZF domains of NEMO
As both NOA and the C-terminal ZF are able to bind ubiquitin, it became important to determine which of these two domains was critical for the high-affinity, K63-specific binding of NEMO to ubiquitin. We first measured the affinity of the NEMO and ABIN2 ZFs for K48 or K63-linked tri-ubiquitin, for K63-linked poly-ubiquitin3–7 and for linear tetra-ubiquitin using fluorescence polarization measurements (Table I, see Materials and methods). The results indicate that neither of the two ZFs shows any preference for K63-linked chains, and that the ABIN2 ZF binds poly-ubiquitin with a higher affinity than the NEMO ZF. For the NOA (aa 298–330), we used a relatively long fragment that includes the CC2 and the LZ domains (aa 215–362), in order to maintain the parallel intermolecular coiled-coil structure, which is required for NOA binding to poly-ubiquitin. Table II represents the affinity of the CC2-LZ domain for K48- or K63-linked poly-ubiquitin3–7, K63 tetra-ubiquitin and for linear tetra- and nona-ubiquitin, measured by a fluorescence competition technique using an F312W mutant of the CC2-LZ peptide as a source of fluorescence (see section Materials and methods; binding curves are presented in Supplementary Figure S3). The values obtained by this technique for the CC2-LZ peptide were similar to those obtained by direct affinity measurement using intrinsic fluorescence of the F312W mutant of the CC2-LZ peptide (we verified that the F312W NEMO mutant was able to complement NEMO-deficient T cells as efficiently as WT NEMO; Supplementary Figure S4), but we did not pursue with the direct technique because the purification of soluble undegraded CC2-LZ-ZF peptide substituted by F312W proved extremely difficult. The results indicate that the CC2-LZ domain binds K48- and K63-linked chains with a similar affinity of 25–45 μM. As neither ZF nor NOA shows any specificity for K63-linked chains, we decided to measure the affinity of a fragment encompassing the CC2-LZ domain, the Pro-rich linker region and the C-terminal ZF (aa 241–412, hereafter called NOAZ) for K48- or K63-linked poly-ubiquitin chains. The affinity of the NOAZ for K63-linked chains increased by a factor of 117 (reaching 0.3 μM) when compared with CC2-LZ, whereas K63/K48 specificity increased by a factor of 23 and reached 32-fold.
Table 2.
Affinity of CC2-LZ and NOAZ for K48-, K63-linked and linear poly-ubiquitin chainsa
| CC2-LZ KD (chain concentration, μM) | NOAZ KD (chain concentration, μM) | Gain of affinity with ZF | |
|---|---|---|---|
| K63-Poly-Ub3–7 | 35 | 0.3 | × 117 |
| K48-Poly-Ub3–7 | 45 | 9.5 | × 4.7 |
| K63/K48 specificity | 1.3 | 32 | |
| K63-Tetra-Ub | 25 | 0.22 | × 114 |
| Linear tetra-ubiquitinb | 2.2 | 1.7 | × 1.3 |
| Linear nona-ubiquitin | 2.0 | 1.9 | × 1.05 |
| ZF, zinc finger; Ub, ubiquitin. | |||
| Italics represent gain of specificity or affinity, whereas bold represents actual affinity values. | |||
| aThe EC50 (defined as the concentration of competitor necessary to displace 50% of the preformed complex) of the CC2-LZ and CC2-LZ-ZF (NOAZ) peptides for K48-, K63-linked or linear poly-ubiquitin chains were measured by fluorescence competition as described under section Materials and methods. The EC50 were transformed into KD after fitting experimental curves obtained with two different concentrations of CC2-LZ(F312W) (0.5 and 3 μM) as described in Supplementary data. The curves corresponding to K48- or K63-linked polyubiquitin3–7, K63 tetra-ubiquitin and linear nona-ubiquitin are shown in Supplementary Figure S3. | |||
| bThe affinity of CC2-LZ and NOAZ for GST-(linear tetra-Ub) was also measured by fluorescence polarization as described under section Materials and methods. Similar KD values of 3 and 3.9 μM were obtained for CC2-LZ and NOAZ, respectively, again showing no gain of affinity with the ZF. | |||
These results demonstrate that presence of both NOA and ZF is required for the specific high-affinity binding of NEMO to K63-linked chains, explaining why mutations located in either domain interfere with NF-κB activation. To verify that this was also true for the chimaeric molecules described above, and in particular that the activity of these molecules was still dependent on the ability of the NOA domain to bind ubiquitin, we introduced the Y301S/D304N mutations in the chimaeric molecule carrying the WRNIP1 ZF, and observed that these mutations strongly interfered with the ability of the chimaeric molecule to complement NEMO-deficient cells (data not shown).
We then asked whether the unique function of the CC2-LZ-linker-ZF region (NOAZ) of NEMO might be to specifically bind K63-linked poly-ubiquitin chains.
The essential, if not unique, function of the NOAZ is to bind K63-linked chains
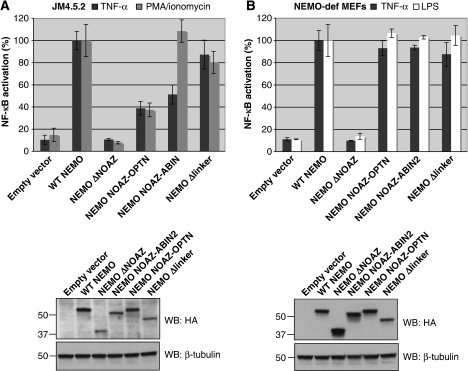
The NOA domain has been found to be conserved in four other proteins besides NEMO, Optineurin and the three ABIN proteins, and it has been demonstrated in all cases to bind poly-ubiquitin (Zhu et al, 2007; Wagner et al, 2008). As mentioned previously, two of these proteins (Optineurin and ABIN2) also exhibit a C-terminal ZF, separated from the NOA by a distance of ∼60 aa (Figure 1). We, therefore, decided to replace the entire region (NOAZ) that includes the two UBDs (aa 300–412) of NEMO by the equivalent region of ABIN2 (aa 306–429) or Optineurin (aa 471–577). To maintain the dimeric coiled-coiled structure of the NOA, we included in the region derived from Optineurin and ABIN2 the equivalent of the NEMO CC2-LZ (structure predictions suggest that the corresponding regions of Optineurin and ABIN2 also adopt a coiled-coil structure, thus potentially allowing the NOA of ABIN2 and Optineurin to bind ubiquitin in a dimeric manner). As shown in Figure 4, replacing the NOAZ of NEMO by that of ABIN2 (NEMO NOAZ-ABIN2) was sufficient to fully complement NF-κB activation by PMA/ionomycin (in JM4.5.2 cells) and LPS or TNF (in NEMO-deficient embryonic fibroblasts (MEFs)). Replacing the NOAZ of NEMO by that of Optineurin (NEMO NOAZ-OPTN) fully complemented the LPS and TNF response in NEMO-deficient MEFs, although it only partially complemented the TNF and PMA/ionomycin response in JM4.5.2 cells. These results indicate that the main function of the C-terminal half of NEMO is to specifically bind K63-linked poly-ubiquitin chains through a new type of bipartite UBD.
