Abstract
Deletions of chromosome 6 are a common abnormality in diverse human malignancies including astrocytic tumours, suggesting the presence of tumour suppressor genes (TSG). In order to help identify candidate TSGs, we have constructed a chromosome 6 tile path microarray. The array contains 1780 clones (778 PACs and 1002 BACs) that cover 98.3% of the published chromosome 6 sequences. A total of 104 adult astrocytic tumours (10 diffuse astrocytomas, 30 anaplastic astrocytomas (AA), 64 glioblastomas (GB)) were analysed using this array. Single copy number change was successfully detected and the result was in general concordant with a microsatellite analysis. The pattern of copy number change was complex with multiple interstitial deletions/gains. However, a predominance of telomeric 6q deletions was seen. Two small common and overlapping regions of deletion at 6q26 were identified. One was 1002 kb in size and contained PACRG and QKI, while the second was 199 kb and harbours a single gene, ARID1B. The data show that the chromosome 6 tile path array is useful in mapping copy number changes with high resolution and accuracy. We confirmed the high frequency of chromosome 6 deletions in AA and GB, and identified two novel commonly deleted regions that may harbour TSGs.
Keywords: Brain tumour, Molecular cytogenetics, Array-CGH, Glioblastoma, Astrocytoma
Introduction
Astrocytic tumours are the most common human primary malignancies of brain. Adult astrocytic tumours are subclassified into 3 malignancy grades, namely diffuse astrocytomas (malignancy grade II, abbreviated as A), anaplastic astrocytomas (malignancy grade III, AA) and glioblastoma (malignancy grade IV, GB) in the World Health Organization (WHO) classification (Kleihues & Cavenee, 2000). Glioblastomas, the most malignant of all, are resistant to the conventional therapy with median survival being less than one year. It has been shown that the p53 pathway (TP53, p14ARF, MDM2) is frequently altered by way of genetic abnormalities of one of its component in all three subtypes, while the RB1 pathway (CDKN2A/B, CDK4, RB1) is simultaneously altered along with the p53 pathway in some AAs and the majority of GBs, but not in As. Amplification, aberrant expression and rearrangements of EGFR as well as mutations/deletions of PTEN are also common among GBs. In addition, genomic abnormalities have been identified in a number of chromosomal regions, suggesting that further oncogenes or tumour suppressor genes (TSGs) are involved (reviewed in (Ichimura et al., 2004)). The long arm of chromosome 6 is one such region. We have previously studied the allelic status of chromosome 6 in 159 astrocytic tumours using 31 microsatellite markers and identified several commonly deleted regions (Miyakawa et al., 2000). However, these regions were too large to pin-point candidate TSGs.
Recently, sequencing of human chromosome 6 has been completed (Consortium, 2004; Mungall et al., 2003). It is approximately 171 Mb in size, representing about 6% of the human genome and 167 Mb of the euchromatin has been sequenced. The sequence encodes at least 1611 genes (http://www.ensembl.org/Homo_sapiens/), some of which have been associated with human disease such as PARK2 (juvenile parkinsonism), SCA1 (spinocerebellar ataxia) and HFE (hemochromatosis) ((Mungall et al., 2003) and http://bioinfo.weizmann.ac.il/cards/index.shtml). A number of human tumours have deletions of various regions on chromosome 6, suggesting the presence of novel tumour suppressor genes (TSGs) ((Acquati et al., 2001) and references therein). Several genes have been isolated as candidate TSGs in these regions, including PDCD2 (6q27) in lymphomas (Steinemann et al., 2003) and UNC93A (6q27) in ovarian cancer (Liu et al., 2002), however neither of these had convincing evidence of tumour-specific mutations. CDKN1A/p21CIP1 (6p21.31) is found mutated in a small fraction of Burkitt's lymphoma and prostate cancers (Bhatia et al., 1995; Gao et al., 1995), however no mutations in CDKN1A have been identified in astrocytic tumours (Koopmann et al., 1995). Thus, to-date a confirmed TSG on chromosome 6 involved in astrocytic tumours has yet to be identified.
Recent advances in microarray-based comparative genomic hybridisation (array-CGH) and the completion of chromosome 6 sequencing allowed us to develop a chromosome 6 tile path array for CGH, to further investigate chromosome 6 copy number alterations in astrocytic tumours (Fiegler et al., 2003; Mungall et al., 2003). Unlike microsatellite analysis (MSA), the array-CGH allows the quantitative assessment of copy number changes independent of naturally occurring, unevenly distributed microsatellite repeat polymorphisms and interpreting allelic imbalance. The findings are directly linked to the sequences, and hence the genes, unlike conventional metaphase-based CGH. Here we report the construction, validation and application of the human chromosome 6 tile path array, covering 98.3% of the published chromosome sequence, to the investigation of copy number abnormalities in 104 adult astrocytic tumours. We confirmed that chromosome 6 deletions are a frequent event in AA and GB, and identified two commonly deleted regions on 6q26.
Results
Validation of the chromosome 6 tile path array
We constructed two versions of the chromosome 6 tile path array (for detail see Materials and Methods). The first version consisted of the chromosome 6 tile path clone set, 38 chromosome X clones and 6 Drosophila BAC clones, while the second version included a set of clones that covers the whole genome in approximately 10 Mb interval (the 10-Mb clone set) in addition to the chromosome 6 tile path set. The second version was used in the majority of the cases (76 cases). All clones are listed in the supplementary table 1.
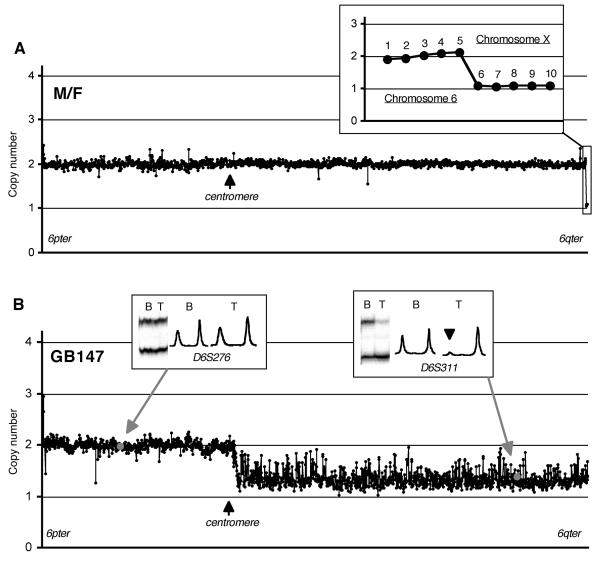
In order to validate the ability of the chromosome 6 CGH-microarray to discriminate single copy number change, a series of 16 hybridisations using normal DNA from 4 individual males, 4 individual females, one mixture of 20 males and one mixture of 20 females in various combinations was performed (Figure. 1A). The mean copy number of the chromosome 6 clones was 2.00 (SD = 0.10). When the copy numbers of five selected chromosome X specific clones (RP11-58H17, RP11-576P23, RP11-446J19, RP11-230E14, RP11-481F23) in normal male DNA were assessed using female DNA as reference, the mean copy number was 1.13 (SD = 0.06), which was consistent with the presence of a single copy of chromosome X in the male. These results indicated that the array used in this study is capable of discriminating a single copy number change (Figure. 1A inset).
