Abstract
Background
Triatomines are vectors of Trypanosoma cruzi, the etiological agent of Chagas disease in Latin America. The most effective vector, Triatoma infestans, has been controlled successfully in much of Latin America using insecticide spraying. Though rarely undertaken, surveillance programs are necessary in order to identify new infestations and estimate the intensity of triatomine bug infestations in domestic and peridomestic habitats. Since hosts exposed to triatomines develop immune responses to salivary antigens, these responses can be evaluated for their usefulness as epidemiological markers to detect infestations of T. infestans.
Methodology/Principal Findings
T. infestans salivary proteins were separated by 2D-gel electrophoresis and tested for their immunogenicity by Western blotting using sera from chickens and guinea pigs experimentally exposed to T. infestans. From five highly immunogenic protein spots, eight salivary proteins were identified by nano liquid chromatography-electrospray ionization-tandem mass spectrometry (nanoLC-ESI-MS/MS) and comparison to the protein sequences of the National Center for Biotechnology Information (NCBI) database and expressed sequence tags of a unidirectionally cloned salivary gland cDNA library from T. infestans combined with the NCBI yeast protein sub-database. The 14.6 kDa salivary protein [gi|149689094] was produced as recombinant protein (rTiSP14.6) in a mammalian cell expression system and recognized by all animal sera. The specificity of rTiSP14.6 was confirmed by the lack of reactivity to anti-mosquito and anti-sand fly saliva antibodies. However, rTiSP14.6 was recognized by sera from chickens exposed to four other triatomine species, Triatoma brasiliensis, T. sordida, Rhodnius prolixus, and Panstrongylus megistus and by sera of chickens from an endemic area of T. infestans and Chagas disease in Bolivia.
Conclusions/Significance
The recombinant rTiSP14.6 is a suitable and promising epidemiological marker for detecting the presence of small numbers of different species of triatomines and could be developed for use as a new tool in surveillance programs, especially to corroborate vector elimination in Chagas disease vector control campaigns.
Author Summary
Chagas disease, caused by Trypanosoma cruzi, is a neglected disease with 20 million people at risk in Latin America. The main control strategies are based on insecticide spraying to eliminate the domestic vectors, the most effective of which is Triatoma infestans. This approach has been very successful in some areas. However, there is a constant risk of recrudescence in once-endemic regions resulting from the re-establishment of T. infestans and the invasion of other triatomine species. To detect low-level infestations of triatomines after insecticide spraying, we have developed a new epidemiological tool based on host responses against salivary antigens of T. infestans. We identified and synthesized a highly immunogenic salivary protein. This protein was used successfully to detect differences in the infestation level of T. infestans of households in Bolivia and the exposure to other triatomine species. The development of such an exposure marker to detect low-level infestation may also be a useful tool for other disease vectors.
Introduction
Control programs for Chagas disease in South America, such as the ‘Southern Cone Initiative’ have relied mainly upon vector control using insecticide spraying [1]. These campaigns have reduced the distribution of T. infestans to an area of 14.6% of the initial endemic area. However, especially the Gran Chaco region (Bolivia, Argentina, Paraguay), Andean Bolivia, western Argentina and a small area in south Peru are now still harbouring significant vector populations, some of these regions with sylvatic foci of T. infestans [1]–[3]. In controlled and Chagas disease free areas, i. e. free of vector-borne transmission, such as Argentina and Uruguay, surveillance activities have been greatly reduced allowing the re-establishment of T. infestans [4]. Moreover, after elimination of domestic vectors, peridomestic or sylvatic bug populations and species persist and may replace the former domestic populations due to changes in the ecological balance [5].
Current methods to assess the prevalence and intensity of triatomine bug infestations in domestic and peridomestic sites involve timed manual collections using an irritant spray or artificial shelter units. These methods are costly, require skilled staff, and usually lack the sensitivity and precision necessary for detection of low-density populations. Additionally, current methods are too expensive for large-scale surveillance campaigns and are not easily adaptable to many peridomestic sites [6]–[8]. Thus, new methodologies are required to detect re-emerging T. infestans populations and for long-term monitoring of previously endemic regions for Chagas disease [9].
Hematophagous arthropods have evolved a wide range of salivary anti-hemostatic compounds such as anti-coagulants, anti-histamines, vasodilators and inhibitors of platelet aggregation, sodium channel blockers, immunosuppressors, pore forming molecules and complement inhibitors that are injected into the host when feeding on blood to overcome host defence mechanisms (hemostasis, inflammation, immunity) [10]–[15]. Salivary proteins can elicit humoral immune-responses in their hosts [16]–[20]. The detection of antibodies to salivary antigens has been used as an epidemiological tool and biological marker of exposure to disease vectors including mosquitoes, ticks, sand flies and tsetse flies [19], [21]–[27]. The humoral immune response to salivary proteins of triatomines were studied in chickens, guinea pigs, mice, rabbits and humans; the latter studies using saliva and focusing on epidemiology [16], [28]–[31].
Recently, we described the anti-saliva immune responses of chickens and guinea pigs which had been experimentally exposed to T. infestans [32]. Antibody responses were detected as soon as two days after the first bug bites. Salivary antigens of 14 and 21 kDa were recognized by all chicken sera and a 79 kDa protein by all guinea pig sera. Sera from animals naturally exposed to triatomines in Bolivia also reacted with these antigens. In the present paper we describe the development of a highly sensitive exposure assay resulting in a specific recombinant salivary antigen to be used as an epidemiological tool for Chagas disease surveillance.
Materials and Methods
Insects: Origin, maintenance, saliva and salivary gland protein collection
Triatoma infestans, T. brasiliensis, T. sordida, Rhodnius prolixus and Panstrongylus megistus were reared at 27±1°C, 60–70% relative humidity, with a 16/8-h light/dark cycle and were fed on chickens [33]. T. infestans originated from a domestic population from Northern Chile, the Cachiyuyo village (29°1′48.90″S, 70°53′55.53″W, 808 m), at the border of the provinces Atacama and Coquimbo [34],[35]. T. brasiliensis and T. sordida were originally collected from a chicken house from Sítio do Cleniro and Bairro Sosó, state of Piauí, Brazil (GAS). R. prolixus originated from San Juan de Arama, Meta, Colombia (obtained from A. D'Alessandro-Bacigalupo) and P. megistus from Minas Gerais, Brazil (obtained from J. Jurberg, Departamento de Entomologia, Instituto Oswaldo Cruz, Rio de Janeiro, Brazil). All experiments were performed with pooled saliva obtained from about 300 fifth instars and adults using capillary pipettes [36]. Typically 0.5–1 µl saliva were obtained from each bug. The saliva was desalted with a 4 kDa cut-off centrifugal concentrator Fugisep-Mini (Intersep), and the protein concentration was determined using a BCA Protein Assay Kit (Perbio Science) according to the manufacturer's instructions. Aliquots of saliva, containing 30 µg protein/µl, were stored at −80°C.
Adult Anopheles freeborni from Marysville (California, USA), Aedes aegypti (Liverpool black eye strain, UK) and Culex quinquefasciatus from Vero Beach (Florida, USA) were maintained at 28°C, 75% relative humidity and a 12/12 h light/dark cycle. They were provided with 10% sucrose solution for maintenance and blood-fed on anesthetized BALB/c mice [37],[38]. Lutzomyia longipalpis (Jacobina strain, Brazil) were reared according to Modi and Tesh [39] with modifications. Briefly, adult sand flies were maintained at 26°C and 70% relative humidity with a 14/10 h light/dark cycle and fed either with 30% fructose solution or on anesthetized C57Bl/6 mice [32].
Immune sera
The early IgG-response and serial challenges of chickens and guinea pigs by T. infestans have been described previously [32]. Briefly, for the early response five chickens were exposed to starved adult T. infestans (5 per chicken) for 1 h and blood samples were taken daily for five days. For serial challenge, 12 chickens and 10 guinea pigs were exposed every two weeks over a period of 1 h or 30 min and for 19 or 23 weeks, respectively, to a low (5 adults) or a high (5 adults and 20 fourth and fifth instars) number of T. infestans. Sera from animals taken prior to the first feeding served as negative control. For the positive control, sera were pooled from chickens which had been used for routine maintenance of triatomines for at least six months.
Groups of three chickens were exposed weekly either to five adults (T. brasiliensis, T. sordida, R. prolixus or P. megistus) or An. freeborni, Ae. aegypti or Cx. quinquefasciatus (approx. 500 insects/animal/expsoure) for one month. Triatomines were allowed to feed 1 h and mosquitoes about 30 min (until about 90–100% had fed). Blood samples were collected before and always five days after exposure (triatomines) or at the end (mosquitoes), and the exposure sera were pooled [32]. Pooled serum was also prepared after one month from five mice exposed weekly for 30–45 min to L. longipalpis (approx. 100 insects/animal/exposure).