Figure 4.
NF-κB activity elicited by NOAZ-swap NEMO derivatives in NEMO-deficient T cells (A) and MEFs (B). NF-κB activation by TNF-α or PMA/ionomycin was measured in NEMO-deficient JM4.5.2 T cells, and by TNF-α or LPS in NEMO-deficient MEFs, following transfection of various NEMO derivatives together with a NF-κB-dependent firefly luciferase reporter construct and a control Renilla luciferase-expressing plasmid. The results shown were obtained from an experiment performed in triplicate. 100% corresponds to the ratio between stimulated and non-stimulated cells following transfection of WT NEMO. It corresponds to 9.6- and 6.8-fold for TNF-α and PMA/ionomycin, respectively, in JM4.5.2 cells, and 9- and 7.7-fold for TNF-α and LPS, respectively, in NEMO-def MEFs. Each condition was repeated at least three times and gave similar results. The expression level of the different constructs was determined in a typical experiment by western blot analysis using an anti-HA antibody (shown below the graphs).
We also verified that these chimaeric molecules still exhibit a specificity for K63-linked chains (Figure 3, lanes 8 and 9). The amount of K63 ladder pulled down by the chimaeric molecule containing the Optineurin NOAZ was similar to that pulled down by WT NEMO (compare lanes 8–3). For the ABIN2 swap, the amounts of both the pulled down K63 ladder and the associated ubiquitinated cellular proteins were significantly higher (lane 9). This, however, cannot be the consequence of the higher affinity of the ABIN2 ZF for ubiquitin relative to the NEMO ZF, as the simple ABIN2 ZF swap (lane 7) did not show such an increased affinity. This suggests that there might be some specificity in the association between the NOA and ZF domains derived from the same protein, unless the linker region also plays a role in the overall affinity.
We thus asked the question of the role of the linker region that separates NOA from ZF. There is little functional information regarding this region in the case of NEMO, but circular dichroism measurements indicated that this region does not form a dimeric coiled-coil, mostly due to the presence of multiple Proline residues (data not shown). However, it includes at least the binding site for the CylD de-ubiquitinase (Saito et al, 2004), as well as a number of predicted or identified phosphorylation sites (Carter et al, 2003, and see http://www.phosphosite.org/proteinAction.do?id=2445&showAllSites=true). We, therefore, tested the ability of a NEMO molecule deleted of the linker region to complement NEMO-deficient cells for NF-κB activation by TNF, PMA/ionomycin or LPS. Figure 4A and B indicates that deletion of the linker region barely interferes with the ability of NEMO to complement NF-κB activation in either NEMO-deficient MEFs or JM4.5.2 (construct NEMO Δlinker). In parallel, pull-down assays (Figure 3, lane 10, and Supplementary Figure S1A, lanes 7 and 12) confirmed that the NOAZ deleted of the linker region binds K63-linked poly-ubiquitin with an apparent affinity and K63 specificity similar to that of WT NOAZ.
Different requirements for binding of NEMO to K63-linked versus linear poly-ubiquitin chains
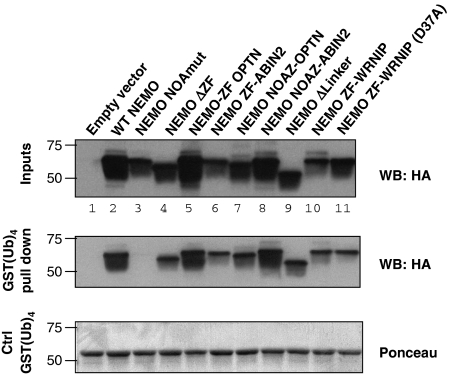
Recent data suggest that NEMO is ubiquitinated using linear poly-ubiquitin chains by the HOIP/HOIL-1 dimeric E3 ligase (Tokunaga et al, 2009). In addition, recent determination, by a combination of X-ray crystallography, NMR and mutagenesis, of the structure of the NOA region complexed with K63 or linear di-ubiquitin (Lo et al, 2009) indicates that the NOA binds to both types of chains, but exhibits higher affinity (by ∼100 fold) for linear di-ubiquitin. However, Lo et al show that the contact points between NOA and either linear or K63-linked chains are almost identical. Another recent report (Rahighi et al, 2009) presents the crystal structure of the NOA bound to linear di-ubiquitin, and proposes that NEMO can bind two linear poly-ubiquitin chains that would run ‘parallel' to the NEMO dimeric coiled-coil. We, therefore, asked whether the ZF also plays a role in the binding of NEMO to linear poly-ubiquitin chains. We first measured the affinity of the ZF for linear poly-ubiquitin chains and observed that it was similar to the affinity for K63- or K48-linked chains (Table I). We then performed pull-down assays using GST-(linear tetra-ubiquitin) and observed that the binding of NEMO to linear chains was strictly dependent on the NOA (Figure 5, compare lane 2 with lane 3, corresponding to the murine Y301S/D304N mutation) but independent of the ZF (compare lane 4 with lane 2). Similar results were obtained with recombinant NEMO-derived peptides (Supplementary Figure S1B). Quantitative measurements confirmed that presence of the ZF did not modify the affinity of NEMO for linear tetra-ubiquitin chains (Table II and Supplementary Figure S3). We obtained a value of ∼2 μM, which was similar to that determined by Lo et al (2009) using ITC and Rahighi et al (2009) using surface plasmon resonance. To test whether the length of the poly-ubiquitin chains could be responsible for the differential requirement for the ZF, we performed pull-down experiments using K63-linked chains of length 5, 6 and 8, as well as linear nona-ubiquitin chains (Supplementary Figure S2), and measured the affinity of NOA and NOAZ for nona-ubiquitin (Table II and Supplementary Figure S3). The results confirm the role of the ZF in NEMO binding to K63-linked chains but not to linear chains, irrespective of the chain length.