Figure. 1.
A. A linear scale plot of copy number at chromosome 6 and chromosome X clones in a normal male DNA (test) against a normal female DNA (reference) normalized using 6p clones. Inset shows a plot for five contiguous chromosome 6 clones (1, RP11-302L19; 2, RP5-1086L22; 3, RP5-894D12; 4, RP1-140C12; 5, RP1-191N21) and five chromosome X clones (6, RP11-58H17; 7, RP11-576P23; 8, RP11-446J19; 9, RP11-230E14; 10, RP11-481F23). B. A linear scale plot of copy numbers of chromosome 6 tile path clones in a glioblastoma (GB147) normalized using 6p clones. This case was shown to have allelic imbalance at all informative microsatellite markers on 6q used in the previous study (Miyakawa et al., 2000). An array-CGH experiment showed reduction of copy number close to 1 in all 6q clones indicating total loss of one copy of 6q. Insets show representative microsatellite results (autoradiographic band patterns and their densitometric profiles) at two loci, one on 6p (D6S276, allelic balance) and the other on 6q (D6S311, allelic imbalance). Allelic imbalance is indicated by an arrowhead. The clones containing the markers in the plot are indicated by grey spots. B, blood; T, tumour.
Chromosome 6 status assessment by microsatellite and array-CGH analysis
In total 104 primary astrocytic tumours (10 A, 30 AA, 64 GB) were subjected to chromosome 6 tile path array-CGH. Among them, 54 tumours were selected based on previously reported MSA data showing allelic imbalance for chromosome 6 loci ((Miyakawa et al., 2000) and see below). Twenty-eight of these were studied using the first version of the array alone, 12 using both versions while the remaining 14 and the 50 randomly selected tumours were examined using the second version alone. The chromosome 6 copy number status for the 54 tumours was determined by comparing the MSA and array-CGH data while the randomly selected additional 50 tumours were assessed solely on the basis of data obtained using the second version of the chromosome 6 array, which included the 10-Mb set clones. The 10-Mb set clones were only used for normalization of the chromosome 6 array and no copy number assessment of the chromosomes represented by this set of clones was made.
Eight of the 54 tumours with MSA data (GB7, GB14, GB82, GB137, GB147, GB148, GB150, AA3) had allelic imbalance at all informative loci on 6q (Miyakawa et al., 2000). These 8 cases invariably showed copy number reduction at all 6q clones, consistent with deletion of 6q (see example in Figure. 1B). Copy number values were however higher in some tumours when compared to those from chromosome X clones in normal male/female DNA hybridisations.
In the light of the above findings, a copy number value lower than 1.7 or higher than 2.3 was first used as an indication of deletion or gain, respectively (i.e., <Mean−3×SD or >Mean+3×SD of chromosome 6 clone copy numbers in normal male/female hybridisations). However, when copy number changes were borderline at a number of consecutive clones, the corresponding region was judged as having deletions/gains affecting either 1) all tumour cells but with a large amount of DNA from normal cells in the tumour tissue or 2) only a subpopulation of tumour cells with the deletions/gains the majority of tumour cells do not have the change.
The calculated copy number fluctuated to a varying degree from region to region and case to case. This is because a) each clone contains a varying amount of repetitive sequence, which in some cases is incompletely suppressed despite an excess of Cot1 DNA in the hybridisation mix, and b) because of experimental artefacts such as scanning-field un-uniformity. To avoid over-interpretation of the data, a copy number change was considered significant only when it was observed in two or more consecutive clones (see Discussion). In addition, 50 individual clones, distributed along chromosome 6, consistently showed discordant copy numbers as compared to their neighbouring clones in control/control and tumour/control hybridisations. Almost all these clones were isolated non-consecutive clones, and it was considered unlikely that these clones represented a genuine tumour-specific change, but rather that they represented a regional copy number polymorphism (Iafrate et al., 2004; Sebat et al., 2004) or cross-hybridisation to a homologous region from another part of the genome. These 50 clones were excluded from the analysis as not being suitable for copy number assessment (see supplementary table 1).
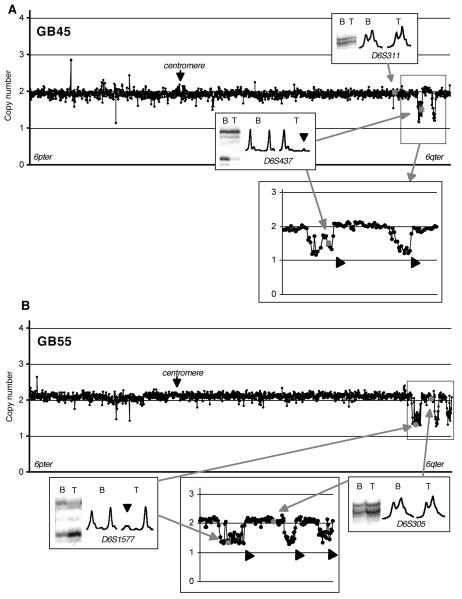
Based on the above principles, the chromosome 6 status of the 54 tumours with MSA data was determined. In the majority of the cases, copy number changes were observed by array-CGH where allelic imbalance was detected by MSA. Examples are shown in Figures. 2 and 3. GB45 was shown to have allelic imbalance at two markers, D6S415 and D6S437, suggesting the presence of a small deletion (Miyakawa et al., 2000). Array-CGH showed that the clones containing these markers had clear copy number reduction, confirming hemizygous deletion and refining it to an approximately 2 Mb region (RP11-230C9 to RP11-507C10) (Figure. 2A). Telomeric to this region, GB45 had another small deletion (RP11-266I4 to RP3-492C2, approximately 1.2 Mb) in a region where no microsatellite markers were used. Array-CGH using the constitutional DNA of GB45 showed no abnormality on chromosome 6, indicating that these deletions are somatic (data not shown). GB55 was judged to have allelic imbalance at D6S1577 and D6S415 (Miyakawa et al., 2000), a finding that was concordant with a small deletion identified by array-CGH (Figure. 2B). Telomeric to this deletion, this case had two additional interstitial deletions.
Figure. 2.
Linear scale plots of copy numbers at chromosome 6 tile path clones in two glioblastomas (A, GB45 and B, GB55) normalized using the 10-Mb set clones. An enlarged plot around the deleted region is shown below each plot. Small interstitial deletions identified by array-CGH are indicated by thick arrows. Insets show microsatellite results at two loci in each case, D6S311 (allelic balance) and D6S437 (allelic imbalance) for GB45 and D6S1577 (allelic imbalance) and D6S305 (allelic balance) for GB55. Allelic imbalance is indicated by an arrowhead. The clones containing the markers in the plot are indicated by grey spots. B, blood; T, tumour.