From September to November 2007, T. infestans were collected at peridomestic sampling sites and blood samples from animals were only taken if either 1–12 bugs (low exposure group) or ≥100 bugs (high exposure group) were collected by 3–5 persons within 30–60 min. Blood samples from 28 chickens (taken from the brachial vein) and 26 guinea pigs (taken from the ear vein) were collected from 16 out of 17 households in the following rural villages in the Department of Cochabamba: Sipe Sipe (17°27′2.78″S, 66°21′38.91″W, 2555 m; 2 out of 3 households colonized), Lipez (17°33′47.12″S, 66°15′27.643″W, 2542 m; 5 households), Chajra Corral (18°1′18.30″S, 64°55′25.157″W, 1796 m; 3 households), Pampas (18°3′26.812″S, 64°54′35.01″W, 1708 m; 3 households), Peña Colorada (18°10′5.288″S, 64°52′0.309″W, 1583 m; 1 household) and Arpita (17°33′51.62″S, 66°4′15.049″W, 718 m; 2 households). The blood was centrifuged at 10,000×g for 10 min at room temperature, and the sera were stored at −20°C.
Ethics statement
All animal procedures described in this study and carried out at the Ruhr-Universität Bochum were approved by the Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen, Recklinghausen, Germany. All animal studies carried out at the The National Institute of Allergy and Infectious Diseases (NIAID) were approved by the Animal Care and Use Committee at NIAID, Bethesda, MD, USA. In Bolivia the blood was taken by veterinarians of the Faculty of Veterinary, Universidad Mayor de San Simón, Cochabamba, Bolivia.
2D-gel electrophoresis and Western blots
Salivary gland proteins of T. infestans were separated by isoelectric focusing (IEF) using a Multiphor II Electrophoresis Unit (GE Healthcare), pH 4–10 [40]. Saliva (40 µg protein) was dissolved in reducing buffer containing 1% ß-mercaptoethanol and 0.5% Ampholine carrier ampholytes, pH 3.5–10 (GE Healthcare). Immobilized pH gradient (IPG) strips (7 cm, pH 4–10) were rehydrated with the protein solution and the proteins focused at 10°C using the following gradient: 0–200 V, 2 mA, 5 W for 5 min, then 200–3500 V, 1 mA, 3 W for 90 min and 3500 V constantly, 1 mA, 3 W for 60 min. For the second dimension electrophoresis, the IPG strips were equilibrated in Laemmli loading buffer [41]. The strips were placed onto 15% SDS-PAGE gels and the proteins separated according to their molecular weight using a Hoefer SE 600 apparatus (GE Healthcare). Ten independent samples of saliva were separated to recognize differences in the protein profile.
Gels were stained either with colloidal Coomassie blue or silver nitrate with modifications to ensure mass spectrometry-compatibility [42],[43]. Molecular weights were calculated with reference to the mobility of standard proteins (Prestained Protein Marker, New England Biolabs) included in one lane of the gel using the software ImageMaster 2D Elite, version 4.3 (GE Healthcare).
Western blot analyses were carried out with replica gels of the 2D-gel electrophoresis using individual sera from chickens and guinea pigs exposed to low and high numbers of triatomines in the long-term exposure study [32]. The Western blot images were compared to silver stained 2D-gels in order to determine the recognized salivary gland proteins.
Protein identification by mass spectrometry (MS)
Salivary proteins of T. infestans which were recognized by immune animal sera in the 2D-Western blot experiments were identified by mass spectrometry. Colloidal Coomassie stained protein spots were decolorised using alternating solution A (20 mM ammonium bicarbonate) and solution B (10 mM ammonium bicarbonate, 50% acetonitrile). Silver stained spots were decolorised with 50 mM sodium thiosulfate and 15 mM potassium hexacyanoferrate (III) followed by washing with solution A and B [44]. Proteins from all spots were trypsin-digested and extracted [44].
The peptides were subject to liquid chromatography-electrospray ionization-tandem mass spectrometry (nanoLC-ESI-MS/MS) analysis with collision induced dissociation experiments using the Ultimate 3000 HPLC system (Dionex LC Packings) coupled online to the HCTultra PTM Discovery System ion trap mass spectrometer (Bruker Daltonics) [45]. For protein identification, the MS output files (mgf format) were stored in the Proteinscape database (Bruker Daltonics) and the MS/MS data were compared to the protein sequences of the NCBI database (Update: 07/05/2007) using both the Mascot and Sequest algorithms. To improve the protein identification the MS/MS data were also compared to expressed sequence tags of a unidirectionally cloned salivary gland cDNA library from T. infestans combined for statistical reasons to the yeast protein sub-database of the NCBI database (TaxID 4932, 07/05/2007) [46]–[48]. The following search parameters were selected: peptide mass accuracy of 0.6 Da (mono-isotopic), fragment mass accuracy of 0.2 Da (mono-isotopic), variable modification due to oxidation of methionine, acrylamide modification of cysteine, two maximal missed cleavage sites in case of incomplete protease digests. As database searches did not consider all possible types of fragment ions, the quality of the fragment ion spectra explaining identified peptides was analysed using the ESI Compass 1.3, DataAnalysis 4.0 software (Bruker Daltonics) and by theoretical fragmentation with the MS-Product software tool (http://prospector.ucsf.edu/cgi-bin/msform.cgi?form=msproduct). For positive protein identification, a minimum of two unique peptides, adequately explained by additional manual interpretation of the respective fragment ion spectra, and at least 5% sequence coverage of the respective protein were required.
Heterologous synthesis and HPLC purification of T. infestans salivary proteins
Complementary DNAs from the T. infestans library encoding the salivary antigens were amplified by PCR, cloned into the VR2001-TOPO plasmid and purified as described previously (for primer sequences see Table S1) [47],[49]. To aid purification, a six histidine-tag coding sequence was added to the C-terminus of the cDNAs by PCR in a single step using reverse primers ending with the histidine-tag and the stop codon. Plasmids were analyzed for their correct insert and the insert orientation by sequencing using a CEQ 2000 DNA sequencer (Beckman Coulter) as previously described [49].
Plasmid purification was carried out using the GenElute HP Endotoxin-Free Plasmid Megaprep Kit (Sigma-Aldrich) according to manufacturer's instructions. DNA was concentrated with a Centricon plus-20 centrifugal filter device (Millipore) of a 100 kDa cut-off. After measurement of the concentration using the NanoDrop spectrophotometer, the DNA solution was sterile filtered using a 0.22 µm Millex-GS Filter Unit (Millipore) and stored at −80°C.
FreeStyle 293-F Cells (Invitrogen) (1×106 cells/ml) were transfected with plasmids coding for salivary gland proteins following the manufacturer's instructions and incubated for 72 h at 37°C, 8% CO2 on a stirrer plate with the propeller of the flask rotating at 135 rpm. Forty-eight hours after transfection, cells were centrifuged and the supernatant filtered through a 0.8 µm filter unit (Nalgene Labware). The supernatant was concentrated using an Amicon ultrafiltration device with a 10 kDa cut-off membrane (Millipore) in presence of Buffer A (20 mM NaH2PO4, 20 mM Na2HPO4, 500 mM NaCl, pH 7.4). After concentration, the expressed recombinant proteins were dialysed using Slide-A-Lyzer Dialysis Cassettes (3 kDA cut-off, Pierce) against PBS at 4°C overnight.
The proteins were purified by HPLC using a HiTrap Chelating HP column (GE Healthcare). The concentrated supernatant was loaded onto a prepared HiTrap Chelating HP column (GE Healthcare) and the column connected to a Summit HPLC System with a P680 HPLC pump and a PDA-100 photodiode array detector (Dionex). The column was washed for at least 30 min with buffer A and the following gradient was used to elute the proteins at a flow rate of 1 ml/min: 0–10 min buffer A, from 10–25 min buffer B (buffer A plus 50 mM imidazole, pH 7.4,), 25–45 min 80% buffer B and 20% buffer C (buffer A plus 500 mM imidazole, pH 7.4), 45–105 min buffer C and 105–106 min buffer A. The elution of the proteins was detected at 280 nm, and protein fractions were collected every minute in a 96 deep well microtiter plate using a Foxy 200 fraction collector (ISCO).
The purity of the fractions containing the recombinant proteins was tested by SDS-PAGE using a 4–12% NuPAGE Novex 4–12% Bis-Tris gel and the XCell SureLock Mini-Cell electrophoresis system (Invitrogen). The gel was silver stained using the SilverQuest silver staining kit (Invitrogen). Only fractions containing the recombinant proteins were pooled and dialysed against PBS, and the purity of the proteins was tested again by SDS-PAGE and silver staining. The protein concentration was measured using the NanoDrop spectrophotometer. Only the 14.6 kDa recombinant salivary protein was obtained in sufficient amount and stored at −80°C.
Deglycosylation of the recombinant salivary protein of T. infestans
The 14.6 kDa recombinant salivary protein of T. infestans was deglycosylated using the enzymatic deglycosylation kit Glyko according to the manufacturer's instructions (ProZyme Inc.). Three aliquots each containing 5 µg recombinant protein were incubated with N-Glycanase PNGase F, a mixture of PNGase F and Sialidase A or a mixture of PNGase F, Sialidase A and O-Glycanase, respectively. After incubation for 3 h at 37°C, the effect of deglycosylation was assessed by SDS-PAGE, and the peptides visualised by SimplyBlue SafeStain solution (Invitrogen).