Figure 5.
Pull-down activity of NEMO derivatives for linear tetra-ubiquitin chains. Pull-down experiments using HA-tagged NEMO derivatives and linear tetra-ubiquitin (GST-(Ub)4). The interactions were set up as described under section Materials and methods: cytosolic extracts of transfected 293T cells were incubated with GST(Ub)4 bound to glutathione agarose beads. After washing, the bound material was eluted with Laemmli buffer and blotted with anti-HA (middle panel). The level of expression of the NEMO derivatives was determined by a direct anti-HA blot (top panel). The level of recovered GST-(Ub)4 was determined by Ponceau staining of the membrane (lower panel).
From these data we can conclude that NEMO binding to linear poly-ubiquitin chains does not require the C-terminal ZF, whereas this domain is necessary for specific high-affinity binding to K63-linked poly-ubiquitin chains. In addition, while the NOA binds linear with a higher affinity than K63-linked poly-ubiquitin (Table II and reference Lo et al, 2009), the reverse is true for the NOAZ (Table II).
Discussion
The results presented here indicate that NEMO, as well as Optineurin and ABIN2, contain a new type of bipartite UBD that specifically binds K63-linked poly-ubiquitin chains with high affinity. This domain that we call NOAZ includes the NOA, a Pro-rich linker region of ∼60 aa (which seems to be dispensable) and the C-terminal ZF. In addition, our data indicate that the requirements for binding of NEMO to K63-linked or linear poly-ubiquitin chains are different, as the ZF is only required for high-affinity binding to K63-linked chains.
The NOA domain is conserved in the three proteins mentioned above, as well as in ABIN1 and ABIN3. However, a composite domain similar to NOAZ cannot be found in ABIN1 and ABIN3, although putative divergent ZF motifs can be identified in these two molecules. Interestingly, ABIN1 has recently been demonstrated to bind K48- and K63-linked poly-ubiquitin chains with a similar affinity (Oshima et al, 2009). Based on our results, it can be predicted that the specificity of binding of ABIN1 and ABIN3 may be different from that of NEMO, Optineurin and ABIN2.
Our recent results (Cordier et al, 2009) indicate that the ZF of NEMO binds ubiquitin through the classical Ile44-centred hydrophobic surface. This UBD binds K48-, K63-linked and linear chains (Table I) with similar affinity. Deletion or mutations of the ZF of NEMO have been shown to strongly interfere with its ability to respond to NF-κB-activating signals (Schwamborn et al, 2000; Aradhya et al, 2001; Courtois et al, 2001; Jain et al, 2001; Makris et al, 2002; Tang et al, 2003; Zhou et al, 2004; Temmerman et al, 2006; Fusco et al, 2008; Hanson et al, 2008) and to be associated with severe pathologies in humans (Fusco et al, 2008; Hanson et al, 2008). More recently we demonstrated that the M415S mutation, which prevents binding of NEMO to ubiquitin (but does not interfere with the structure of the ZF), severely impairs NF-κB activation by TNF and that the M407V mutation, which is associated with the severe IP pathology (Smahi et al, 2000), also interferes with binding of the ZF to ubiquitin, as well as with activation of NF-κB by TNF in an ex vivo complementation assay (Cordier et al, 2009). These results suggest that binding to ubiquitin is an important function of the ZF. To further address this point, we replaced the ZF of NEMO by a series of progressively more distant ubiquitin-binding ZFs. We first chose the ZF of Optineurin, which is highly similar to that of NEMO, and the ZF of ABIN2, which is slightly more distant but belongs to the same subgroup, and which we demonstrated to bind ubiquitin with an affinity higher than that of the NEMO ZF. Whereas deletion of the NEMO ZF strongly interferes with NF-κB activation by TNF, PMA/ionomycin and LPS, adjunction of either of these two heterologous ZFs restored NF-κB activation. We then replaced the NEMO ZF by that of the DNA repair protein WRNIP1, a more distant ZF belonging to a different family (UBZ) of UBDs, and obtained a similar result, the resulting chimaeric molecule being even more active than WT NEMO. Importantly, mutation of an Asp residue in the ZF of WRNIP1 known to interfere with binding to ubiquitin (mutation D37A; Bish and Myers, 2007) in the context of the NEMO-WRNIP1 chimaera strongly reduced the activity of this molecule in the complementation assay. These data indicate that the ability of the NEMO C-terminus to bind ubiquitin is necessary and sufficient for NF-κB activation by several stimuli.
These three ZFs contact ubiquitin through the classical hydrophobic surface centred on Ile44. To determine whether this is an important parameter, we replaced the ZF of NEMO by the ubiquitin-binding ZF of the guanine nucleotide exchange factor Rabex5, which has been shown to contact ubiquitin through a new surface centred around Asp58 (Lee et al, 2006; Penengo et al, 2006). Interestingly, the chimaeric molecule was as efficient as the WRNIP1 chimaera, indicating that there is some degree of flexibility in the manner the NEMO ZF binds ubiquitin, possibly as a consequence of the presence of the flexible linker region.
Pull-down experiments indicated that the chimaeric molecules that contain the WRNIP1 and Rabex5 ZFs bound K63 chains with a higher affinity than WT NEMO (Figure 3 and data not shown), possibly explaining the more efficient complementation ability observed in NEMO-deficient cells (Figure 2). However, measurements of the affinities of the NEMO and WRNIP1 ZFs for ubiquitin does not provide an obvious explanation for these differences: the affinity of the WRNIP1 ZF for K48- or K63-linked chains has been measured to be ∼20 μM (Crosetto et al, 2008), and we calculated that of WT NEMO to be about the same (Table I; the affinity of the Rabex5 ZF has only be calculated for mono-ubiquitin and found to be 12 or 22 μM depending on the authors; Lee et al, 2006; Penengo et al, 2006).