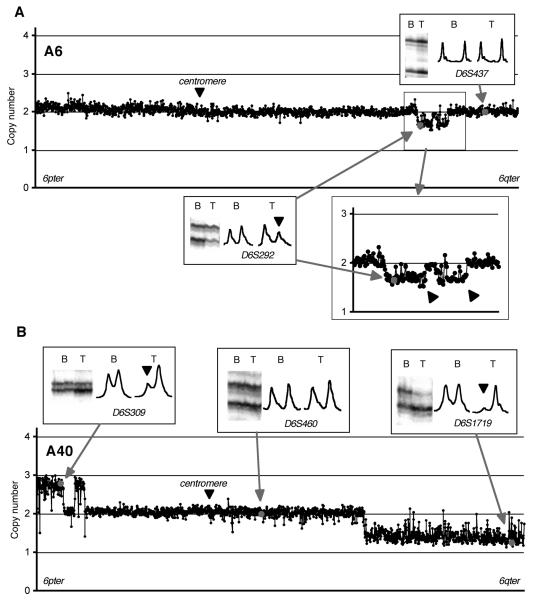
Figure. 3.
Linear scale plots of copy numbers at chromosome 6 tile path clones in two diffuse astrocytomas (A, A6 and B, A40) normalized using the 10-Mb set clones. Two closely located deletions are indicated by thick arrows in an enlarged plot around the deletions in A6 below the plot (A). Insets show microsatellite results, D6S292 (allelic imbalance) and D6S437 (allelic balance) for A6, and D6S309 (allelic imbalance), D6S460 (allelic imbalance) and D6S1719 (allelic imbalance) for A40. Note that the ratio of the signal intensities of the two bands at D6S292 in A6 tumour DNA or D6S309 in A40 tumour DNA as compared to that of their corresponding blood DNA was about 50%, compared to near 100% disappearance of the larger band at D6S1719 in A40 tumour DNA (indicated by an arrowhead). Because the amount of DNA loaded in blood and tumour are not exactly the same, it would not be possible to determine whether the imbalance was due to reduction of one allele or increase of the other solely by MSA.
The diffuse astrocytoma, A6, had allelic imbalance at a single marker D6S292. In this case both alleles were clearly visible with the intensity of the smaller allele being approximately 50% less than that of the larger allele (a pattern referred as “partial allelic imbalance” hereafter, see Figure. 3A). The array-CGH data showed copy number reduction in the region including the clone containing the D6S292 sequences (CTA-31J9 to RP11-164A17, 11 Mb), although the mean copy number in this region was 1.67 (SD=0.12), being considerably higher than that of chromosome X clones (mean=1.13, SD=0.06). The intensity ratio of the deleted allele in the tumour compared to blood at D6S292 was 0.53, suggesting that half of the cells in the tumour piece did not have the deletion (either normal cells or a subpopulation of tumour cells).
In some cases, allelic imbalance turned out to represent copy number gain. A40 had allelic imbalance at two loci (D6S174 and D6S309) on 6p24, as well as at D6S1657 and all informative loci telomeric to this including D6S1719 (Miyakawa et al., 2000). The microsatellite band pattern at D6S309 in A40 showed partial allelic imbalance, a pattern very similar to that of D6S292 in A6 (compare D6S292 in Figure. 3A and D6S309 in Figure. 3B), while at D6S1719 the larger allele almost completely disappeared in the tumour DNA. Array-CGH demonstrated that there was an increase in copy number of the distal 6p region encompassing the D6S309 locus while a large portion of the terminal 6q region containing all microsatellite markers that showed allelic imbalance had a clear deletion. We concluded that A40 had copy number gains in the telomeric 6p region containing D6S309 and deletion in the large telomeric 6q region containing D6S1719.
These two cases, A6 and A40, demonstrate that while gain and loss may be difficult to distinguish by MSA, array-CGH can clearly identify single copy number change even in a tissue with a heterogeneous cell population as long as approximately 50% of the cells in the specimen have the abnormality.
Of the 10 tumours where MSA showed imbalance at all informative loci, 7 tumours showed a decreased copy number value from all clones on chromosome 6, a finding consistent with monosomy. The other three tumours did not show copy number changes at any chromosome 6 clone. This was interpreted as being the result of loss and re-duplications or polyploidy.
Eight tumours (AA15, AA19, AA57, AA90, AA105, GB12, GB29 and GB133) showed partial allelic imbalance at multiple consecutive loci by MSA in the previous study (Miyakawa et al., 2000). Array-CGH detected regions of copy number changes, either loss or gain, in the corresponding regions. However, the regions of deletion or gain detected by array-CGH appeared larger than the ones defined by MSA, including in some cases neighbouring regions harbouring the markers judged conservatively as showing allelic balance in the previous study. The pattern of partial allelic imbalance in all these cases suggested the presence of a high proportion of normal cells or subpopulations in the tumour tissue. A careful re-estimation of the MSA data showed subtle allelic imbalances judged as insignificant at these loci in the previous study. Therefore, it was considered that the discrepancy between MSA and array-CGH in these eight cases was resolved.
In 3 cases, small regions of partial allelic imbalance observed in MSA were not detected as copy number changes (GB44, D6S276; GB229, D6S1657, D6S407, D6S1572; AA65, D6S1719, D6S297, D6S1590, D6S281. See supplementary table 2 and (Miyakawa et al., 2000)). Possible explanations are either loss accompanied by re-duplication of the corresponding region, polyploidy or the region of copy number change being too small (below the detection size of the corresponding clone, GB44 had allelic imbalance only at D6S276). Array-CGH was unable to define a region of chromosome 6 altered in these 3 cases and therefore their chromosome 6 status remains unclear.
In addition to the 54 cases described above, a random series of 50 astrocytic tumours (8 A, 16 AA, 26 GB) without MSA data were analysed using the array (version 2). Copy number changes in these tumours were solely determined by array-CGH using the 10-Mb clones for normalization. Of these, 27 tumours (8 A, 4 AA, 15 GB) showed no copy number changes at any locus on chromosome 6. Four tumours (2 AA, 2 GB) showed monosomy 6 and one AA (AA42) showed trisomy 6. All the other 18 tumours (9 AA, 9 GB) had copy number changes resulting either in deletion, gain or amplification, of various regions of chromosome 6 (Table 1).
Table 1.