ELISA
Concentrations of anti-saliva IgG in pooled chicken and guinea pig sera from the long-term T. infestans exposure study were measured by ELISA using 0.5 µg recombinant salivary protein per well of a 96-well microtitre plate (Immunolon, Nunc, Wiesbaden, Germany) as previously described [32]. Individual serum samples from Bolivia were tested either using 0.5 µg recombinant salivary protein or crude saliva of T. infestans per well as the antigen. Responses to challenge with other hematophagous insects to measure cross-reactivity used capture ELISA with recombinant salivary gland protein of T. infestans as described above.
Data analysis
Analysis of data was performed using SigmaStat 3.1 (Systat Software Inc.). Comparison of responses to bug exposure in chickens and guinea pigs in the long-term study was carried out using a Friedman Repeated Measures Analysis of Variance on Ranks (One Way RM ANOVA) with a Pairwise Multiple Comparison Procedure (Tukey test) as these data were normally distributed. In the other experiments, depending on the distribution of the ELISA test data, either an Unpaired t-test or a Mann-Whitney Rank Sum test was carried out to compare ELISA assay results using the recombinant salivary protein or the crude saliva of T. infestans analysed with sera from former laboratory studies or field collections sites in Bolivia [32]. The level of significance for all tests was p≤0.05.
Initially, the amino acid sequence of the protein was compared to the NCBI protein database to identify species with putative similar proteins. Found sequences were aligned using Clustal X 2.0.3, and graphically displayed using BioEdit 7.0.9.0 [50],[51]. The presence of a signal secretion peptide was predicted by SignalP 3.0 [52].
Results
Salivary antigens of T. infestans recognized by host antibodies
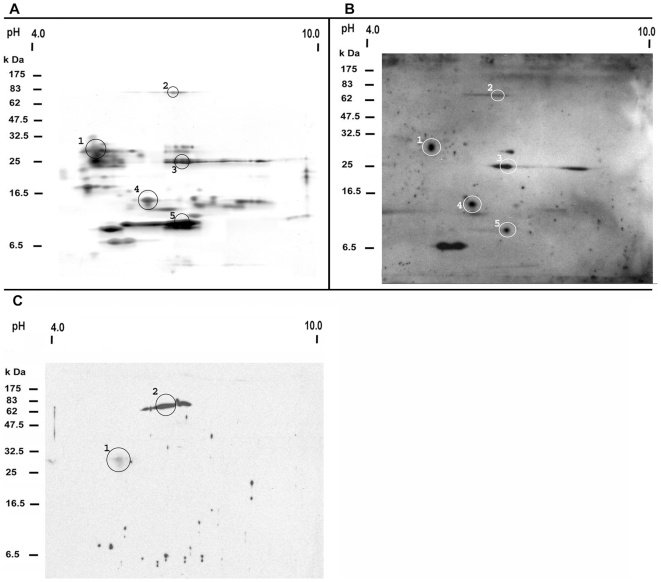
2D-gel electrophoreses of T. infestans salivary proteins revealed consistently (n = 10) that most proteins were present between pH 4.6 and 7.3 and between 25 and 31 kDa (Fig. 1A). In 2D-Western blots using serum from T. infestans challenged chickens and guinea pigs, four different proteins (protein spots 1, 3, 4 and 5) of 30, 26, 14 and 12 kDa reacted with sera of all T. infestans challenged chickens (Fig. 1B), while a 79 kDa protein (spot 2) was recognized by 8 out of 10 challenged animals. Sera from all challenged guinea pigs reacted with a 79 kDa protein (spot 2) and weakly recognized a 30 kDa protein (spot 1) (Fig. 1C).
Figure 1. Immunogenic proteins in T. infestans saliva.
Salivary proteins were resolved by 2D-gel electrophoresis (A) and 2D-Western blot (B, C) of the saliva of T. infestans. The molecular weight standard and the pI range (from pH 4 to 10) are included in each panel. Protein spots which were recognized by all chicken (B) and all guinea pig sera (C) exposed to T. infestans are encircled and numbered (1–5 or 1–2) on the gel and blot. Corresponding protein spots of the 2D-gel and the 2D-Western blot are labelled with the same number.
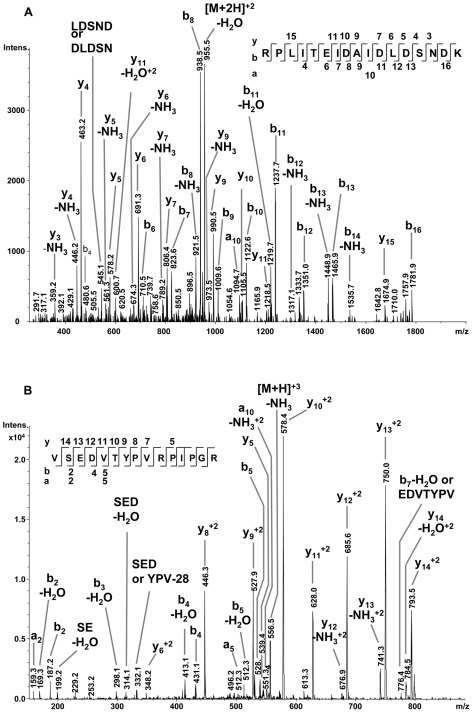
Partial peptide sequences from these five protein spots were identified by mass spectrometry and compared to the NCBI database and to a salivary gland cDNA library from T. infestans. Eight different protein matches were identified within these spots (Table 1). As an example, Fig. 2A and 2B present MS/MS spectra of two unique peptides of the 14.6 kDa salivary secreted protein [gi|149689094] from spot 4. In all cases there was a difference between theoretical and measured pI, and also the theoretical molecular weights derived from the cDNA sequence differed from the molecular weight determined by the 2D-gel; protein spot 4 contained not only a protein of 14.6 kDa but also a 21.4 kDa salivary protein. This also applies to all other protein spots except for protein spot 1 (Table 1).
Table 1. Protein identification of salivary proteins of T. infestans by mass spectrometry.
| Spot no. | Proteinno. | Data-base# | Accession number | Protein name | Protein family | MW* (kDa) | pI* (kDa) | MS/MS analysis | ||
| Mascot/Sequest score | Protein sequence coverage (%) | Peptide sequence | ||||||||
| 1 | 1 | NCBI | gi|149898926 | truncated pallidipin-like | triabin | 29.5/30 | 4.2/5.2 | 77.2/7.5 | 25.8 | EQVEDGEEGALIEK |
| lipocalin precursor | 77.7/6.9 | FFEGDWSLTYSTR | ||||||||
| 49.7/8.1 | HYGLTMDEFITR | |||||||||
| 40.2/9.5 | KEQVEDGEEGALIEK | |||||||||
| 42.9/8 | TDEEYVK | |||||||||
| 37.8/7 | VTESTINR | |||||||||
| 31.8/8.4 | YGSMVGFDPK | |||||||||
| 2 | dbEST | gi|148469123 | truncated pallidipin-like | triabin | 29.5/30 | 4.3/5.2 | 96.3/9.3 | 36.7 | EQVEEGEEGALIEK | |
| lipocalin precursor | 57.5/6.8 | FFEGDWSLTYSTK | ||||||||
| 56.7/6.2 | GGNGAQEEKGETGAQEEK | |||||||||
| 49.7/8.1 | HYGLTMDEFITR | |||||||||
| 73.9/7.6 | LIEYGQNQCQR | |||||||||
| 42.9/8 | TDEEYVK | |||||||||
| 37.8/6.2 | VTESTINR | |||||||||
| 57.6/10.8 | YGSMTGFEPNR | |||||||||
| 2 | 3 | NCBI | gi|34481604 | 79 kDa salivary apyrase | 5′nucleotidase | 62.5/79 | 9.5/6.4 | 52.2/9.7 | 14.0 | GANLIFAVGHSGINIDK |
| precursor | 43.9/13.1 | GVNIAVIGYMTPDTK | ||||||||
| 37.6/5.9 | LTLLHTNDMHSR | |||||||||
| N.F./4.3 | VFIPNVLK | |||||||||
| 62.8/13.6 | WTDVPITLINGGSIR | |||||||||
| 4 | dbEST | gi|148468017 | truncated 79 kDa | 5′nucleotidase | 18.8/79 | 9.5/6.4 | 44/5.4 | 43.4 | EQVYEEVDLK | |
| salivary apyrase | 83/8.5 | GKGSGEFLQFSGLK | ||||||||
| precursor | 42.8/5.9 | GSGEFLQFSGLK | ||||||||
| 28.4/7.5 | IIISKPLPTLNLNNPK | |||||||||
| 30.9/4.5 | LSMPGSTLK | |||||||||
| 59.5/11.8 | YLEQMSPVYTGLQAR | |||||||||
| 24.8/N.F. | YYTIAEDK | |||||||||
| 3 | 5 | dbEST | gi|148468913 | truncated unknown | 18.2/26 | 6.3/6.6 | 33.1/4.8 | 84.7 | AGIEIQK | |
| salivary protein | 31.6/7.9 | AGKDLYK | ||||||||
| N.F./4.4 | DIEKGWNK | |||||||||
| 27.6/5.2 | DVGEELEK | |||||||||
| N.F./5.8 | EIEKGFEDVGSELEK | |||||||||
