Abstract
MYC is an oncogene involved in cell cycle regulation, cell growth arrest, cell adhesion, metabolism, ribosome biogenesis, protein synthesis, and mitochondrial function. It has been described as a key element of several carcinogenesis processes in humans. Many studies have shown an association between MYC deregulation and gastric cancer. MYC deregulation is also seen in gastric preneoplastic lesions and thus it may have a role in early gastric carcinogenesis. Several studies have suggested that amplification is the main mechanism of MYC deregulation in gastric cancer. In the present review, we focus on the deregulation of the MYC oncogene in gastric adenocarcinoma carcinogenesis, including its association with Helicobacter pylori (H pylori) and clinical applications.
Keywords: MYC, Gastric adenocarcinoma, Gastric preneoplastic lesions, Gastric carcinogenesis, Helicobacter pylori
INTRODUCTION
A temporal decline in gastric cancer (GC) incidence has been seen in several countries, including Brazil[1,2]. However, this cancer causes nearly one million deaths a year worldwide and is still a serious public health cancer[3], especially in the Pará State, Northern Brazil, where mortality rates are higher than the national average rate[2]. GC is usually diagnosed at advanced stages and the single curative therapy available requires surgical resection[4].
Over 95% of gastric malignancies are adenocarcino-mas[5]. They are subdivided into two main histological types: well-differentiated or intestinal-type, and undifferentiated or diffuse-type[6]. Intestinal-type gastric tumors predominate in high-risk geographic areas whereas diffuse-type tumors are more common in low-risk areas[7].
The identification of peculiar genetic characteristics of gastric tumors may help predict prognosis of GC patients and allow more accurate therapeutic approaches. Genetic analyses of GC suggest that there occur structural and functional alterations of several oncogenes and tumor suppressor genes, as well as genetic instability[8]. Additionally, GC has been an interesting carcinogenesis model. Evidence suggests that intestinal- and diffuse-type gastric carcinomas develop through distinct genetic pathways due to different genetic alterations identified in these histological types[9,10].
MYC (C-MYC) oncogene has been described as a key element of several carcinogenesis processes in humans[11]. In the present review, we focus on the deregulation of the MYC oncogene in gastric carcinogenesis.
MYC AND CANCER
MYC gene was found to be the cellular homolog of retroviral v-myc oncogene about 30 years ago[12-14]. It is located on chromosomal region 8q24.1, has 3 exons[15,16] and encodes a nuclear phosphoprotein[17].
MYC has to heteromerize with MAX, a protein expressed constitutively, to acquire DNA-binding activity. MYC/MAX dimmers are made viable by a basic region helix-loop-helix leucine-zipper motif (bHLH-Zip), conserved sequences in the carboxyl terminus of both proteins. MYC/MAX dimmers bind to E-box sequence CACGTG in the promoters of specific target genes and stimulate their transcription[18].
MYC has an effect on up to about 15% of genes in genomes of many organisms, from flies to humans[19]. Groups of genes involved in cell cycle regulation, metabolism, ribosome biogenesis, protein synthesis, and mitochondrial function are over-represented in the Myc target gene network.
MYC also consistently represses genes involved in cell growth arrest and cell adhesion[20]. Dominguez-Sola et al[21] recently showed that Myc interacts with the pre-replicative complex and localizes to early sites of DNA synthesis. Thus, it also has a direct role in the control of DNA replication.
MYC regulates transcription from its targets through several mechanisms, including recruitment of histone acetylases, chromatin modulating proteins, basal transcription factors and DNA methyltransferase[22-26].
Protein products of MYC target genes go on to mediate the downstream effects of MYC on cell biology. MYC is then rapidly degraded, and the pathway switches to a transcriptionally repressive state when MAX dimerizes with a group of related bHLH-Zip proteins, the MAD family, that act as MYC antagonists[27] (Figure 1).
Figure 1.
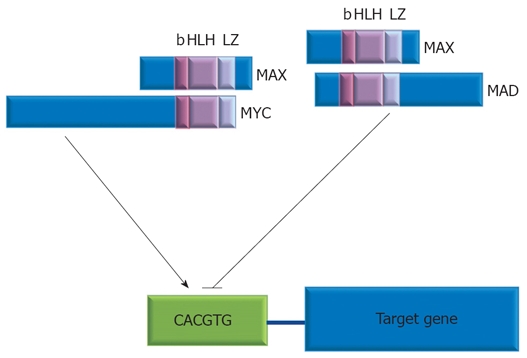
Activation of MYC target genes by the interaction between MYC:MAX or their repression by MAX:MAD. Domains that are common to each protein and are involved in heterodimerization are shown; b: Basic region; HLH: Helix-loop-helix; LZ: Leucine zipper. E-box sequence is shown in green.