Summary of chromosome 6 alteration in astrocytic tumours
| n | MSA | Normal | ND | Any copy number changes |
Monosomy | Trisomy | Partial copy number changes | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||
| n | (%) | Total | 6p loss |
6p gain |
6q loss |
6q gain |
Amp | |||||||
| All cases |
||||||||||||||
| A | 10 | 2 | 8 | 0 | 2 | na | 0 | 0 | 2 | 0 | 1 | 2 | 0 | 0 |
| AA | 30 | 14 | 4 | 1 | 25 | na | 4 | 1 | 20 | 4 | 6 | 16 | 3 | 1 |
| GB | 64 | 38 | 15 | 2 | 47 | na | 9 | 0 | 38 | 7 | 3 | 35 | 5 | 1 |
| Total | 104 | 54 | 27 | 3 | 74 | na | 13 | 1 | 60 | 11 | 10 | 53 | 8 | 2 |
| Unselected series |
||||||||||||||
| A | 8 | 0 | 8 | 0 | 0 | 0% | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AA | 16 | 0 | 4 | 0 | 12 | 75% | 2 | 1 | 9 | 4 | 3 | 5 | 1 | 0 |
| GB | 26 | 0 | 15 | 0 | 11 | 42% | 2 | 0 | 9 | 4 | 0 | 7 | 0 | 1 |
| Total | 50 | 0 | 27 | 0 | 23 | 46% | 4 | 1 | 18 | 8 | 3 | 12 | 1 | 1 |
MSA, cases examined by microsatellite analysis and selected based on their findings. ND, not determined (see Results). Amp, amplification. na, not applicable because some tumours were selected based on the previously known chromosome 6 alterations.
The data from all 104 primary astrocytic tumours (10 A, 30 AA, 64 GB) studied are summarized in Table 1 and Figure 4. In the random series of 50 tumours, the incidence of chromosome 6 alterations was 0% in A, 75% in AA and 42% in GB (Table 1). The chromosome 6 raw data for all 104 tumours is listed in supplementary table 2.
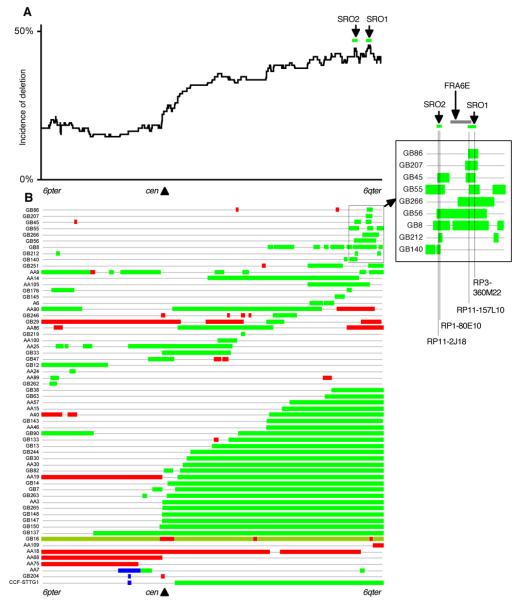
Figure. 4.
A. The number of cases that had deletion at each clone and plotted along the entire length of chromosome 6 (see text). B. A diagram of chromosome 6 copy number alterations. Tumours with either monosomy, trisomy or no copy number changes are not shown. Green bar, deletion. Pale green bar, loss and re-duplication. Red bar, gain. Blue bar, amplification. Inset shows a magnified view of selected tumours with small deletions on 6q25-27. The most frequently deleted regions (SRO1, between RP11-157L10 and RP3-360M22 and SRO2, between RP11-2J18 and RP1-80E10) are indicated by green lines and the borders shown by dotted lines. The FRA6E region is indicated by a grey line.
Common regions of copy number alterations
A number of small interstitial deletions were observed throughout the chromosome (Figure. 4B). In some tumours these overlapped, forming a number of clusters of small commonly deleted regions. Such clustering of commonly deleted regions was particularly evident in the telomeric region of 6q (6q25-q27). As several tumours had multiple isolated regions of deletions or gains, defining the exact borders of each minimum overlapping region is difficult. In order to assist elucidating possible target regions of deletions, the chromosome 6 status was first determined at each clone and then the incidence of deletion at each clone in all tumours studied was calculated and plotted against the clones (Figure. 4A).
Several peaks of deletion were identified. The highest incidence of deletion was shared by a number of small interstitial deletions in cases GB86, GB207, GB45, GB55, GB266, GB56, GB251, AA9 and GB8. By comparing the deleted regions among these 9 tumours that had small deletions involving those peaks, a smallest region of overlap (SRO1, Figure. 4A and inset) was defined as being a 1002 Kb region between RP11-157L10 (centromeric border of a deletion in GB55) and RP3-360M22 (telomeric border of a deletion in GB45). The second most frequently deleted region was observed between RP11-230C9 and RP11-96F3. This region was shared by small interstitial deletions in GB45, GB55, GB56, GB8, GB212 and GB140, as well as the cases with larger deletions. The smallest region of overlap (SRO2, Figure. 4A and inset) was defined as being a 199 kb region between RP11-2J18 (centromeric border of a deletion in GB212) and RP1-80E10 (telomeric border of a deletion in GB140).
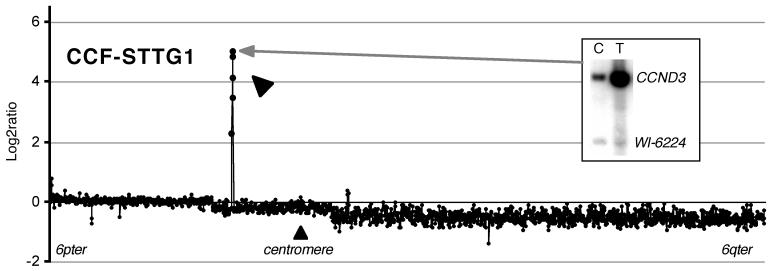
Two overlapping amplifications were found in 6p12-p21 (AA7 and GB204, Figure. 4B). In GB204, the clone RP11-238N22 (at least 110 kb in size) showed a copy number of 6.7. This clone covers the same region as RP5-973N23, which contains the cyclin D3 (CCND3) gene together with three other genes. For comparison, a glioblastoma cell line CCF-STTG1, known to have CCND3 amplification (Buschges et al., 1999), was examined by array-CGH. CCF-STTG1 showed amplification at 5 consecutive clones (approximately 570 kb region) including RP11-238N22, the copy number ranging from 9.4 to 70.7 (Figure. 4 and 5). Amplification of CCND3 in CCF-STTG1 was confirmed by Southern hybridisation (Figure. 5, inset). Minimum overlapping regions of copy number gain, except for the amplified region described above, were not further investigated because of the small numbers of gains.
Figure. 5.
A log2 plot of test/reference ratio at chromosome 6 tile path clones in a glioblastoma cell line (CCF-STTG1) normalized using 6p clones. An amplified region is indicated by a thick arrow. Inset shows Southern hybridisation data of CCF-STTG1 using a CCND3 specific probe indicating amplification of CCND3 (Ichimura et al., 1996).
Discussion
We report the construction of a human chromosome 6 tile path array and its use in investigating chromosome 6 alterations in human astrocytic tumours. All clones for the tile path array were mapped and sequenced at the Wellcome Trust Sanger Institute (Mungall et al., 2003). The majority of the array clones were selected from the Golden Path, making sequence and annotation of the clones easily retrievable from public databases such as the Ensembl genome browser (www.ensembl.org/Homo_sapiens). Where these clones were not available, alternative clones were selected from the underlying map (Bentley et al., 2001). Clones from the HLA homozygous consanguineous cell line were used to cover the MHC region representing a single haplotype (Stewart et al., 2004).