| 36.3/5.8 | EIIEKIENGWK | |||||||||
| N.F./7.9 | FGKDIEK | |||||||||
| N.F./6.6 | FGKDIEKGWNK | |||||||||
| 24.1/5.6 | FGKEIEK | |||||||||
| 42.1/9.1 | FGKEIEKGFEDVGSELEK | |||||||||
| N.F./11.4 | FKFNNPIPK | |||||||||
| 30.9/8.1 | FNNPIPK | |||||||||
| 67.7/7.8 | FSHHHEHAPESEGIITISIE | |||||||||
| 32.6/10 | DGNLKGFEDVGSELEK | |||||||||
| 53.3/7 | GFEDVGSELEKYGK | |||||||||
| 38.1/6.2 | GFEDVGSELEKYGKEVER | |||||||||
| 46.1/9.2 | GFKDVGEELEK | |||||||||
| N.F./8 | GFKDVGEELEKVGK | |||||||||
| N.F./10.8 | IEEGFRDFGQK | |||||||||
| N.F./5 | IENGWK | |||||||||
| N.F./4.5 | IENGWKQFEKDTK | |||||||||
| N.F./5.1 | KIEEGFR | |||||||||
| 26.1/9.7 | KIEEGFRDFGQK | |||||||||
| 44.4/5.8 | YGKEVER | |||||||||
| 4 | 6 | NCBI | gi|149898816 | salivary lipocalin | triabin | 21.4/14 | 7.6/6.1 | 26.9/N.F. | 37.2 | CTSFQDR |
| 29.5/10.4 | DATNAYDAVCR | |||||||||
| 31.1/N.F. | DYALAYR | |||||||||
| 56.6/N.F. | DYALAYRCTSFQDR | |||||||||
| 35.1/15.3 | KPYTTTCSSVISPIPYGTIR | |||||||||
| 38.9/4.6 | SVHTGNLVLLQR | |||||||||
| 53/5.2 | TETADGSKATAGLK | |||||||||
| 7 | NCBI | gi|149689094 | salivary secreted protein | 14.6/14 | 10.3/6.1 | N.F./7.8 | 24.8 | RPLITEIDAIDLDSNDK | ||
| 43.4/8.3 | VSEDVTYPVRPIPGR | |||||||||
| 5 | 8 | NCBI | gi|149689054 | salivary lipocalin | triabin | 20.0/10 | 9.2/6.6 | N.F./4.9 | 44.1 | AKTDFSAK |
| 66/13.2 | DDNYLVLSR | |||||||||
| 57.5/7.7 | FFTGTWFVSHVQK | |||||||||
| 93.3/11.3 | GYKDDNYLVLSR | |||||||||
| N.F./8.7 | RFFTGTWFVSHVQK | |||||||||
| 46.2/9.8 | TSGGQVIPASLK | |||||||||
| 62.6/7.3 | TSSTVCQTFTASTPSEGK | |||||||||
| 64.7/8.6 | YIVEYTYQSHTGEQR | |||||||||
After nanoLC-ESI-MS/MS analysis the MS/MS data were compared to the protein sequences of the NCBI database (Update: 07/05/2007) using both the Mascot and Sequest algorithms and to expressed sequence tags (ESTs) of a unidirectionally cloned salivary gland cDNA library from T. infestans combined for statistical reasons to the yeast protein sub-database of the NCBI database (TaxID 4932, 07/05/2007). All assembled ESTs of the salivary gland protein database are published in the NCBI dbEST. Calculations (e.g. molecular weight (MW), isoelectric point (pI) etc.) for proteins of the cDNA library correspond to the mature salivary protein. Thus, only nucleotide sequences from the start (including the signal peptide) to the stop codon were considered. Not found Mascot or Sequest scores are indicated (N.F.).
*: Theoretical MW/calculated MW and theoretical pI/calculated pI from 2D-PAGE analysis.
Figure 2. NanoLC-ESI-MS/MS analysis of the 14.6 kDa salivary secreted protein of T. infestans (gi|149689094).
MS/MS spectra are shown for the two unique peptides of the identified protein: (A) RPLITEIDAIDLDSNDK with the doubly charged precursor ion mass/charge ratio (m/z) 964.5 Da and (B) VSEDVTYPVRPIPGR with the doubly charged precursor ion m/z 562.6 Da. The y-(C-terminus of the peptide), b-(N-terminus of the peptide) and a-(N-terminus of the peptide) fragment ions are highlighted in the spectra and at the peptide sequences.
Expression of recombinant salivary protein of T. infestans
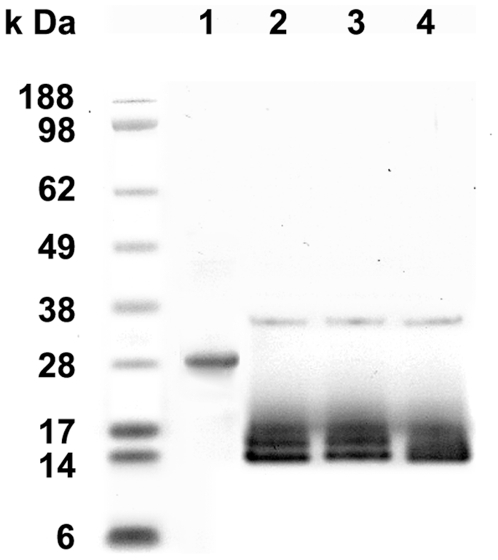
Four out of eight identified salivary proteins of T. infestans were selected for recombinant expression. These were the truncated 79 kDa salivary apyrase precursor (spot 2), the truncated unknown salivary protein (spot 3), the salivary lipocalin and the salivary secreted protein (both in spot 4). Of these four proteins, the 14.6 kDa salivary secreted protein of T. infestans (spot 4) was successfully produced as recombinant protein in the mammalian expression system in sufficient amount and purified by HPLC (Fig. 3). All other proteins were expressed at very low levels and were not used for further analyses. SDS-PAGE analyses of the recombinant 14.6 kDa T. infestans salivary protein (rTiSP14.6) (Fig 3, lane 1) gave an apparent molecular weight of 28 kDa. Enzymatic deglycosylation resulted in the expected molecular weight of 14 kDa on SDS-PAGE (Fig. 3, lanes 2–4).
Figure 3. The salivary secreted protein of T. infestans (gi|149689094) after purification and deglycosylation.
The His-tagged recombinant protein (rTiSP14.6) was purified by HPLC (lane 1) and deglycosylated with PNGase F (lane 2), PNGase F and Sialidase A (lane 3) or PNGase F, Sialidase A and O-glycanase (lane 4). The molecular weight standard is given on the left in the graph.
Antibody response in experimental challenge to rTiSP14.6
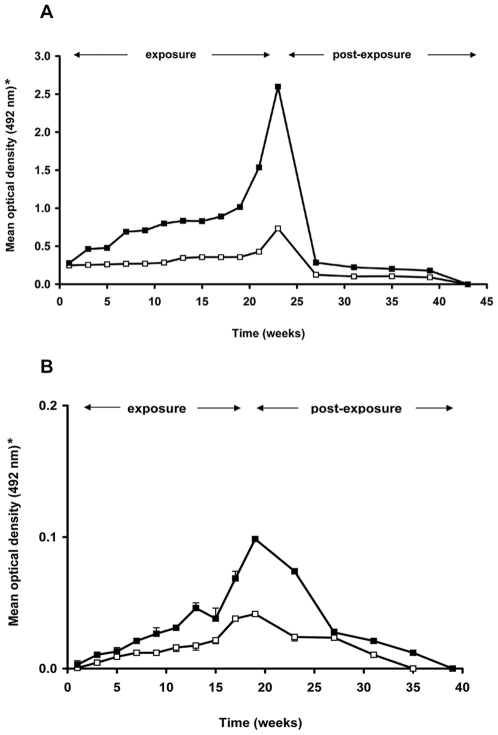
The earliest antibody reaction with rTiSP14.6 was detectable from the second day onwards after a single challenge of five bugs per chicken (n = 5, mean O.D.492 nm = 0.032). In the long-term exposure study, chickens had detectable responses in the low (O.D.492 nm = 0.249) and high (O.D.492 nm = 0.279) exposure groups, at the first time point of sera collection (five days after the first exposure) (Fig. 4A). Subsequently, responses increased up to O.D.492 nm = 0.734 and O.D.492 nm = 2.598 for the low and high exposure groups, respectively, at the end of the 19 or 23 week exposure period. Overall, the antibody reaction measured with rTiSP14.6 was significantly stronger in high exposure versus low exposure chicken sera (One Way RM ANOVA, p≤0.001). The serum reactivity declined rapidly during the first month post-exposure to O.D.492 nm = 0.125 (low exposure) and O.D.492 nm = 0.285 (high exposure). Afterwards, the serum reactivity remained stable for three months and then declined to zero. For all guinea pig sera, only a very weak reaction occurred in the low (maximum mean O.D.492 nm = 0.042) and high (maximum mean O.D.492 nm = 0.100) exposure groups, possessing no statistically significantly differences (p>0.05) (Fig. 4B). The intensity of the reaction also decreased rapidly after 20 weeks of exposure during four months post-exposure time.