MYC expression might be regulated transcriptionally (initiation and elongation), post-transcriptionally (mRNA stability and translation) or post-translationally (protein stability)[28].
MYC is generally recognized as an important regulator of proliferation, growth, differentiation and apoptosis[29,30]. Therefore, it is also accepted that the deregulation of MYC expression is a major event in cancer pathogenesis or progression. Deregulated expression of a wild-type MYC protein is sufficient to lead to cellular transformation in vitro and tumorigenesis in vivo[31].
Recent studies have also found that MYC oncoprotein, in addition to its directly transforming role, can mediate genomic instability via the induction of reactive oxygen species and by promoting whole chromosome instability leading to tetraploidy and aneuploidy. MYC’s ability to promote chromosomal instability is closely linked to its function as a transcriptional regulator[32]. Our research group reported higher frequency of tetraploid clones in GC cell line[33] and aneuploid cells in primary gastric tumor[34,35].
Oncogenic alterations of MYC are commonly induced by events such as point mutations, gene amplification, chromosomal translocation, viral insertion at the MYC locus, and resistance of MYC protein to ubiquitin-mediated proteolysis and enhanced transcription or translation by other oncogenic signaling pathways[30].
MYC AND GASTRIC CARCINOGENESIS
MYC overexpression has been described in over 40% of GC[36]. We found that MYC protein was expressed in all cases of both intestinal- and diffuse-type gastric adenocarcinoma samples of individuals from Northern Brazil[37]. Table 1[38-59] shows the proportion of cases with MYC aberration in several GC studies.
Table 1.
Several MYC studies in gastric cancer
| Reference | Cause of MYC deregulation | Increased MYC | Number of cases | Rate (%) of cases with MYC deregulation |
| [38] | Overexpression | Protein | 88 | 55 |
| [39] | Overexpression | Protein | 213 | 23.5 |
| [40] | Amplification/Overexpression | DNA/Protein | 31/51 | 12.9/41.2 |
| [41] | Overexpression | RNA | 51 | 68.6 |
| [42] | Amplification | DNA | 23 | 26 |
| [43] | Amplification | DNA | 21 | 48 |
| [44] | Amplification | Protein | 154 | 15.5 |
| [45] | Overexpression | Protein | 48 advanced/28 early | 50/42 |
| [46] | Overexpression | Protein | 98 advanced/45 early | 28/34 |
| [47] | Amplification | DNA | 51 | 24 |
| [48] | Amplification | DNA | 10 | 30 |
| [49] | Overexpression | Protein | 42 advanced/77 early | 40.5/15.6 |
| [50] | Amplification/Overexpression | DNA/Protein | 23 | 26 |
| [51] | Overexpression | Protein | 30 advanced/ 6 early | 63.3/50 |
| [52] | Overexpression | Protein | 35 advanced/74 early | 34/16 |
| [53] | Overexpression | Protein | 84 | 88.1 |
| [54] | Overexpression | Protein | 63 | 52.4 |
| [55] | Overexpression | Protein | 65 | 61.5 |
| [56] | Amplification | DNA/Protein | 11 | 100 |
| [37] | Amplification/Overexpression | DNA/Protein | 7 | 100 |
| [57] | Overexpression | Protein | 204 | 43 |
| [58] | Overexpression | Protein | 71 | 42.3 |
| [59] | Amplification/Overexpression | DNA/Protein | 5 early | 100 |
Several studies have shown the association between MYC expression and histopathologic characteristics. Xu et al[51] and Yang et al[54] described a significantly higher expression of MYC in intestinal-type than in diffuse-type GC.
Kozma et al[50] and Yang et al[54] reported that higher MYC expression was associated with the presence of metastasis. Onoda et al[41] also found MYC mRNA levels were higher in metastatic than in primary lesions. Han et al[45] described that patients with high levels of MYC expression had poor disease-free survival. Therefore, MYC expression may represent an aggressive phenotype of GC.