The NEMO ZF includes a conserved Lys residue, which has been suggested to be an important site of ubiquitination, and whose mutation interferes with NF-κB activation by certain stimuli such as TCR (Sun et al, 2004; Zhou et al, 2004, 2005). The structure of the NEMO ZF indicates that when complexed with ubiquitin, Lys399 is no longer accessible (Cordier et al, 2009 and data not shown), suggesting that binding of ZF to ubiquitin and ubiquitination of Lys399 are mutually exclusive. This suggests the possibility that ubiquitination of NEMO on this Lys residue might interfere with its ability to interact with ubiquitin. It is, however, difficult, within the current stage of our understanding of the role of NEMO ubiquitination, to determine whether this represents a negative (through preventing NEMO from binding to its normal targets) or a positive event (allowing NEMO and the IKK complex to detach from the receptor in order to become active, as has been recently described for CD40-dependent NF-κB activation (Matsuzawa et al, 2008) and suggested to take place in the case of TNF (Zhang et al, 2000)). In the latter publication, the de-ubiquitinase A20, a negative regulator of NF-κB activation, seems to increase the recruitment of the signalosome to the TNF receptor, in accordance with the idea that NEMO ubiquitination might represent a positive event.
Regarding the NOA, our unpublished results (Grubisha et al, 2009) indicate that the CC2-LZ region that encompasses the NOA domain forms a dimeric intermolecular coiled-coil, which is able to contact two ubiquitin molecules, one on each side of the coiled-coil. A good correlation could be observed between some of the contact points with ubiquitin we identified and human pathologies, including D311, which contacts H68 of ubiquitin and whose mutation is associated with EDA-ID in male patients (Doffinger et al, 2001). Rather unexpectedly based on the current literature, the NOA domain did not show any preference for binding to K48 or K63-linked chains (Table II and Supplementary Figure S3). Whereas the ZF exhibited the same lack of specificity (Table I), we demonstrated that the presence of both the NOA and ZF domains resulted in high-affinity K63-specific binding to poly-ubiquitin chains; compared with the CC2-LZ domain, the adjunction of the ZF results in an ∼100-fold increase in affinity and a 30-fold increase in the preference for K63-linked versus K48-linked chains. This bipartite UBD, we call NOAZ, includes a Pro-rich linker region (aa 340–393) located between the C-terminus of the coiled-coiled LZ region and the ZF, and which is predicted to be poorly structured (data not shown). Interestingly, among the five known proteins that contain a NOA domain (NEMO, Optineurin and ABIN1–3), three of them (NEMO, Optineurin and ABIN2) also contain a NOAZ domain, with a relatively conserved C-terminal ZF and a non-conserved ∼50-aa linker region. The NEMO NOAZ can be replaced by the NOAZ of Optineurin or ABIN2 without losing the ability to complement NEMO-deficient cells for the NF-κB response to at least three types of signals (TNF, PMA/ionomycin and LPS). Therefore, the NOAZ represents a new type of bipartite UBD, whose modular nature is required to generate specificity for K63-linked chains. In addition, we also demonstrated that the presence of the linker region is not absolutely required, at least within the limits of the relatively crude assays we use, as its deletion in the context of NEMO does not interfere with the ability of the resulting deleted molecule to complement NEMO-deficient cells, nor to bind to K63-linked chains with an affinity similar to that of the WT molecule. The presence of multiple phosphorylation sites within this linker region, as well as presence of a putative binding site for the CylD de-ubiquitinase, suggests it might be a regulatory domain, although the role of these phosphorylation events is currently unknown. It will be interesting to determine whether these modifications regulate the ability of NEMO to bind ubiquitin.
In the absence of an X-ray structure of a complex between NOAZ and K63 poly-ubiquitin chains, we can only speculate about the organization of such a complex. A recently published study proposes a structure for the complex between the NOA (within the CC2-LZ domain) and K63-linked or linear di-ubiquitin, obtained by a combination of X-ray crystallography, NMR and mutagenesis (Lo et al, 2009). Lo et al propose that a poly-ubiquitin chain (whether linear or K63-linked) wraps around the dimeric NOA coiled-coil, contacting both sides of the CC2-LZ with two consecutive ubiquitins. In this case the poly-ubiquitin chain would run ‘perpendicular' to the CC2-LZ domain. In addition, Lo et al report that the affinity of the NOA for linear di-ubiquitin (1.4 μM) is ∼100-fold higher than for K63-linked di-ubiquitin (131 μM). More recently an X-ray structure of the NOA domain bound to linear di-ubiquitin has been obtained (Rahighi et al, 2009), associated with a mutagenesis analysis that suggests that NEMO binding to this type of chain may be physiologically relevant. Rahighi et al demonstrate that a dimeric NOA presents two binding sites for ubiquitin on each side of the dimer, suggesting an organization with two poly-ubiquitin chains running ‘parallel' to the CC2-LZ. This organization is consistent with the model we propose for K63-linked chains (Grubisha et al, 2009), except that in addition to the dimeric NOA, the two ZFs would contact two additional ubiquitin molecules. Rahighi et al (2009) also show that the C-terminal/N-terminal junction between the two adjacent ubiquitins is a critical determinant of the high-affinity specific binding of NOA to linear poly-ubiquitin chains. What is unclear at the moment is whether the NOA can also contact two consecutive ubiquitin molecules (on each side of the dimeric molecule) in the context of K63-linked chains.
Interestingly, our study reveals that the ZF of NEMO is not required for high-affinity binding to linear poly-ubiquitin chains (Figure 5, Supplementary Figure S1B, S2, S3 and Table II), although we cannot determine at the moment whether it can bind simultaneously with the NOA to this type of chain. One may, thus, wonder why it is required to bind to K63-linked chains. One simple explanation might be that the affinity of the NOA for linear ubiquitin is higher than that of the ZF, therefore minimizing the contribution of the latter, whereas the affinity of the NOA for K63-linked chains is slightly weaker than that of the ZF. As to why the combination of NOA and ZF shows stronger affinity for K63-linked than for K48-linked chains, the explanation will have to await the determination of the structure of the NOAZ complexed with K63-linked chains, the obvious next challenge raised by our study.