Figure 4. Evolution of serum IgG reactions to rTiSP14.6 during exposure and post-exposure period.
Reactivity to rTiSP14.6 was measured in sera from chickens (A) and guinea pigs (B) using ELISA. (*) Data presented are mean and standard deviation of optical densities of two series of measurements (assayed in triplicate) using sera from the low (open squares) and high (closed squares) exposure groups (exposure to 5 or 25 T. infestans per feeding, respectively).
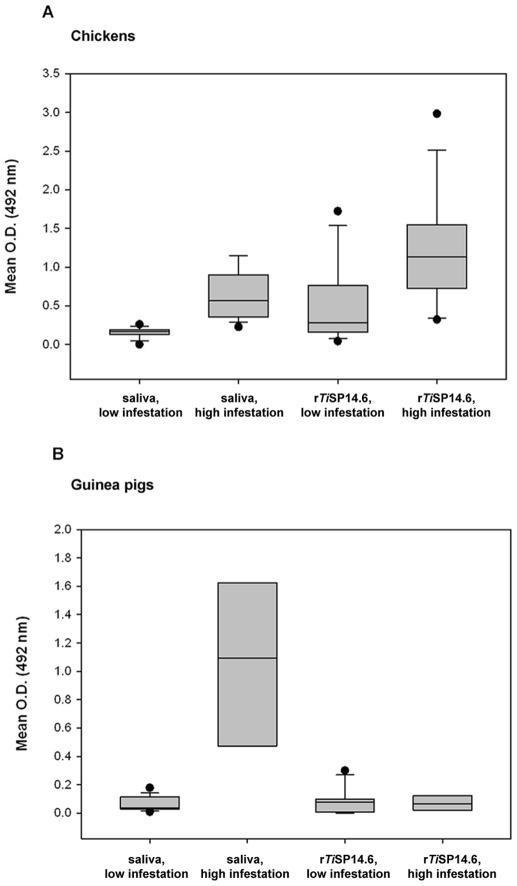
Antibody responses to crude saliva and rTiSP14.6 of T. infestans were measured using sera from peridomestic hosts from villages with low and high T. infestans infestation densities (Fig. 5, for detailed antibody reactivities see Table S2). In the case of chickens, a significantly higher serum antibody response to both crude saliva (Mann-Whitney Rank Sum test, p = 0.001) and rTiSP14.6 (Mann-Whitney Rank Sum test, p = 0.006) was detected in the high infestation households as compared to the low-infestation households (Fig. 5A). In the case of guinea pigs, this difference was only manifested in the serum antibody response to crude saliva (Mann-Whitney Rank Sum test, p = 0.001) (Fig. 5B).
Figure 5. Reactions of T. infestans saliva and rTiSP14.6 with chicken and guina pig sera from Bolivia.
Chicken (A) and guinea pig (B) sera were collected in the Department of Cochabamba in households with a low (1–12 bugs) or high (≥100 bugs) infestation levels of triatomines from September to November 2007. Every ELISA assay was repeated and the final optical density determined by calculating the mean O.D.492 nm of the triplicate wells in two experiments, respectively, and subtracting the O.D.492 nm of the negative control. Boxes show median and 25th and 75th percentiles of mean ELISA optical densities for the different antigens and exposure groups. Whiskers show 10th and 90th percentiles. Dots represent outliers.
Specificity of rTiSP14.6 for estimating triatomine challenge
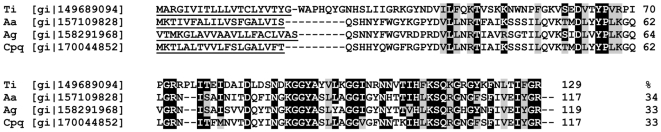
The value of rTiSP14.6 as a marker of triatomine challenge would be compromised if salivary antigens eliciting cross-reactive antibodies are expressed in other blood feeding insects feeding on the same host. A search of the NCBI database was therefore undertaken using the rTiSP14.6 sequence and similar peptide sequences were found in Ae. aegypti (34% identity), An. gambiae (33% identity) and Cx. pipiens quinquefasciatus (33% identity) (Fig. 6). Challenge experiments with these hematophagous insects and other triatomine species were then undertaken to determine if cross-reactive antibodies were elicited. No antibody responses to rTiSP14.6 were detected in sera from chickens exposed to An. freeborni (chickens, n = 3), Ae. aegypti (chickens, n = 3) and Cx. quinquefasciatus (chickens, n = 3) and in sera from mice (n = 5) exposed to L. longipalpis. Serum samples from chickens (n = 3) exposed only once to T. brasiliensis (mean O.D.492 nm = 0.075), T. sordida (mean O.D.492 nm = 0.078), R. prolixus (mean O.D.492 nm = 0.063) and P. megistus (mean O.D.492 nm = 0.093) all reacted with rTiSP14.6. Comparing reactions of rTiSP14.6 with sera from chickens exposed to other triatomines and the pooled serum of chickens to T. infestans the latter was 2.4 fold higher.
Figure 6. Alignment of predicted amino acid sequences of salivary proteins of T. infestans and mosquitoes.
Similar peptide sequences of T. infestans (Ti, rTiSP14.6), Ae. aegypti (Aa), An. gambiae (Ag) and Cx. pipiens quinquefasciatus (Cpq) in the NCBI database were aligned using Clustal X 2.0.3. Accession numbers are shown in front of the sequence, percentages of sequence identities at the end. Signal peptides were predicted using Signal P 3.0 and are underlined at the beginning of the sequence. Identical residues with rTiSP14.6 are marked by a black background and similar residues by a grey background [52].
Discussion
In this study, we used immune sera to identify and characterize immunogenic salivary proteins of T. infestans and made the first steps towards developing a novel immune-epidemiological tool to assess domestic and peridomestic infestations by T. infestans. In Western blots, sera of challenged chickens recognized salivary antigens of 30, 26, 14 and 12 kDa, and a 79 kDa antigen was recognized by sera from challenged guinea pigs. Differences between the molecular weights and pIs of these proteins estimated from the 2D-gels and the theoretical characteristics derived from the MS and cDNA sequences were most likely due to post-translational modifications, e.g. proteolytic cleavage, partial glycosylation, protein-protein complex formation [53].
In MS analyses, four proteins were identified as lipocalins. These comprise 55% of the putative secreted salivary proteins of T. infestans [47] and are also the most abundant secretory proteins in the saliva of T. brasiliensis and R. prolixus [54],[55]. Lipocalins are typically small extracellular proteins that possess a highly conserved eight-stranded antiparallel ß-barrel which forms a central hydrophobic ligand binding cavity. These secreted proteins are carriers of ligands involved in the retinol and pheromone transport, cryptic coloration, olfaction, prostaglandin synthesis and regulation of immune responses as well as the mediation of cell homoeostasis [56]–[58]. The salivary protein pallidipin, previously characterized in T. pallidipennis, is a lipocalin [59] and was highly immunoreactive at 30 kDa in the Western blots. This protein is an inhibitor of collagen-induced platelet aggregation and inhibits the release of ATP from platelets. Its effect on blood platelets is reversible, and it does not influence other types of platelet aggregation [59].
The 79 kDa salivary apyrase precursor from spot 2 is a member of the 5′-nucleotidase family and also a platelet aggregation inhibitor by hydrolysis of ATP or ADP to AMP [15], [60]–[63]. This glycosylated protein is frequently found in the saliva of different blood-feeding insects and ticks and was recently described in T. infestans [60],[64]. The apyrase was also detected in T. brasiliensis, but at 10-fold lower expression levels than in T. infestans [55],[65]. R. prolixus also possesses a similar apyrase [15],[66].
Four of eight identified proteins were selected for expression studies. The truncated 79 kDa salivary apyrase precursor [gi|148468017] of protein spot 2 was chosen because it was recognized by all guinea pig sera. The truncated unknown salivary protein [gi|148468913], the salivary lipocalin [gi|149898816] and the salivary secreted protein [gi|149689094] of spots 3 and 4 were chosen because they were recognized by chicken sera. Furthermore, the salivary lipocalin and the salivary secreted protein from protein spot 4 were available as full-length cDNA clones from the T. infestans cDNA library [47], increasing the likelihood that the expressed recombinant proteins would present the relevant epitopes. The salivary secreted protein was expressed successfully and named rTiSP14.6. RTiSP14.6 was insensitive to sialidase A and O-glycanase, but sensitive to peptide-N-glycosidase F. Therefore, the apparent 28 kDa MW was due to glycosylation with N-linked oligosaccharides. (In a subsequent MS analysis of rTiSP14.6, two types of glycans were identified for one N-glycosylated site of rTiSP14.6, biantennary and triantennary glycans linked to asparagine 105: NeuAcMan5GlcNac4, Man8GlcNAc2). Three N-glycosylated and no O-glycosylated sites were predicted for rTiSP14.6 using the NetGlyc 1.0 and NetOGlyc 3.1 Server (detailed data not presented) [67],[68]. Such glycosylations are known for other T. infestans salivary proteins [16]. Differences in the glycosylation of rTiSP14.6 from the native salivary secreted protein [gi|149689094] are probably due to it being expressed in a mammalian system rather than in its native insect salivary gland cell.