MYC overexpression has also been seen in early GC when tumor invasion is confined to the mucosa or submucosa regardless of the presence of lymph node metastasis[40,41,45,46,52,54,59]. Yang et al[54] found a significantly higher expression of MYC in advanced GC than in early stage GC. However, Onoda et al[41] reported that MYC expression was found to be more frequent and stronger in early than in advanced lesions. Other studies have not found this same difference.
Several studies demonstrated an increased MYC expression in pre-cancerous gastric lesions and its increased expression also has been associated with Helicobacter pylori (H pylori) infection. H pylori is defined as a carcinogen factor to gastric carcinoma infection by the International Agency for Research on Cancer (IARC)[60]. Chronic gastritis caused by H pylori infection may progress to intestinal metaplasia and even to GC[61,62].
Tatsuta et al[63] evaluated MYC mRNA expression by in situ hybridization in 31 elevated gastric lesions. Patients who had borderline lesions with and without MYC overexpression were followed up with repeated endoscopic examinations and gastric biopsies. The authors reported that well-differentiated elevated-type adenocarcinomas were detected in 46% of patients with elevated lesions that presented MYC overexpression during a follow-up period of about 15 mo (range, 2-32 mo) and that no cancers were found in patients with elevated lesions without MYC overexpression. These sample groups were significantly different. Therefore, MYC overexpression may provide a valuable tool for distinguishing between adenomas and well-differentiated elevated-type adenocarcinomas.
Xu et al[51] noticed that MYC protein expression increased progressively as follows: chronic active gastritis, gastric ulcer, mild nonclassic proliferation, severe non-classic proliferation, early GC, and progressive GC.
Lan et al[53] found that MYC expression was higher in GC than in chronic gastritis, intestinal metaplasia and dysplasia. MYC expression was higher in type III intestinal metaplasia with H pylori compared to the same metaplasia without infection and the positive rate in dysplasia with H pylori was higher than that without infection. Zhang et al[55] also reported that MYC expression was higher in chronic atrophic gastritis with severe intestinal metaplasia than that with mild intestinal metaplasia. In chronic atrophic gastritis with severe intestinal metaplasia, MYC expression was higher in cases with H pylori infection than in those without infection. Higher MYC expression was also found in GC with H pylori infection than in that without infection. Thus, MYC expression was coordinately up-regulated in H pylori infected GC and chronic atrophic gastritis with severe intestinal metaplasia. Authors have suggested that H pylori infection may affect MYC expression in gastric diseases, especially in chronic atrophic gastritis.
Several studies have shown that patients with preneoplastic and neoplastic gastric epithelial lesions are more likely to be infected by cagA positive strains. H pylori cagA is one of the most virulent strains of H pylori. Increased cancer risk is described in individuals infected by cagA-positive H pylori strains compared with those infected by cagA-negative H pylori strains and, in general, in those living in areas with a high rate of cagA-positive H pylori strains[64]. Yang et al[54] compared MYC expression in gastric tissues (intestinal metaplasia, dysplasia and GC) with and without H pylori cagA. These authors found that MYC expression was significantly higher in those lesions of type III intestinal metaplasia and dysplasia II-III with cagA than in those without cagA. Nardone et al[64] also suggested that the increased prevalence of MYC expression was in agreement with the high prevalence of cagA positivity seen in the population studied.
Kim et al[65] investigated the expression of MYC protein and mRNA in 22 patients with chronic gastritis who had been successfully treated for H pylori. Two endoscopic antral biopsies were taken before and 2 mo after H pylori eradication. The proportion of gastric antral epithelial cells expressing MYC protein was significantly lower after H pylori eradication. MYC mRNA expression was not changed by H pylori eradication. H pylori may affect cell cycle progression and carcinogenesis through post-translational effects on specific gene expression. Nardone et al[64] also found that MYC expression disappeared after H pylori eradication.
In vitro studies have also confirmed that H pylori can affect MYC expression. Yang et al[66] described that H pylori induces apoptosis in human gastric adenocarcinoma cells mediated by an increased expression of MYC mRNA.
Epstein-Barr virus (EBV) is another infectious agent thought to contribute to cancerous transformation of human host cells. EBV infection is seen in about 10% of gastric adenocarcinoma cases[49,58,67]. Ishii et al[49] found MYC expression in early stages of EBV-positive GC was higher than that of EBV-negative GC, while MYC expression in advanced stages of EBV-positive GC was lower than that of EBV-negative tumors. It was inferred that EBV might cause the host cell to induce MYC expression in early cancer development, but then negatively affect MYC expression in advanced stages of cancers, making them less likely to have a natural regression via apoptosis. Lima et al[58] also reported MYC low expression in EBV-positive GC samples. However, Luo et al[67] have not found any correlation between EBV and MYC expression in GC, suggesting that EBV does not inhibit MYC expression in advanced stages of EBV-positive gastric cancer.