Interestingly, two recent papers elucidate the reason for specific binding of the Rap80 protein to K63-linked chains (Sato et al, 2009; Sims and Cohen, 2009): this protein contains two UIM domains separated by a seven-amino-acid linker, and the authors demonstrate that the high-affinity binding to K63-linked chains of this composite domain is dependent on the length and structure of the linker. The situation of NEMO seems to be different as the linker is long and unstructured (and its length varies between NEMO, Optineurin and ABIN2), and the way the ZF contacts ubiquitin does not seem to be important.
Finally a recent paper (Tokunaga et al, 2009) proposes that NEMO is ubiquitinated using linear poly-ubiquitin chains by the HOIP/HOIL-1 dimeric E3 ligase. Therefore, one of the putative ubiquitinated molecules recognized by NEMO might be NEMO itself. Assuming NEMO binding to both K63-linked and linear poly-ubiquitin chains is physiologically relevant, one way of functionally separating the two activities would be to specifically abolish one or the other. Interfering specifically with binding to K63-linked chains can be achieved by mutating or deleting the ZF. Indeed the ample evidence regarding the importance of the ZF for NEMO activity and the existence of mutations found in patients that interfere with the ability of the ZF to bind ubiquitin confirm that NEMO binding to K63-linked chains is physiologically relevant. Another interesting possibility of interfering with the activity of the ZF consists in the ubiquitination of its conserved Lysine residue, which as discussed above would interfere with its ability to bind ubiquitin, and, therefore, would prevent binding of NEMO to K63-linked chains but not to linear chains.
On the other hand, specifically interfering with binding to linear chains depends on the existence of specific contact points between the NOA and linear chains that would differ from the contact points with K63-linked chains. Lo et al (2009) suggest that mutation of R319 (corresponding to murine R312) specifically affects binding to linear chains but not to K63-linked chains. On the other hand Rahighi et al (2009) propose that the distal binding site on the NOA, which includes R309/R312/E313, is unique to linear chains. Therefore, based on these observations it should be possible to separate the two specificities of NEMO, helping to understand their respective biological functions.
Materials and methods
Cell culture, transient transfection, and reagents
Human 293T cells (obtained from the American Type Culture Collection) and NEMO-deficient MEFs (NEMO−/−; Schmidt-Supprian et al, 2000)) were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% foetal bovine serum, penicillin (50 U/ml) and streptomycin (50 μg/ml). NEMO-deficient Jurkat T cells (JM4.5.2; Harhaj et al, 2000) were cultured in RPMI medium supplemented with 10% foetal bovine serum and antibiotics as above. Transient transfections were performed by the calcium phosphate method for 293T. NEMO-deficient cell lines were transfected using Fugene HD (Roche) according to the manufacturer's instructions. Recombinant human TNF-α was obtained from R&D Systems, recombinant mouse TNF-α was from Apotech Corporation and phorbol-12-myristate-13-acetate (PMA), LPS (from Salmonella typhimurium) and ionomycine were from Sigma.
Cloning and mutagenesis
HA-tagged mouse NEMO cloned in pCMV vector (Yamaoka et al, 1998) was used as a backbone to derive all the variants described in this study. Replacement of the NEMO ZF by that of Optineurin has been described previously (Schwamborn et al, 2000). NEMO-ΔZF was obtained by introducing a stop codon after amino acid 386 of NEMO by PCR site-directed site directed mutagenesis using Phusion-HS (Fermentas) as DNA polymerase. NEMO-ZFmut (C406A/H410A) was obtained by site-directed mutagenesis. For replacement of the NEMO ZF with other ZF domains, an EcoRI site was introduced at position 1158 (amino acid 386) of the mouse NEMO cDNA by site-directed mutagenesis. The EcoRI/XbaI fragment was then replaced by the ZF-coding DNAs of the other proteins, either amplified by PCR or chemically synthethized. Coordinates of the ZFs are aa 405–430 of mouse ABIN2; aa 15–44 of human WRNIP1 and aa 17–42 of human Rabex5. Point mutants NEMO Y301S D304N, NEMO-ZF(WRNIP1) Y301S D304N and NEMO-ZF(WRNIP1 D37A) were obtained by site-directed mutagenesis. For the exchange of the NOAZs, an existing EcoRV site on mNEMO located at the beginning of the NEMO NOA domain (position 895, amino acid 300) was used to excise the C-terminal part of NEMO and replace it by the C-terminal part of human Optineurin (aa 471–577) and human ABIN2 (aa 306–430) obtained by PCR. The NEMO Δ-linker was generated by introduction of EcoRI sites at positons 1016 and 1158 of mNEMO, resulting after EcoRI digestion and re-ligation in a deletion of aa 339–386. NEMO Δ-NOAZ was obtained by EcoRV–XbaI digestion, fill-in and re-ligation, resulting in truncation of mNEMO after amino acid 300. Human OPTN-containing plasmids have been described previously (Schwamborn et al, 2000). Human and mouse ABIN2-containing plasmids were obtained from Dr Rudi Beyaert (VIB Gent, Belgium).
For cloning into bacterial expression vectors, the C-terminal part of mNEMO as well as NEMO Y301S D304N mutant (aa 215–412), NEMO Δ-ZF (aa 215–386) and NEMO Δ-linker (Δ339–386) were amplified by PCR and subcloned into pET-28 expression vector (Novagen) in frame with a 6 × Histidine-tag. All the constructs mentioned above were fully verified by sequencing.
Ubiquitin binding
Transiently transfected 293T cells expressing HA-tagged NEMO derivatives were lysed in binding buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 0.2 % Triton X-100, 1 mM EDTA, 0.5 mM DTT, 800 μM N-ethyl-maleimide, 200 μM iodoacetamide, and a mixture of protease inhibitors (Complete, Roche) and phosphatase inhibitors (Sigma). For analysis of binding to linear poly-ubiquitin chains, cleared supernatants were incubated with GST(Ub)4 bound to glutathione agarose beads for 1 h at 4°C. Beads were then washed three times in binding buffer and the bound material was eluted with Laemmli buffer. For binding to K48 or K63 poly-ubiquitin chains, NEMO was first immunoprecipitated from cleared lysates using the 12CA5 anti-HA antibody, followed by protein G-agarose bead precipitation. After three washes in binding buffer, beads were divided in two aliquots and incubated with 1 μg of either K48 or K63 ubiquitin chains (length 3–7; Boston Biochem, Cambridge, MA) for 1 h at 4°C in binding buffer. Beads were then washed three times with the same buffer and the bound material was eluted with Laemmli buffer. For binding using recombinant NEMO, 1 μg of bacterially produced 6 × His-tagged NEMO derivative was first bound to nickel-activated Sepharose beads (GE Healthcare) in a buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 0.2% Triton X-100 and 20 mM imidazole. Bound NEMO polypeptides were incubated in the same buffer with 1 μg of either GST(Ub)4, K48 or K63 poly-ubiquitin for 1 h at 4°C. After three washes, bound material was eluted with Laemmli buffer. After SDS–PAGE separation and western blotting, membranes were probed with an anti-HA antibody (HA-11; Covance) to detect NEMO, an anti-ubiquitin antibody coupled to the horseradish peroxidase (P4D1-HRP; Santa Cruz) to detect K48 and K63 ubiquitin chains, and an anti-GST antibody (Sigma) to detect GST(Ub)4. Recombinant NEMO proteins were detected using an anti-NEMO monoclonal antibody (antibody B24; homemade).