The suitability of rTiSP14.6 as a tool for the immunological assay of exposure to T. infestans was demonstrated by several results. 1) The serum reactivity to rTiSP14.6 increased with exposure time and gave a strong signal. 2) The sensitivity of rTiSP14.6 was high enough to react with sera from chickens in low-infested households. 3) The weaker antibody reaction to rTiSP14.6 in sera of guinea pigs than in chickens which also occurred in the field study, is no strong disadvantage, since chickens are found in all areas of the country, while guinea pigs only occur in limited areas restricted by the availability of alfalfa (Medicago sativa) which cannot be cultivated in the dry regions [69]. Since chickens are also present near or inside the houses in other countries of Latin America, rTiSP14.6 is an excellent exposure marker for T. infestans (but see point 5 below). 4) Sera from hosts exposed to non-triatomine vectors (mosquito and sand fly species) failed to react with rTiSP14.6, confirming the specificity of rTiSP14.6 as an immune marker for triatomine challenge. Since in the previous investigation, cross reactions were also not found using bed bugs and ticks, false positive results in an epidemiological survey of triatomine distribution are less likely [32]. 5) A major advantage is the reaction of rTiSP14.6 with sera of chickens challenged with T. brasiliensis, T. sordida, R. prolixus and P. megistus. Other triatomine species are capable of replacing T. infestans, and after control programs they may invade and colonize peridomestic sites; for example T. brasiliensis and P. megistus have replaced T. infestans in Brazil and T. guasayana may substitute for T. infestans in Argentina [1],[70],[71]. Although Rhodnius and Triatoma/Panstrongylus do not belong to the same tribe (Rhodniini: monophyletic or paraphyletic origin and Triatomini: polyphyletic or paraphyletic origin) within the polyphyletic Triatominae [72],[73], the native protein form of the cross reactive rTiSP14.6 seems to be ubiquitous in the different triatomine families and probably is an evolutionary conserved protein (orthologous) in the Triatominae. These results are consistent with previous Western blot results showing all these species elicited antibodies to a 14 kDa salivary protein [32]. Therefore, rTiSP14.6 is not T. infestans specific, but is suitable for detecting infestations by at least five species of triatomines. There seems to be a stronger reaction with sera challenged with T. infestans than with other triatomines which might allow a differentiation from these triatomine species using chicken sera. However, this needs to be verified in further investigations. Testing the more widespread use of rTiSP14.6 as an exposure marker for triatomines other than T. infestans can best be evaluated by sera from chickens living in regions where other triatomines are found. Within such a survey, sera from other animals including dogs and cats, which are frequent domestic reservoir hosts of T. cruzi, can also be tested [4]. In a recent study, T. infestans showed a feeding preference for dogs over chickens and cats [74].
In conclusion, the aim of this study was to develop an epidemiological marker to detect low-level infestation of T. infestans in endemic countries after insecticide control activities. Out of eight different highly immunogenic T. infestans salivary proteins, a 14.6 kDa salivary protein of unknown function was successfully expressed recombinantly. It reacted strongly with sera from chickens exposed to a low number of T. infestans in both the laboratory and field. This recombinant protein will enable the immunological detection of low numbers of different triatomines, not only T. infestans, and hence could be developed as a sensitive surveillance tool to warn of exposure risk to triatomines and thereby for Chagas disease. The development of recombinant salivary proteins as epidemiological tools may also be useful for the control of other vectors such as mosquitoes, ticks, tsetse flies and sand flies and to understand the epidemiology of the diseases that they transmit [22], [75]–[78].
Supporting Information
Portuguese translation of the abstract by Dr. Eloi S. Garcia
(0.02 MB DOC)
Spanish translation of the abstract by Dr. Ricardo E. Gürtler.
(0.02 MB DOC)
Primer sequences used for PCR amplification of cDNAs from the T. infestans library encoding four salivary proteins.
(0.03 MB DOC)
Antibody reactions of chicken and guinea pig sera from Bolivia to saliva and rTiSP14.6 of T. infestans.
(0.10 MB DOC)
Acknowledgments
The authors are very grateful to Sabine Kindermann, Holger Schlierenkamp, Van My Pham and John Andersen for their technical assistance and to Christian Meiser for the introduction to the technology for obtaining the saliva of Triatoma infestans. We thank Dr. José Jurberg (Departamento de Entomologia, Instituto Oswaldo Cruz, Rio de Janeiro, Brazil) for providing colonies of triatomines for this study; the veterinarians of the Faculty of Veterinary, Universidad Mayor de San Simón, Cochabamba, Bolivia, for taking blood from guinea pigs and chickens; and unknown reviewers and Dr. Ricardo E. Gürtler for critical suggestions to the manuscript. We also thank Dr. Eloi S. Garcia and Dr. Ricardo E. Gürtler for their Portuguese and Spanish translations of the abstract, respectively.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by grants from the German Academic Exchange Service (DAAD), Bonn, Germany (short-term scholarship), the Boehringer Ingelheim Fonds (BIF), Heidesheim, Germany (travel award), and the ‘Deutsche Forschungsgemeinschaft’ (project Scha 339/13-1). The support by the Humboldt Foundation to NMM is gratefully acknowledged, and AS was funded by the University of Aberdeen, Scotland (Sixth Century Scholarship). This work was also in part supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schofield CJ, Jannin J, Salvatella R. The future of Chagas disease control. Trends Parasitol. 2006;22:583–588. doi: 10.1016/j.pt.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Gorla DE. Variables ambientales registradas por sensores remotos como indicadores de la distribución geográfica de Triatoma infestans. Ecología Austral. 2002;12:117–127. [Google Scholar]
- 3.Noireau F, Cortez MG, Monteiro FA, Jansen AM, Torrico F. Can wild Triatoma infestans foci in Bolivia jeopardize Chagas disease control efforts? Trends Parasitol. 2005;21:7–10. doi: 10.1016/j.pt.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Gürtler RE, Kitron U, Cecere MC, Segura EL, Cohen JE. Sustainable vector control and management of Chagas disease in the Gran Chaco, Argentina. Proc Natl Acad Sci U S A. 2007;104:16194–16199. doi: 10.1073/pnas.0700863104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cecere MC, Gürtler RE, Canale DM, Chuit R, Cohen JE. Effects of partial housing improvement and insecticide spraying on the reinfestation dynamics of Triatoma infestans in rural northwestern Argentina. Acta Trop. 2002;84:101–116. doi: 10.1016/s0001-706x(02)00183-3. [DOI] [PubMed] [Google Scholar]
- 6.Gürtler RE, Vázquez Prokopec GMV, Ceballos LA, Lund Petersen C, Salomon OD. Comparison Between Two Artificial Shelter Units and Timed Manual Collections for Detecting Peridomestic Triatoma infestans (Hemiptera: Reduviidae) in Rural Northwestern Argentina. J Med Entomol. 2001;38:429–436. doi: 10.1603/0022-2585-38.3.429. [DOI] [PubMed] [Google Scholar]
- 7.Rabinovich JE, Gürtler RE, Leal JA, Feliciangeli D. Density estimates of the domestic vector of Chagas disease, Rhodnius prolixus Stål (Hemiptera: Reduviidae), in rural houses in Venezuela. Bull World Health Organ. 1995;73:347–357. [PMC free article] [PubMed] [Google Scholar]
- 8.Vazquez-Prokopec GM, Ceballos LA, Salomón OD, Gürtler RE. Field Trials of an Improved Cost-effective Device for Detecting Peridomestic Populations of Triatoma infestans (Hemiptera: Reduviidae) in Rural Argentina. Mem Inst Oswaldo Cruz. 2002;97:971–977. doi: 10.1590/s0074-02762002000700008. [DOI] [PubMed] [Google Scholar]
- 9.Dias JCP. Southern Cone Initiative for the elimination of domestic populations of Triatoma infestans and the interruption of transfusional Chagas disease. Historical aspects, present situation, and perspectives. Mem Inst Oswaldo Cruz. 2007;102(Suppl 1):11–18. doi: 10.1590/s0074-02762007005000092. [DOI] [PubMed] [Google Scholar]