A number of chromosome 6 arrays have been reported. Some of them included chromosome 6 in an array covering the entire genome in various intervals (Cowell et al., 2004; Fiegler et al., 2003; Ishkanian et al., 2004; Snijders et al., 2001; Tagawa et al., 2004; Vissers et al., 2003), including a tile path array which covers the majority of the whole genome covering approximately 95% of sequenced region of chromosome 6 (Ishkanian et al., 2004). Others focus on a part or whole of the chromosome, including a 6q23 regional array (Chibon et al., 2004), a 6p25 array (Ekong et al., 2004) and a 0.5-Mb array of entire chromosome 6 (Tchinda et al., 2004). The chromosome 6 tile path array used in this study, with 98.3% of published sequences being included, is the most comprehensive chromosomal 6 array constructed to-date.
We used different normalization methods for the two versions of the array used. In the first version, which included chromosome 6 and X clones, the median signal intensity ratio of the clones on the chromosomal arm (6p or 6q) where the microsatellite data showed allelic balance were used to normalize the whole array. Only tumours with chromosome 6 microsatellite data were analysed using this version. We then developed the second version of chromosome 6 array that contained an additional 270 clones from all autosomes spaced at approximately 10 Mb intervals (10-Mb clone set), that were then used to normalize these chromosome 6 array clones. This array allowed robust normalization without prior microsatellite data and was used to analyse the majority of the tumours, with or without microsatellite data. The normalization in the second version assumes that the median copy number of all autosomes is approximately 2, i.e., the tumour is near-diploid. Conventional cytogenetic analysis has shown that at least 80% of astrocytic tumours are near-diploid (Bigner et al., 1988). However, where a tumour is polyploid, this normalization will not result in the correct assessment of copy number. This problem is common to all CGH analyses including the conventional metaphase CGH, and can only be addressed by calibrating the absolute copy number by other methods, e.g., interphase FISH, and adjusting the normalization accordingly.
The ability of our chromosome 6 array to discriminate single copy number change was confirmed by comparing it with MSA data previously obtained from the same tumour series. Among 1159 MSA data points (the number of informative markers multiplied by the number of cases) verified by array-CGH, 96% were concordant (see examples in Figures 1, 2, 3 and 5). In some cases, array-CGH detected very subtle but convincing deletions in large contiguous stretches of sequence where only some of, but not all, informative microsatellite markers were judged as having allelic imbalance in the previous study (Miyakawa et al., 2000). The discordance in these cases was due to a conservative estimation of MSA in the earlier study (Miyakawa et al., 2000).
We have previously identified overlapping homozygous deletions on chromosome 22 in two glioblastomas using our chromosome 22 tile path array constructed and used in the same manner, demonstrating that this technology allows the discrimination of homozygous deletion from other changes (Seng et al., 2005). In this study we observed no homozygous deletions. We conclude that there were no homozygous deletions of a size that could be detected on chromosome 6 in the tumour series examined.
The resolution of a chromosome tile path array is dependent on the size of the individual clone at each locus, the density of clones and how the analysis is carried out. The clones used in this study generally have insert sizes of between 100-300 kb, although the exact size is not always known. A copy number change affecting a region significantly smaller than the size of a clone would give a signal change difficult to distinguish from signal variation caused by variable amounts of repetitive sequence incompletely suppressed, cross-hybridisation of homologous sequences from other regions of the genome, deletion/insertion polymorphisms, or hybridisation artefacts. In a tile path array in which a set of overlapping clones are used, deletions/gains as large as or larger than a single clone may also involve neighbouring clones. It is thus unlikely that a deletion/gain of such a size would be represented only by a single clone. Accordingly, we judged a region as having a convincing copy number change only when the shift was observed in two or more consecutive clones, thus avoiding over-interpretation of changes at single clones.
Among the randomly selected cases, 0% of A, 75% of AA and 42% of GB had chromosome 6 alterations. This would suggest that chromosome 6 changes are associated with progression of A to AA and are not significant in oncogenesis of A. The lower incidence in the GB's indicates that there are other genetic pathways to GB than through AA with chromosome 6 abnormalities. It is well known that primary GB and secondary GB differ in their patterns of genetic abnormalities (reviewed in (Ichimura et al., 2004)).
The pattern of chromosome 6 deletion is complicated (Figure. 4). However, when the incidence of deletion at each clone was calculated, several most frequently deleted regions could be identified. The most frequently deleted region, SRO1 (see Figure. 4 inset and Results), contained two genes, PACRG and QKI. PACRG (the PARK2 co-regulated gene) is located immediately telomeric to PARK2 in a head-to-head arrangement sharing a bi-directional promoter, thus it is transcriptionally co-regulated with PARK2 (West et al., 2003). PACRG is known to suppress cell death induced by the accumulation of unfolded Pael receptor (Pael-R), a substrate of Parkin (Imai et al., 2003). Parkin, the protein product of PARK2, which is often mutated in autosomal recessive juvenile parkinsonism, is a protein-ubiquitin E3 ligase that targets substrate proteins for proteasomal degradation (Shimura et al., 2000). Homozygous deletions of PARK2 have been identified in ovarian tumours and lung cancer cell lines (Cesari et al., 2003), however, no tumour specific biallelic inactivation of PACRG has been reported. QKI (quaking homolog, KH domain RNA binding (mouse)) has several isoforms produced by alternative transcripts and has been shown to be involved in the regulation of central nervous system (CNS) myelinogenesis in mice (Hardy et al., 1996), presumably by regulating alternative splicing of other myelin-specific genes (Wu et al., 2002). QKI is abundantly expressed in human brain, and expression of some of its alternative transcripts have been found to be downregulated in human astrocytic gliomas, although no somatic mutations or homozygous deletions have been identified (Li et al., 2002).
Interestingly, the FRA6E region, the third most frequently observed common fragile site that spans approximately a 3.6 Mb region on 6q26, lies centromeric to SRO1, partially overlapping with it ((Denison et al., 2003) and Figure. 4 inset). Chromosomal abnormalities, e.g., deletions, amplifications and translocations, are frequently observed at fragile sites. Several candidate TSGs have been identified at some of the common fragile sites (reviewed in (Richards, 2001)). The clustering of deletion breakpoints around SRO1 observed in this study may be associated with FRA6E. Although the role of fragile sites in brain tumours remains to be determined, these deletions appear to be tumour-specific and therefore warrant further investigation.
The second most frequently deleted region, SRO2 (see Figure. 4 inset and Results), contains ARID1B (AT rich interactive domain 1B (SWI1-like)). ARID1B is a member of the ARID family of DNA-binding proteins and a subunit of human SWI/SNF-related complexes, which are involved in chromatin remodelling and thus control of gene transcription (Wang et al., 2004). Protein products of several cancer-related genes including BRCA1 are considered to contribute to tumorigenesis through interaction with the SWI/SNF complex (Roberts & Orkin, 2004). SMARCB1/SNF5/INI1, a gene encoding for another component of the SWI/SNF-complex located at 22q11, is mutated in rare but aggressive CNS tumours, i.e., atypical teratoid/rhabdoid tumours, that occur in children (Versteege et al., 1998). Mutations in any of the other components of the SWI/SNF complex in tumours have not been reported.