Cell stimulation and luciferase assays
NEMO-deficient JM4.5.2 cells were transfected in 96-well plate (1.105 cells/well) using 0.375 μl Fugene HD (Roche) and 250 ng DNA per point (75 ng Igκ–firefly luciferase reporter, 25 ng thymidine kinase–Renilla luciferase reporter, 10 ng NEMO or empty vector and 140 ng empty vector). Forty-two hours after transfection, cells were stimulated with 10 ng/ml human TNF-α or 50 ng/ml PMA plus 1 μg/ml ionomycine for 4 h at 37°C. Firefly and Renilla luciferase activities were then measured using the Dual Reporter Assay system (Promega), according to the manufacturer's instruction, and a luminometer (Centro XS, Berthold) equipped with automated injection. NEMO-deficient MEFs were plated on 24-well plates (5 × 104 cells/well) and transfected the next day using 1 μl Fugene HD (Roche) and 250 ng DNA per point as described above. Forty-two hours after transfection, cells were stimulated with 10 ng/ml mouse recombinant TNF-α or 15 μg/ml LPS for 4 h at 37°C. Quantification of firefly and Renilla luciferase activities were determined as described above. Relative NF-κB activation was determined as the ratio of the luciferase activity and Renilla activity. Standard deviations were calculated from the values obtained from experiments performed in triplicate. For each condition, experiments, performed in triplicate, were repeated at least three times and gave similar results.
Fluorescence spectroscopy
Fluorescence measurements were recorded with a PTI QuantaMaster spectrofluorometer (PTI, Lawrenceville, NJ) equipped with a thermostated cuvette holder. All experiments were performed with a 0.3-cm path length quartz cuvette at 25°C. The excitation wavelength was 296 nm to reduce the contribution of the tyrosyl residues to the total fluorescence. Intrinsic fluorescence emission measurement was recorded at 347 nm. Bandwidths of excitation and emission monochromators were set at 2 and 4 nm, respectively. Proteins were diluted to the desired concentrations in 25 mM Tris–HCl containing, 150 mM KCl, 0.2 mM DTE, 0.1 mM DDM at pH 7.5 (buffer A), and allowed to equilibrate for 1 h at room temperature prior to analysis. The F312W mutation was introduced to allow fluorescence measurement, and we verified that this mutation did not interfere with the affinity for poly-ubiquitin chains nor with the ability of NEMO(F312W) to complement NEMO-deficient cells (data not shown and Supplementary Figure S4). In all titrations, the progressive dilution during the course of titration never exceeded 10%, and each fluorescence reading was collected for 1 min after a 2-min period of equilibration. Dissociation constants (KD) were calculated as described in (Cordier et al, 2009), and refer to chain concentration. For competition experiments, the intrinsic fluorescence of the CC2-LZ(F312W) mutant (0.5 or 3 μM) was recorded alone and after adding either K63- or K48-linked poly-Ub3–7, K63-tetra-ubiquitin or linear tetra- or nona-ubiquitin (1.5 or 3 μM) to preform the complex. This led to a fractional quenching of 28 or 40% depending on the concentration of poly-Ub. The intrinsic fluorescence was then measured after adding various concentrations (0.15–30 μM) of the CC2-LZ or NOAZ peptides. The dissociation constants (KD) of the CC2-LZ and NOAZ peptides were calculated as described in the Supplementary data. To assess the binding of CC2-LZ or NOAZ domain to GST-linear tetra-ubiquitin, the N-terminally Cy5-labeled CC2-LZ (residues 253–336; 0.2 μM) was incubated with the CC2-LZ or NOAZ domain (5 μM) in buffer A and allowed to equilibrate for 30 min to preform heterodimers. The mixture was then incubated with a variable concentration of GST-linear tetra-ubiquitin (2–20 μM). Fluorescence polarization measurements were taken on a Tecan microplate reader infiniteR F500 (Tecan France S.A.S) using a 635-nm excitation filter and a 665-nm emission filter. Binding curves were fitted as described by Cordier et al (2009).
To measure the affinity of the ZFs derived from NEMO and ABIN2 for poly-ubiquitin chains, NEMO and ABIN2 ZF peptides labelled at their N terminus with fluorescein were purchased from Biopeptide Co. (San Diego, CA) and New England Peptide (Gardner, MA), respectively. The sequence of the NEMO ZF peptide (>98%) has been previously described by Cordier et al (2009). The sequence of the ABIN2 ZF peptide (> 95%) is: F-GDLQCPHCLQCFSDEQGEELLRHVAECCQ. Fluorescence polarization measurements were taken in Buffer A containing 0.6 mM ZnCl2 using a concentration of peptide of 0.1 μM and a variable concentration of poly-Ub (Boston Biochem, length 3–7) or linear tetra-ubiquitin. Measurements were taken using a 485-nm excitation filter and a 535-nm emission filter. I44A mono-ubiquitin was used as control to verify that the increase of fluorescence polarization signal results from specific poly-Ub binding. No deviation of polarization signal was observed using a variable concentration of I44A mono-Ub from 0.1 to 800 μM.