- 10.Ribeiro JMC, Garcia ES. Platelet antiaggregating activity in the salivary secretion of the blood sucking bug Rhodnius prolixus. Experientia. 1981;37:384–386. doi: 10.1007/BF01959876. [DOI] [PubMed] [Google Scholar]
- 11.Hellmann K, Hawkins RI. Prolixin-S and Prolixin-G; Two Anticoagulants from Rhodnius prolixus Stål. Nature. 1965;207:265–267. doi: 10.1038/207265a0. [DOI] [PubMed] [Google Scholar]
- 12.Pereira MH, Souza MEL, Vargas AP, Martins MS, Penido CM, et al. Anticoagulant activity of Triatoma infestans and Panstrongylus megistus saliva (Hemiptera/Triatominae). Acta Trop. 1996;61:255–261. doi: 10.1016/0001-706x(96)00007-1. [DOI] [PubMed] [Google Scholar]
- 13.Ribeiro JMC, Walker FA. High affinity histamine-binding and antihistaminic activity of the salivary nitric oxide-carrying heme protein (nitrophorin) of Rhodnius prolixus. J Exp Med. 1994;180:2251–2257. doi: 10.1084/jem.180.6.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ribeiro JMC, Marinotti O, Gonzales R. A salivary vasodilator in the blood-sucking bug, Rhodnius prolixus. Br J Pharmacol. 1990;101:932–936. doi: 10.1111/j.1476-5381.1990.tb14183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarkis JJF, Guimarães JA, Ribeiro JMC. Salivary apyrase of Rhodnius prolixus. Kinetics and purification. Biochem J. 1986;233:885–891. doi: 10.1042/bj2330885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volf P, Grubhoffer L, Hošek P. Characterisation of salivary gland antigens of Triatoma infestans and antigen-specific serum antibody response in mice exposed to bites of T. infestans. Vet Parasitol. 1993;47:327–337. doi: 10.1016/0304-4017(93)90033-j. [DOI] [PubMed] [Google Scholar]
- 17.Volf P, Rohoušová I. Species-specific antigens in salivary glands of phlebotomine sandflies. Parasitology. 2001;122 Pt 1:37–41. doi: 10.1017/s0031182000007046. [DOI] [PubMed] [Google Scholar]
- 18.Gomes RB, Brodskyn C, de Oliveira CI, Costa J, Miranda JC, et al. Seroconversion against Lutzomyia longipalpis Saliva Concurrent with the Development of anti-Leishmania chagasi Delayed-Type Hypersensitivity. J Infect Dis. 2002;186:1530–1534. doi: 10.1086/344733. [DOI] [PubMed] [Google Scholar]
- 19.Cornelie S, Remoue F, Doucoure S, Ndiaye T, Sauvage FX, et al. An insight into immunogenic salivary proteins of Anopheles gambiae in African children. Malar J. 2007;6:75. doi: 10.1186/1475-2875-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inokuma H, Ohno K, Onishi T. Is the detection of anti-Rhipicephalus sanguineus (Rs24p) antibodies a valuable epidemiological tool of tick infestation in dogs? Vet Res. 2000;31:365–369. doi: 10.1051/vetres:2000126. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz BS, Nadelman RB, Fish D, Childs JE, Forseter G, et al. Entomologic and Demographic Correlates of Anti-Tick Saliva Antibody in a Prospective Study of Tick Bite Subjects in Westchester County, New York. Am J Trop Med Hyg. 1993;48:50–57. doi: 10.4269/ajtmh.1993.48.50. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz BS, Ford DP, Childs JE, Rothman N, Thomas RJ. Anti-tick Saliva Antibody: A Biologic Marker of Tick Exposure That Is a Risk Factor for Lyme Disease Seropositivity. Am J Epidemiol. 1991;134:86–95. doi: 10.1093/oxfordjournals.aje.a115996. [DOI] [PubMed] [Google Scholar]
- 23.Lane RS, Moss RB, Hsu YP, Wei T, Mesirow ML, et al. Anti-arthropod saliva antibodies among residents of a community at high risk for Lyme disease in California. Am J Trop Med Hyg. 1999;61:850–859. doi: 10.4269/ajtmh.1999.61.850. [DOI] [PubMed] [Google Scholar]
- 24.Poinsignon A, Cornelie S, Remoue F, Grébaut P, Courtin D, et al. Human/Vector Relationships During Human African Trypanosomiasis: Inital Screening of Immunogenic Salivary Proteins Of Glossina Species. Am J Trop Med Hyg. 2007;76:327–333. [PubMed] [Google Scholar]
- 25.Barral A, Honda E, Caldas A, Costa J, Vinhas V, et al. Human Immune Response to Sand Fly Salivary Gland Antigens: A Useful Epidemiological Marker? Am J Trop Med Hyg. 2000;62:740–745. doi: 10.4269/ajtmh.2000.62.740. [DOI] [PubMed] [Google Scholar]
- 26.Hostomska J, Rohoušová I, Volfová V, Stanneck D, Mencke N, et al. Kinetics of Canine Antibody Response to Saliva of the Sand Fly Lutzomyia longipalpis. Vector Borne Zoonotic Dis. 2008;8:443–450. doi: 10.1089/vbz.2007.0214. [DOI] [PubMed] [Google Scholar]
- 27.Rohoušová I, Ozensoy S, Ozbel Y, Volf P. Detection of species-specific antibody response of humans and mice bitten by sand flies. Parasitology. 2005;130:493–499. doi: 10.1017/s003118200400681x. [DOI] [PubMed] [Google Scholar]
- 28.Hecht MM, Bussacos AC, Lozzi SP, Santana JM, Teixeira AR. Triatoma infestans chooses to feed upon immune prey. Am J Trop Med Hyg. 2006;75:893–900. [PubMed] [Google Scholar]
- 29.Nascimento RJ, Santana JM, Lozzi SP, Araujo CN, Teixeira AR. Human IgG1 and IgG4: the main antibodies against Triatoma infestans (Hemiptera: Reduviidae) salivary gland proteins. Am J Trop Med Hyg. 2001;65:219–226. doi: 10.4269/ajtmh.2001.65.219. [DOI] [PubMed] [Google Scholar]
- 30.Barbosa SE, Diotaiuti L, Braga EM, Pereira MH. Variability of the salivary proteins of 20 Brazilian populations of Panstrongylus megistus (Hemiptera: Reduviidae: Triatominae). Acta Trop. 2004;92:25–33. doi: 10.1016/j.actatropica.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 31.Machado de Assis GF, Martins Braga E, Diotaiuti L, de Figueiredo Gontijo N, de Lana M, et al. Subclases de IgG anti-antIgenos salivares de triatominos para estimación del contacto humano-vector: Resultados preliminares de un estudio realizado en el municipio de Berilo, Minas Gerais, Brasil. 2007. pp. 88–91. Conference proceedings of the Taller del Cono Sur “Actualizacion de la tripanosomiasis americana”. Asunción, Paraguay, 25–27 April 2007: 88–91.
- 32.Schwarz A, Sternberg JM, Johnston V, Medrano-Mercado N, Anderson JM, et al. Antibody responses of domestic animals to salivary antigens of Triatoma infestans as biomarkers for low-level infestation of triatomines. Int J Parasitol. 2009;39:1021–1029. doi: 10.1016/j.ijpara.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaub GA. Trypanosoma cruzi: quantitative studies of the development of two strains in small intestine and rectum of the vector Triatoma infestans. Exp Parasitol. 1989;68:260–273. doi: 10.1016/0014-4894(89)90108-2. [DOI] [PubMed] [Google Scholar]
- 34.Schaub GA, Schottelius J. Identification of trypanosomes isolated from Reduviidae from North Chile. Z Parasitenkd. 1984;70:3–9. doi: 10.1007/BF00929569. [DOI] [PubMed] [Google Scholar]
- 35.Kollien AH, Schaub GA. The Development of Trypanosoma cruzi (Trypanosomatidae) in the Reduviid Bug Triatoma infestans (Insecta): Influence of Starvation. J Eukaryot Microbiol. 1998;45:59–63. doi: 10.1111/j.1550-7408.1998.tb05070.x. [DOI] [PubMed] [Google Scholar]
- 36.Amino R, Tanaka AS, Schenkman S. Triapsin, an unusual activatable serine protease from the saliva of the hematophagous vector of Chagas disease Triatoma infestans (Hemiptera: Reduviidae). Insect Biochem Mol Biol. 2001;31:465–472. doi: 10.1016/s0965-1748(00)00151-x. [DOI] [PubMed] [Google Scholar]
- 37.Dimopoulos G, Seeley D, Wolf A, Kafatos FC. Malaria infection of the mosquito Anopheles gambiae activates immune-responsive genes during critical transition stages of the parasite life cycle. EMBO J. 1998;17:6115–6123. doi: 10.1093/emboj/17.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dimopoulos G, Richman A, Müller HM, Kafatos FC. Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proc Natl Acad Sci U S A. 1997;94:11508–11513. doi: 10.1073/pnas.94.21.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Modi GB, Tesh RB. A simple technique for mass rearing Lutzomyia longipalpis and Phlebotomus papatasi (Diptera: Psychodidae) in the laboratory. J Med Entomol. 1983;20:568–569. doi: 10.1093/jmedent/20.5.568. [DOI] [PubMed] [Google Scholar]
- 40.Görg A, Obermaier C, Boguth G, Harder A, Scheibe B, et al. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 2000;21:1037–1053. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1037::AID-ELPS1037>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 41.Laemmli UK. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 42.Neuhoff V, Stamm R, Eibl H. Clear background and highly sensitive protein staining with Coomassie Blue dyes in polyacrylamide gels: A systematic analysis. Electrophoresis. 1985;6:427–448. [Google Scholar]