Copy number gains of one or more regions were observed in 19 tumours including one trisomy 6. There was no obvious region specifically targeted for copy number gains except the region around the Cyclin D3 gene (CCND3) amplified in one glioblastoma (GB204) and one anaplastic astrocytoma (AA7) (see Figures 4 and 5). Cyclin D3, like Cyclin D1 and D2, binds to CDK4 and forms a kinase complex that can phosphorylate RB when activated, thus disrupting the RB1 pathway. Amplification and overexpression of CCND3 has been reported in a small subset of glioblastomas as well as in the glioblastoma cell line CCF-STTG1 (Figure. 5) (Buschges et al., 1999). CCND3 has also been identified as a target of translocation in lymphomas resulting in juxtaposition of CCND3 and IGH (Immunoglobulin heavy chain) resulting in overexpression of CCND3 (Sonoki et al., 2001). Our data confirmed that CCND3 is one target on chromosome 6 in astrocytic tumours, although the frequency is low. No other regions of amplification were found.
A number of regions deleted or gained other than those mentioned above were observed in this study (see Figure. 4B). Whether all those regions with copy number abnormalities indicate targeted genes awaits further investigation.
In conclusion, we confirmed a frequent occurrence of chromosome 6 alterations in anaplastic astrocytomas and glioblastomas by combining a high resolution/coverage chromosome 6 tile path array and MSA analysis. We refined the deletion map and identified two small regions of overlapping deletions on 6q26 as well as amplifications encompassing the CCND3 gene. The results should help further elucidation of the genes on chromosome 6 involved in the oncogenesis and progression of astrocytic tumours.
Materials and Methods
Tumour materials and DNA extraction
Collection and handling of tumour tissues and the patients' blood samples have been described previously (Ichimura et al., 2000). The study was approved by the Ethical Committee of the Karolinska Hospital (No. 91:16) and Cambridge Local Research Ethics Committee, Cambridge, UK (Ref. LREC 03/115). The histopathological diagnosis of the tumours was carried out according to the WHO classification (Kleihues & Cavenee, 2000). A total of 104 astrocytic tumours including 10 diffuse astrocytomas (prefixed as A in the research number), 30 anaplastic astrocytomas (AA) and 64 glioblastomas (GB) were subjected to chromosome 6 array-CGH analysis. CCF-STTG1, a glioblastoma cell line known to have CCND3 amplification at 6p21 (Buschges et al., 1999), was also studied. The majority of the tumours have been included in previous reports, each tumour having a consistent unique research number with a prefix representing diagnosis (see above) (Ichimura et al., 2000; Liu et al., 2000; Miyakawa et al., 2000). DNA from tumour and blood was extracted as described (Ichimura et al., 1996). Southern hybridisation with a CCND3 probe to confirm CCND3 amplification was performed as described (Ichimura et al., 1996).
Construction of chromosome 6 tile path array
The set of large-insert bacterial clones used in the construction of the microarray were the same set used in the mapping and sequencing of chromosome 6 at the Wellcome Trust Sanger Institute, Hinxton, UK. Clones for the chromosome 6 tile path set were primarily selected from the published Golden Tile Path (Mungall et al., 2003). Where those tile path clones were not available, alternative clones covering the same region were selected from the underlying map (Bentley et al., 2001). In the major histocompatibility complex (MHC) region on 6p21.3, a minimally overlapping set of bacterial artificial chromosome (BAC) clones from the HLA homozygous consanguineous cell line PGF, representing a single MHC haplotype ((Stewart et al., 2004) and http://www.sanger.ac.uk/HGP/Chr6/MHC) was used. As a result, a total of 1780 clones (778 P1-derived artificial chromosomes (PACs) and 1002 BACs were used for analysis that altogether covered 98.3% of the published tile path, leaving at most 28 gaps (total 2.8 Mb) (Mungall et al., 2003). The 10-Mb clone set consisted of 270 BAC clones distributed along all autosomes at an average interval of 10.5 Mb (see supplementary table 1).
The chromosome 6 tile path array was constructed according to the published protocol (Fiegler et al., 2003). Briefly, clone DNA was extracted using a modified alkalilysis method (microprepping) and used as template for DOP-PCR with three different primers (Fiegler et al., 2003). Each DOP-PCR product was subsequently amplified using a 5'-amine-modified universal primer (Fiegler et al., 2003). The three amino-PCR products for each clone were mixed and printed in duplicate onto a CodeLink slide (Amersham Biosciences, Little Chalfont, UK) using a MicroGrid II robot (Genomic Solutions, Huntingdon, UK) in 4×4 subarrays.
Labelling and CGH hybridisation
Tumour and reference genomic DNA (400 ng each) were labelled with either Cy5-dCTP and Cy3-dCTP (Amersham Biosciences, Little Chalfont, UK) using a BioPrime kit (Invitrogen, Paisley, UK) as described (Fiegler et al., 2003). A mixture of normal blood DNA from either 20 males (for female test DNA) or 20 females (for male test DNA) was used as reference. The purified DNA from the test and reference samples were mixed and ethanol precipitated with 45 μg of Cot-1 DNA (Roche Diagnostics, Mannheim, Germany), dissolved in 40 μl of hybridisation buffer (50% formamide, 10% dextran sulphate, 0.1% Tween 20, 2x SSC, 10 mM Tris pH 7.4), denatured and incubated at 37°C for 2 hours before being applied to the array. After 48 hours of hybridisation at 37°C, the arrays were washed in 1x PBS/0.05% Tween20 at room temperature for 15 min twice, 50% formamide/2xSSC at 42°C for 30 min once and 1x PBS/0.05% Tween20 at room temperature for 15min. The arrays were then scanned and quantified using a GenePix Pro 4.1 on a GenePix 4100A personal scanner (Axon Instruments, Union City, CA).
Array-CGH data analysis
Normalization was performed in two ways. In the first version of the array, the chromosome 6 arm that showed no or only small regions of allelic imbalance in previous MSA data (Miyakawa et al., 2000) was used for normalization. The median of the ratio of test/reference spot intensity, after subtraction of local background, of all clones in either 6p or 6q in each subarray was used as the normalization factor. Only cases previously examined by MSA were studied using this version of the array. In the second version, the median of test/reference spot intensity ratios of all autosomal clones in the 10-Mb clone set in each subarray was used as a normalization factor under the assumption that the tumour is diploid (see Discussion). In both versions, the test/reference ratio of each clone was divided by the normalization factor for each subarray, an average of the duplicate calculated and multiplied by 2 to indicate a copy number value (normal = 2). Twelve tumours with known MSA data were examined by both versions and the results were reproducible. Spots were excluded either when the intensity of a clone in the reference channel was less than twice the average of the six Drosophila BAC spot intensities or when the duplicates differed more than 10% from their average. The calculated copy numbers were plotted in a linear scale. When a high copy number amplification was observed, the test/reference ratio was also plotted in a log2 scale. All analyses were performed using Microsoft Excel.