Supplementary Material
Supplementary Information
Review Process File
Acknowledgments
We are grateful to M Kaminska for the pcDNA3-F312W NEMO mutant, to I Dikic for the GST-(Ub)4 construct, to S Polo for the Rabex5 cDNA, to R Beyaert for the ABIN2 cDNA and to T Kamitani for the GST-UbC1 construct. The JM4.5.2 NEMO-deficient cell line was a gift of SC Sun, and the NEMO-deficient MEFs were from M Pasparakis. We thank Robert Weil for critical reading of the paper. This work was supported in part by Canceropole Ile de France, Contract no. 2007 to MV and AI, by Fondation BNP Paribas to MV, and by European Community Network of Excellence RUBICON Project no. LSHC-CT-2005-018683 to EL and AI.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aradhya S, Courtois G, Rajkovic A, Lewis AL, Levy M, Israël A, Nelson DL (2001) Atypical forms of incontinentia pigmenti in males result from mutations of a cytosine tract in exon 10 of NEMO (IKKg). Am J Hum Genet 68: 765–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnéris C, Ageichik AV, Cronin N, Wallace B, Collins M, Boshoff C, Waksman G, Barrett T (2008) Crystal structure of a vFlip–IKKgamma complex: insights into viral activation of the IKK signalosome. Mol Cell 30: 620–631 [DOI] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I (2005) Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310: 1821–1824 [DOI] [PubMed] [Google Scholar]
- Bish RA, Fregoso OI, Piccini A, Myers MP (2008) Conjugation of complex polyubiquitin chains to WRNIP1. J Proteome Res 7: 3481–3489 [DOI] [PubMed] [Google Scholar]
- Bish RA, Myers MP (2007) Werner helicase-interacting protein 1 binds polyubiquitin via its zinc finger domain. J Biol Chem 282: 23184–23193 [DOI] [PubMed] [Google Scholar]
- Bomar MG, Pai MT, Tzeng SR, Li SS, Zhou P (2007) Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase eta. EMBO Rep 8: 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter RS, Pennington KN, Ungurait BJ, Ballard DW (2003) In vivo identification of inducible phosphoacceptors in the IKK{gamma}/NEMO subunit of human I{kappa}B Kinase. J Biol Chem 278: 19642–19648 [DOI] [PubMed] [Google Scholar]
- Chen ZJ (2005) Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol 7: 758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordier F, Grubisha O, Traincard F, Véron M, Delepierre M, Agou F (2009) The zinc finger of NEMO is a functional ubiquitin-binding domain. J Biol Chem 284: 2902–2907 [DOI] [PubMed] [Google Scholar]
- Cordier F, Vinolo E, Veron M, Delepierre M, Agou F (2008) Solution structure of NEMO zinc finger and impact of an anhidrotic ectodermal dysplasia with immunodeficiency-related point mutation. J Mol Biol 377: 1419–1432 [DOI] [PubMed] [Google Scholar]
- Courtois G, Smahi A, Israel A (2001) NEMO/IKK gamma: linking NF-kappa B to human disease. Trends Mol Med 7: 427–430 [DOI] [PubMed] [Google Scholar]
- Crosetto N, Bienko M, Hibbert RG, Perica T, Ambrogio C, Kensche T, Hofmann K, Sixma TK, Dikic I (2008) Human Wrnip1 is localized in replication factories in a ubiquitin-binding zinc finger-dependent manner. J Biol Chem 283: 35173–35185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doffinger R, Smahi A, Bessia C, Geissmann F, Feinberg J, Durandy A, Bodemer C, Kenwrick S, Dupuis-Girod S, Blanche S, Wood P, Rabia SH, Headon DJ, Overbeek PA, Le Deist F, Holland SM, Belani K, Kumararatne DS, Fischer A, Shapiro R et al. (2001) X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-kappaB signaling. Nat Genet 27: 277–285 [DOI] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ (2006) Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell 22: 1–13 [DOI] [PubMed] [Google Scholar]
- Fusco F, Pescatore A, Bal E, Ghoul A, Paciolla M, Lioi MB, D'Urso M, Rabia SH, Bodemer C, Bonnefont JP, Munnich A, Miano MG, Smahi A, Ursini MV (2008) Alterations of the IKBKG locus and diseases: an update and a report of 13 novel mutations. Hum Mutat 29: 595–604 [DOI] [PubMed] [Google Scholar]
- Grubisha O, Kaminska M, Duquerroy S, Fontan E, Cordier F, Haouz A, Raynal B, Chiaravalli J, Delepierre M, Israël A, Véron M, Agou F (2009) DARPin-assisted crystallography of the CC2-LZ domain of NEMO reveals a coupling between dimerization and ubiquitin binding (submitted) [DOI] [PubMed]
- Häcker H, Karin M (2006) Regulation and function of IKK and IKK-related kinases. Sci STKE 2006: re13. [DOI] [PubMed] [Google Scholar]
- Hanson EP, Monaco-Shawver L, Solt LA, Madge LA, Banerjee PP, May MJ, Orange JS (2008) Hypomorphic nuclear factor-kappaB essential modulator mutation database and reconstitution system identifies phenotypic and immunologic diversity. J Allergy Clin Immunol 122: 1169–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harhaj EW, Good L, Xiao G, Uhlik M, Cvijic ME, Rivera-Walsh I, Sun SC (2000) Somatic mutagenesis studies of NF-kappa B signaling in human T cells: evidence for an essential role of IKK gamma in NF-kappa B activation by T-cell costimulatory signals and HTLV-I Tax protein. Oncogene 19: 1448–1456 [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S (2008) Shared principles in NF-kappaB signaling. Cell 132: 344–362 [DOI] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP (2005) Ubiquitin-binding domains. Nat Rev Mol Cell Biol 6: 610–621 [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G (2006) Ubiquitin-binding domains. Biochem J 399: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel A (2006) NF-kappaB activation: nondegradative ubiquitination implicates NEMO. Trends Immunol 27: 395–397 [DOI] [PubMed] [Google Scholar]
- Jain A, Ma CA, Liu S, Brown M, Cohen J, Strober W (2001) Specific missense mutations in NEMO result in hyper-IgM syndrome with hypohydrotic ectodermal dysplasia. Nat Immunol 2: 223–228 [DOI] [PubMed] [Google Scholar]