- 43.Jungblut PR, Seifert R. Analysis by high-resolution 2-dimensional electrophoresis of differentiation-dependent alterations in cytosolic protein pattern of HL-60 leukemic-cells. J Biochem Biophys Methods. 1990;21:47–58. doi: 10.1016/0165-022x(90)90044-d. [DOI] [PubMed] [Google Scholar]
- 44.Schaefer H, Marcus K, Sickmann A, Herrmann M, Klose J, et al. Identification of phosphorylation and acetylation sites in αA-crystallin of the eye lens (Mus musculus) after two-dimensional gel electrophoresis. Anal Bioanal Chem. 2003;376:966–972. doi: 10.1007/s00216-003-1983-1. [DOI] [PubMed] [Google Scholar]
- 45.Helling S, Vogt S, Rhiel A, Ramzan R, Wen L, et al. Phosphorylation and Kinetics of Mammalian Cytochrome c Oxidase. Mol Cell Proteomics. 2008;7:1714–1724. doi: 10.1074/mcp.M800137-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blüggel M, Bailey S, Körting G, Stephan C, Reidegeld KA, et al. Towards data management of the HUPO Human Brain Proteome Project pilot phase. Proteomics. 2004;4:2361–2362. doi: 10.1002/pmic.200400867. [DOI] [PubMed] [Google Scholar]
- 47.Assumpção TC, Francischetti IMB, Andersen JF, Schwarz A, Santana JM, et al. An insight into the sialome of the blood-sucking bug Triatoma infestans, a vector of Chagas' disease. Insect Biochem Mol Biol. 2008;38:213–232. doi: 10.1016/j.ibmb.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chamrad DC, Koerting G, Gobom J, Thiele H, Klose J, et al. Interpretation of mass spectrometry data for high-throughput proteomics. Anal Bioanal Chem. 2003;376:1014–1022. doi: 10.1007/s00216-003-1995-x. [DOI] [PubMed] [Google Scholar]
- 49.Oliveira F, Kamhawi S, Seitz AE, Pham VM, Guigal PM, et al. From transcriptome to immunome: Identification of DTH inducing proteins from a Phlebotomus ariasi salivary gland cDNA library. Vaccine. 2006;24:374–390. doi: 10.1016/j.vaccine.2005.07.085. [DOI] [PubMed] [Google Scholar]
- 50.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 52.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved Prediction of Signal Peptides: SignalP 3.0. Journal of Molecular Biology. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 53.Helmerhorst EJ, Oppenheim FG. Saliva: a Dynamic Proteome. J Dent Res. 2007;86:680–693. doi: 10.1177/154405910708600802. [DOI] [PubMed] [Google Scholar]
- 54.Ribeiro JMC, Andersen J, Silva Neto MAC, Pham VM, Garfield MK, et al. Exploring the sialome of the blood-sucking bug Rhodnius prolixus. Insect Biochem Mol Biol. 2004;34:61–79. doi: 10.1016/j.ibmb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 55.Santos A, Ribeiro JMC, Lehane MJ, Gontijo NF, Veloso AB, et al. The sialotranscriptome of the blood-sucking bug Triatoma brasiliensis (Hemiptera, Triatominae). Insect Biochem Mol Biol. 2007;37:702–712. doi: 10.1016/j.ibmb.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flower DR. The lipocalin protein family: structure and function. Biochem J. 1996;318(Pt 1):1–14. doi: 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Flower DR, North ACT, Sansom CE. The lipocalin protein family: structural and sequence overview. Biochim Biophys Acta. 2000;1482:9–24. doi: 10.1016/s0167-4838(00)00148-5. [DOI] [PubMed] [Google Scholar]
- 58.Urade Y, Hayaishi O. Prostaglandin D synthase: structure and function. Vitam Horm. 2000;58:89–120. doi: 10.1016/s0083-6729(00)58022-4. [DOI] [PubMed] [Google Scholar]
- 59.Noeske-Jungblut C, Krätzschmar J, Haendler B, Alagon A, Possani L, et al. An inhibitor of Collagen-induced Platelet Aggregation from the Saliva of Triatoma pallidipennis. J Biol Chem. 1994;269:5050–5053. [PubMed] [Google Scholar]
- 60.Faudry E, Lozzi SP, Santana JM, D'Souza-Ault M, Kieffer S, et al. Triatoma infestans Apyrases Belong to the 5′-Nucleotidase Family. J Biol Chem. 2004;279:19607–19613. doi: 10.1074/jbc.M401681200. [DOI] [PubMed] [Google Scholar]
- 61.Faudry E, Santana JM, Ebel C, Vernet T, Teixeira ARL. Salivary apyrases of Triatoma infestans are assembled into homo-oligomers. Biochem J. 2006;396:509–515. doi: 10.1042/BJ20052019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ribeiro JMC, Francischetti IMB. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu Rev Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- 63.Champagne DE, Nussenzveig RH, Ribeiro JMC. Purification, Partial Characterization, and Cloning of Nitric Oxide-carrying Heme Proteins (Nitrophorins) from Salivary Glands of the Blood-sucking Insect Rhodnius prolixus. J Biol Chem. 1995;270:8691–8695. doi: 10.1074/jbc.270.15.8691. [DOI] [PubMed] [Google Scholar]
- 64.Faudry E, Rocha PS, Vernet T, Lozzi SP, Teixeira ARL. Kinetics of expression of the salivary apyrases in Triatoma infestans. Insect Biochem Mol Biol. 2004;34:1051–1058. doi: 10.1016/j.ibmb.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 65.Ribeiro JMC, Schneider M, Isaias T, Jurberg J, Galvão C, et al. Role of salivary antihemostatic components in blood feeding by triatomine bugs (Heteroptera). J Med Entomol. 1998;35:599–610. doi: 10.1093/jmedent/35.4.599. [DOI] [PubMed] [Google Scholar]
- 66.Ribeiro JMC, Garcia ES. The salivary and crop apyrase activity of Rhodnius prolixus. J Insect Physiol. 1980;26:303–307. [Google Scholar]
- 67.Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- 68.Gupta R, Jung E, Brunak S. Prediction of N-glycosylation sites in human proteins. 2004. (in prep)
- 69.Galindo SA. Costos de alimentación en cuyes en dos poblaciones de produccion carnica en campo y estación experimental. 2003. Master thesis. Universidad Mayor de San Simón, Facultad de Ciencias Agrícolas y Pecuarias, Cochabamba, Bolivia. Available: http://contentdm.lib.byu.edu/cdm4/document.php?CISOROOT=/Benson&CISOPTR=8039. Accessed 5 March 2009.
- 70.Vazquez-Prokopec GM, Cecere MC, Canale DM, Gürtler RE, Kitron U. Spatiotemporal Patterns of Reinfestation by Triatoma guasayana (Hemiptera: Reduviidae) in a Rural Community of Northwestern Argentina. J Med Entomol. 2005;42:571–581. doi: 10.1093/jmedent/42.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pereira MH, Gontijo NF, Guarneri AA, Sant' Anna MRV, Diotaiuti L. Competitive displacement in Triatominae: the Triatoma infestans success. Trends Parasitol. 2006;22:516–520. doi: 10.1016/j.pt.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 72.Schofield CJ, Galvão C. Classification, evolution, and species groups within the Triatominae. Acta Trop. 2009;110:88–100. doi: 10.1016/j.actatropica.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 73.de Paula AS, Diotaiuti L, Galvão C. Systematics and biogeography of Rhodniini (Heteroptera: Reduviidae: Triatominae) based on 16S mitochondrial rDNA sequences. J Biogeogr. 2007;34:699–712. [Google Scholar]
- 74.Gürtler RE, Ceballos LA, Ordóñez-Krasnowski P, Lanati LA, Stariolo R, et al. Strong Host-Feeding Preferences of the Vector Triatoma infestans Modified by Vector Density: Implications for the Epidemiology of Chagas Disease. PLoS Negl Trop Dis. 2009;3:e447. doi: 10.1371/journal.pntd.0000447. doi: 10.1371/journal.pntd.0000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rohoušová I, Volf P. Sand fly saliva: effects on host immune response and Leishmania transmission. Folia Parasitol. 2006;53:161–171. [PubMed] [Google Scholar]
- 76.Remoue F, Cisse B, Ba F, Sokhna C, Herve JP, et al. Evaluation of the antibody response to Anopheles salivary antigens as a potential marker of risk of malaria. Trans R Soc Trop Med Hyg. 2006;100:363–370. doi: 10.1016/j.trstmh.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 77.Drakeley CJ, Corran PH, Coleman PG, Tongren JE, McDonald SLR, et al. Estimating medium- and long-term trends in malaria transmission by using serological markers of malaria exposure. Proc Natl Acad Sci U S A. 2005;102:5108–5113. doi: 10.1073/pnas.0408725102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Billingsley PF, Baird J, Mitchell JA, Drakeley C. Immune interactions between mosquitoes and their hosts. Parasite Immunology. 2006;28:143–153. doi: 10.1111/j.1365-3024.2006.00805.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Portuguese translation of the abstract by Dr. Eloi S. Garcia
(0.02 MB DOC)
Spanish translation of the abstract by Dr. Ricardo E. Gürtler.
(0.02 MB DOC)
Primer sequences used for PCR amplification of cDNAs from the T. infestans library encoding four salivary proteins.
(0.03 MB DOC)
Antibody reactions of chicken and guinea pig sera from Bolivia to saliva and rTiSP14.6 of T. infestans.
(0.10 MB DOC)