Supplementary Material
Acknowledgement
We would like to thank the Mapping Core, Map Finishing and Microarray Facility groups of the Wellcome Trust Sanger Institute, Hinxton, UK, for initial clone supply and verification, the Centre for Microarray resources in the Department of Pathology, University of Cambridge, for printing of the arrays, and David A. Carter for excellent technical assistance. This work was supported by grants from Cancer Research UK, The Jacqueline Seroussi Memorial Foundation for Cancer Research, Samantha Dickson Research Trust, CAMPOD, the Ludwig Institute for Cancer Research and the Wellcome Trust.
Abbreviations
- CGH
comparative genomic hybridisation
- TSG
tumour suppressor gene
- MSA
microsatellite analysis
- PAC
P1-derived artificial chromosome
- BAC
bacterial artificial chromosome
Footnotes
Supplementary information
The following supplementary information is available at the journal's website:
Supplementary Table 1. List of clones for the chromosome 6 tile path array
Supplementary Table 2. Chromosome 6 copy number status in 104 astrocytic tumours
References
- Acquati F, Morelli C, Cinquetti R, Bianchi MG, Porrini D, Varesco L, Gismondi V, Rocchetti R, Talevi S, Possati L, Magnanini C, Tibiletti MG, Bernasconi B, Daidone MG, Shridhar V, Smith DI, Negrini M, Barbanti-Brodano G, Taramelli R. Oncogene. 2001;20:980–988. doi: 10.1038/sj.onc.1204178. [DOI] [PubMed] [Google Scholar]
- Bentley DR, Deloukas P, Dunham A, French L, Gregory SG, Humphray SJ, Mungall AJ, Ross MT, Carter NP, Dunham I, Scott CE, Ashcroft KJ, Atkinson AL, Aubin K, Beare DM, Bethel G, Brady N, Brook JC, Burford DC, Burrill WD, Burrows C, Butler AP, Carder C, Catanese JJ, Clee CM, Clegg SM, Cobley V, Coffey AJ, Cole CG, Collins JE, Conquer JS, Cooper RA, Culley KM, Dawson E, Dearden FL, Durbin RM, de Jong PJ, Dhami PD, Earthrowl ME, Edwards CA, Evans RS, Gillson CJ, Ghori J, Green L, Gwilliam R, Halls KS, Hammond S, Harper GL, Heathcott RW, Holden JL, Holloway E, Hopkins BL, Howard PJ, Howell GR, Huckle EJ, Hughes J, Hunt PJ, Hunt SE, Izmajlowicz M, Jones CA, Joseph SS, Laird G, Langford CF, Lehvaslaiho MH, Leversha MA, McCann OT, McDonald LM, McDowall J, Maslen GL, Mistry D, Moschonas NK, Neocleous V, Pearson DM, Phillips KJ, Porter KM, Prathalingam SR, Ramsey YH, Ranby SA, Rice CM, Rogers J, Rogers LJ, Sarafidou T, Scott DJ, Sharp GJ, Shaw-Smith CJ, Smink LJ, Soderlund C, Sotheran EC, Steingruber HE, Sulston JE, Taylor A, Taylor RG, Thorpe AA, Tinsley E, Warry GL, Whittaker A, Whittaker P, Williams SH, Wilmer TE, Wooster R, et al. Nature. 2001;409:942–943. doi: 10.1038/35057165. [DOI] [PubMed] [Google Scholar]
- Bhatia K, Fan S, Spangler G, Weintraub M, O'Connor PM, Judde JG, Magrath I. Cancer Res. 1995;55:1431–1435. [PubMed] [Google Scholar]
- Bigner SH, Mark J, Burger PC, Mahaley MS, Jr., Bullard DE, Muhlbaier LH, Bigner DD. Cancer Res. 1988;48:405–411. [PubMed] [Google Scholar]
- Buschges R, Weber RG, Actor B, Lichter P, Collins VP, Reifenberger G. Brain Pathol. 1999;9:435–442. doi: 10.1111/j.1750-3639.1999.tb00532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari R, Martin ES, Calin GA, Pentimalli F, Bichi R, McAdams H, Trapasso F, Drusco A, Shimizu M, Masciullo V, D'Andrilli G, Scambia G, Picchio MC, Alder H, Godwin AK, Croce CM. Proc Natl Acad Sci U S A. 2003;100:5956–5961. doi: 10.1073/pnas.0931262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibon F, Mariani O, Derre J, Mairal A, Coindre JM, Guillou L, Sastre X, Pedeutour F, Aurias A. Genes Chromosomes Cancer. 2004;40:32–37. doi: 10.1002/gcc.20012. [DOI] [PubMed] [Google Scholar]
- Consortium IHGS. Nature. 2004;431:931–945. [Google Scholar]
- Cowell JK, Matsui S, Wang YD, LaDuca J, Conroy J, McQuaid D, Nowak NJ. Cancer Genet Cytogenet. 2004;151:36–51. doi: 10.1016/j.cancergencyto.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Denison SR, Callahan G, Becker NA, Phillips LA, Smith DI. Genes Chromosomes Cancer. 2003;38:40–52. doi: 10.1002/gcc.10236. [DOI] [PubMed] [Google Scholar]
- Ekong R, Jeremiah S, Judah D, Lehmann O, Mirzayans F, Hung YC, Walter MA, Bhattacharya S, Gant TW, Povey S, Wolfe J. Hum Mutat. 2004;24:76–85. doi: 10.1002/humu.20059. [DOI] [PubMed] [Google Scholar]
- Fiegler H, Carr P, Douglas EJ, Burford DC, Hunt S, Scott CE, Smith J, Vetrie D, Gorman P, Tomlinson IP, Carter NP. Genes Chromosomes Cancer. 2003;36:361–374. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- Gao X, Chen YQ, Wu N, Grignon DJ, Sakr W, Porter AT, Honn KV. Oncogene. 1995;11:1395–1398. [PubMed] [Google Scholar]
- Hardy RJ, Loushin CL, Friedrich VL, Jr., Chen Q, Ebersole TA, Lazzarini RA, Artzt K. J Neurosci. 1996;16:7941–7949. doi: 10.1523/JNEUROSCI.16-24-07941.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Ichimura K, Bolin MB, Goike HM, Schmidt EE, Moshref A, Collins VP. Cancer Res. 2000;60:417–424. [PubMed] [Google Scholar]
- Ichimura K, Ohgaki H, Kleihues P, Collins VP. J Neurooncol. 2004;70:137–160. doi: 10.1007/s11060-004-2747-2. [DOI] [PubMed] [Google Scholar]
- Ichimura K, Schmidt EE, Goike HM, Collins VP. Oncogene. 1996;13:1065–1072. [PubMed] [Google Scholar]
- Imai Y, Soda M, Murakami T, Shoji M, Abe K, Takahashi R. J Biol Chem. 2003;278:51901–51910. doi: 10.1074/jbc.M309655200. [DOI] [PubMed] [Google Scholar]
- Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, Coe BP, Snijders A, Albertson DG, Pinkel D, Marra MA, Ling V, MacAulay C, Lam WL. Nat Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- Kleihues P, Cavenee WK. Pathology and Genetics of Tumours of the Nervous System. IARC Press; Lyon: 2000. [Google Scholar]
- Koopmann J, Maintz D, Schild S, Schramm J, Louis DN, Wiestler OD, von Deimling A. Br J Cancer. 1995;72:1230–1233. doi: 10.1038/bjc.1995.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZZ, Kondo T, Murata T, Ebersole TA, Nishi T, Tada K, Ushio Y, Yamamura K, Abe K. Jpn J Cancer Res. 2002;93:167–177. doi: 10.1111/j.1349-7006.2002.tb01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Ichimura K, Pettersson EH, Goike HM, Collins VP. J Neuropathol Exp Neurol. 2000;59:1087–1093. doi: 10.1093/jnen/59.12.1087. [DOI] [PubMed] [Google Scholar]
- Liu Y, Dodds P, Emilion G, Mungall AJ, Dunham I, Beck S, Wells RS, Charnock FM, Ganesan TS. BMC Genet. 2002;3:20. doi: 10.1186/1471-2156-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa A, Ichimura K, Schmidt EE, Varmeh-Ziaie S, Collins VP. Br J Cancer. 2000;82:543–549. doi: 10.1054/bjoc.1999.0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall AJ, Palmer SA, Sims SK, Edwards CA, Ashurst JL, Wilming L, Jones MC, Horton R, Hunt SE, Scott CE, Gilbert JG, Clamp ME, Bethel G, Milne S, Ainscough R, Almeida JP, Ambrose KD, Andrews TD, Ashwell RI, Babbage AK, Bagguley CL, Bailey J, Banerjee R, Barker DJ, Barlow KF, Bates K, Beare DM, Beasley H, Beasley O, Bird CP, Blakey S, Bray-Allen S, Brook J, Brown AJ, Brown JY, Burford DC, Burrill W, Burton J, Carder C, Carter NP, Chapman JC, Clark SY, Clark G, Clee CM, Clegg S, Cobley V, Collier RE, Collins JE, Colman LK, Corby NR, Coville GJ, Culley KM, Dhami P, Davies J, Dunn M, Earthrowl ME, Ellington AE, Evans KA, Faulkner L, Francis MD, Frankish A, Frankland J, French L, Garner P, Garnett J, Ghori MJ, Gilby LM, Gillson CJ, Glithero RJ, Grafham DV, Grant M, Gribble S, Griffiths C, Griffiths M, Hall R, Halls KS, Hammond S, Harley JL, Hart EA, Heath PD, Heathcott R, Holmes SJ, Howden PJ, Howe KL, Howell GR, Huckle E, Humphray SJ, Humphries MD, Hunt AR, Johnson CM, Joy AA, Kay M, Keenan SJ, Kimberley AM, King A, Laird GK, Langford C, Lawlor S, Leongamornlert DA, Leversha M, et al. Nature. 2003;425:805–811. doi: 10.1038/nature02055. [DOI] [PubMed] [Google Scholar]
- Richards RI. Trends Genet. 2001;17:339–345. doi: 10.1016/s0168-9525(01)02303-4. [DOI] [PubMed] [Google Scholar]
- Roberts CW, Orkin SH. Nat Rev Cancer. 2004;4:133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Seng TJ, Ichimura K, Liu L, Tingby O, Pearson DM, Collins VP. Genes Chromosomes Cancer. 2005;43:181–193. doi: 10.1002/gcc.20181. [DOI] [PubMed] [Google Scholar]
- Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K, Suzuki T. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B, Kimura K, Law S, Myambo K, Palmer J, Ylstra B, Yue JP, Gray JW, Jain AN, Pinkel D, Albertson DG. Nat Genet. 2001;29:263–264. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- Sonoki T, Harder L, Horsman DE, Karran L, Taniguchi I, Willis TG, Gesk S, Steinemann D, Zucca E, Schlegelberger B, Sole F, Mungall AJ, Gascoyne RD, Siebert R, Dyer MJ. Blood. 2001;98:2837–2844. doi: 10.1182/blood.v98.9.2837. [DOI] [PubMed] [Google Scholar]
- Steinemann D, Gesk S, Zhang Y, Harder L, Pilarsky C, Hinzmann B, Martin-Subero JI, Calasanz MJ, Mungall A, Rosenthal A, Siebert R, Schlegelberger B. Genes Chromosomes Cancer. 2003;37:421–426. doi: 10.1002/gcc.10231. [DOI] [PubMed] [Google Scholar]
- Stewart CA, Horton R, Allcock RJ, Ashurst JL, Atrazhev AM, Coggill P, Dunham I, Forbes S, Halls K, Howson JM, Humphray SJ, Hunt S, Mungall AJ, Osoegawa K, Palmer S, Roberts AN, Rogers J, Sims S, Wang Y, Wilming LG, Elliott JF, de Jong PJ, Sawcer S, Todd JA, Trowsdale J, Beck S. Genome Res. 2004;14:1176–1187. doi: 10.1101/gr.2188104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa H, Tsuzuki S, Suzuki R, Karnan S, Ota A, Kameoka Y, Suguro M, Matsuo K, Yamaguchi M, Okamoto M, Morishima Y, Nakamura S, Seto M. Cancer Res. 2004;64:5948–5955. doi: 10.1158/0008-5472.CAN-03-4056. [DOI] [PubMed] [Google Scholar]
- Tchinda J, Dijkhuizen T, Vlies Pv P, Kok K, Horst J. Br J Haematol. 2004;126:495–500. doi: 10.1111/j.1365-2141.2004.05082.x. [DOI] [PubMed] [Google Scholar]
- Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, Delattre O. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- Vissers LE, de Vries BB, Osoegawa K, Janssen IM, Feuth T, Choy CO, Straatman H, van der Vliet W, Huys EH, van Rijk A, Smeets D, van Ravenswaaij-Arts CM, Knoers NV, van der Burgt I, de Jong PJ, Brunner HG, van Kessel AG, Schoenmakers EF, Veltman JA. Am J Hum Genet. 2003;73:1261–1270. doi: 10.1086/379977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Nagl NG, Wilsker D, Van Scoy M, Pacchione S, Yaciuk P, Dallas PB, Moran E. Biochem J. 2004;383:319–325. doi: 10.1042/BJ20040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Lockhart PJ, O'Farell C, Farrer MJ. J Mol Biol. 2003;326:11–19. doi: 10.1016/s0022-2836(02)01376-1. [DOI] [PubMed] [Google Scholar]
- Wu JI, Reed RB, Grabowski PJ, Artzt K. Proc Natl Acad Sci U S A. 2002;99:4233–4238. doi: 10.1073/pnas.072090399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.