- Lee S, Tsai YC, Mattera R, Smith WJ, Kostelansky MS, Weissman AM, Bonifacino JS, Hurley JH (2006) Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat Struct Mol Biol 13: 264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YC, Lin SY, Rospigliosi CC, Conze DB, Wu CJ, Ashwell JD, Eliezer D, Wu H (2009) Structural basis for recognition of diubiquitins by NEMO. Mol Cell 33: 602–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makris C, Roberts JL, Karin M (2002) The carboxyl-terminal region of IkappaB kinase gamma (IKKgamma) is required for full IKK activation. Mol Cell Biol 22: 6573–6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa A, Tseng PH, Vallabhapurapu S, Luo JL, Zhang W, Wang H, Vignali DA, Gallagher E, Karin M (2008) Essential cytoplasmic translocation of a cytokine receptor-assembled signaling complex. Science 321: 663–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima S, Turer E, Callahan J, Chai S, Advincula R, Barrera J, Shifrin N, Lee B, Yen B, Woo T, Malynn B, Ma A (2009) ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature 457: 906–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, Di Fiore PP, Polo S, Schneider TR (2006) Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell 124: 1183–1195 [DOI] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I (2009) Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell 136: 1098–1109 [DOI] [PubMed] [Google Scholar]
- Rushe M, Silvian L, Bixler S, Chen LL, Cheung A, Bowes S, Cuervo H, Berkowitz S, Zheng T, Guckian K, Pellegrini M, Lugovskoy A (2008) Structure of a NEMO/IKK-associating domain reveals architecture of the interaction site. Structure 16: 798–808 [DOI] [PubMed] [Google Scholar]
- Saito K, Kigawa T, Koshiba S, Sato K, Matsuo Y, Sakamoto A, Takagi T, Shirouzu M, Yabuki T, Nunokawa E, Seki E, Matsuda T, Aoki M, Miyata Y, Hirakawa N, Inoue M, Terada T, Nagase T, Kikuno R, Nakayama M et al. (2004) The CAP-Gly domain of CYLD associates with the proline-rich sequence in NEMO/IKKgamma. Structure 12: 1719–1728 [DOI] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S (2009) Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J 28: 2461–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidereit C (2006) IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene 25: 6685–6705 [DOI] [PubMed] [Google Scholar]
- Schmidt-Supprian M, Bloch W, Courtois G, Addicks K, Israël A, Rajewsky K, Pasparakis M (2000) NEMO/IKK gamma-deficient mice model incontinentia pigmenti. Mol Cell 5: 981–992 [DOI] [PubMed] [Google Scholar]
- Schwamborn K, Weil R, Courtois G, Whiteside ST, Israël A (2000) Phorbol esters and cytokines regulate the expression of the NEMO-related protein, a molecule involved in a NF-kappa B-independent pathway. J Biol Chem 275: 22780–22789 [DOI] [PubMed] [Google Scholar]
- Sebban H, Yamaoka S, Courtois G (2006) Posttranslational modifications of NEMO and its partners in NF-kappaB signaling. Trends Cell Biol 16: 569–577 [DOI] [PubMed] [Google Scholar]
- Sims JJ, Cohen RE (2009) Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell 33: 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smahi A, Courtois G, Vabres P, Yamaoka S, Heuertz S, Munnich A, Israël A, Heiss NS, Klauck S, Kioschis P, Wiemann S, Poustka A, Esposito T, Bardaro T, Gianfrancesco F, Ciccodicola A, D'Urso M, Woffendin H, Jakins T, Donnai D et al. (2000) Genomic rearrangement in NEMO impairs NF-kB activation and is a cause of incontinentia pigmenti. Nature 405: 466–472 [DOI] [PubMed] [Google Scholar]
- Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ (2004) The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell 14: 289–301 [DOI] [PubMed] [Google Scholar]
- Tang ED, Wang CY, Xiong Y, Guan KL (2003) A role for NF-kappaB essential modifier/IkappaB kinase-gamma (NEMO/IKKgamma) ubiquitination in the activation of the IkappaB kinase complex by tumor necrosis factor-alpha. J Biol Chem 278: 37297–37305 [DOI] [PubMed] [Google Scholar]
- Temmerman ST, Ma CA, Borges L, Kubin M, Liu S, Derry JM, Jain A (2006) Impaired dendritic-cell function in ectodermal dysplasia with immune deficiency is linked to defective NEMO ubiquitination. Blood 108: 2324–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, Yamamoto M, Akira S, Takao T, Tanaka K, Iwai K (2009) Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol 11: 123–132 [DOI] [PubMed] [Google Scholar]
- Wagner S, Carpentier I, Rogov V, Kreike M, Ikeda F, Lohr F, Wu CJ, Ashwell JD, Dotsch V, Dikic I, Beyaert R (2008) Ubiquitin binding mediates the NF-kappaB inhibitory potential of ABIN proteins. Oncogene 27: 3739–3745 [DOI] [PubMed] [Google Scholar]
- Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD (2006) NEMO is a sensor of Lys 63-linked polyubiquitination and functions in NF-kappaB activation. Nat Cell Biol 8: 398–406 [DOI] [PubMed] [Google Scholar]
- Yamaoka S, Courtois G, Bessia C, Whiteside ST, Weil R, Agou F, Kirk HE, Kay RJ, Israël A (1998) Complementation cloning of NEMO, a component of the IkB kinase complex essential for NF-kB activation. Cell 93: 1231–1240 [DOI] [PubMed] [Google Scholar]
- Zhang SQ, Kovalenko A, Cantarella G, Wallach D (2000) Recruitment of the IKK signalosome to the p55 TNF receptor: RIP and A20 bind to NEMO (IKKgamma) upon receptor stimulation. Immunity 12: 301–311 [DOI] [PubMed] [Google Scholar]
- Zhou H, Du MQ, Dixit VM (2005) Constitutive NF-kappaB activation by the t(11;18)(q21;q21) product in MALT lymphoma is linked to deregulated ubiquitin ligase activity. Cancer Cell 7: 425–431 [DOI] [PubMed] [Google Scholar]
- Zhou H, Wertz I, O'Rourke K, Ultsch M, Seshagiri S, Eby M, Xiao W, Dixit VM (2004) Bcl10 activates the NF-kappaB pathway through ubiquitination of NEMO. Nature 427: 167–171 [DOI] [PubMed] [Google Scholar]
- Zhu G, Wu CJ, Zhao Y, Ashwell JD (2007) Optineurin negatively regulates TNFalpha- induced NF-kappaB activation by competing with NEMO for ubiquitinated RIP. Curr Biol 17: 1438–1443 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File