MECHANISMS OF MYC DEREGULATION IN GASTRIC CANCER
Copy number gains are frequently detected along chromosome 8 in gastric tumors[43,48,56,68-73]. Suzuki et al[43] described that chromosome 8 copy number was significantly higher in differentiated than undifferentiated types of GC. Our research group found 8q24.1 gain, where MYC is located, exclusively in intestinal subtype with metastasis by comparative genome hybridization (CGH)[72]. However, Koo et al[48] reported that amplifications in 8q region were more common in diffuse-type cancer.
Some studies have showed an association between MYC amplification and GC[42-44,48]. We have also previously seen MYC amplification in intestinal adenocarcinoma by dual-color fluorescence in situ hybridization (FISH), such as homogeneously staining chromosomal regions and double minutes, supporting our CGH results[56]. Our findings support that these two histological GC types follow different genetic pathways.
Our research group also found that all five early GC cases with MYC overexpression also had three signal to MYC gene by FISH assay, varying between 13% and 26% of cells/case[59]. Suzuki et al[43] found MYC amplification in all 6 early GC cases studied, varying between 19% and 89% of cells/case, and this rate was not significantly difference from that found in advanced GC samples. These findings suggest that MYC amplification can be a critical event to gastric carcinogenesis.
MYC translocation is frequently described in Burkitt’s lymphoma. Few studies have also found translocation of the MYC locus associated with gastric carcinogenesis. Yamashita et al[74] identified chromosomal translocations involved in 8q24 breakpoint by spectral karyotyping (SKY) analysis of established GC cell lines and cancerous ascitic fluids. In a previous study, our findings suggested that translocations can be related to diffuse-type GC using FISH assay[37,56].
Epigenetic events play a significant role in cancer development and progression. DNA methylation is the most studied epigenetic alteration. Some studies also have demonstrated that MYC hypomethylation, which leads to its activation, is significantly more common in GC samples than non-cancerous tissues[75,76]. Fang et al[77] and Weng et al[78] suggest that folate level reduction is associated with upregulation of MYC expression and its promoter hypomethylation in GC.
FUTURE PERSPECTIVES
Proto-oncogenes have a major role not only in cancer development, but also in cancer therapies[79]. MYC alteration is seen in the early gastric carcinogenesis progress. The detection of MYC locus amplification may be used as an auxiliary tool to GC diagnosis and as a predictor of GC aggressiveness.
MYC also could be used as a therapeutical target. Several experimental studies showed that MYC inactivation suppresses tumors in animal models, suggesting MYC as a molecular target in cancer treatment[80-83].
Chen et al[84] evaluated the effect of MYC expression inhibition by recombinant antisense MYC adenovirus (Ad-ASc-myc) infected SGC7901 human gastric carcinoma cells, which have MYC gene amplification, in the proliferation, apoptosis and growth processes of human gastric tumors in nude mice. It was found that MYC expression inhibition may strongly inhibit cell growth and induce apoptosis in SGC7901 cells. Proliferation of Ad-ASc-myc-infected SGC7901 cells was reduced by 44.1%. Studies involving tumorigenicity in nude mice and experimental therapy in nude mice model using Ad-ASc-myc also support these findings. These studies also suggest that Ad-ASc-myc overexpression may result in the elimination of tumor cells via apoptosis and proliferation inhibition, and therefore reduce tumor burden.
Inhibiting MYC expression can be a potential tool for GC treatment in tumors with MYC overexpression. MYC’s therapy target may help identifying more specific and less toxic therapeutic agents[30].
Footnotes
Supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, DQC, MACS and RRB) and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, MFL)
Peer reviewer: Anna Linda Zignego, Professor of Medicine, MASVE Center, Department of Internal Medicine, University of Florence, School of Medicine, Viale Morgagni 85, 50134, Florence, Italy
S- Editor Zhong XY L- Editor Li M E- Editor Ma WH
References
- 1.Terry MB, Gaudet MM, Gammon MD. The epidemiology of gastric cancer. Semin Radiat Oncol. 2002;12:111–127. doi: 10.1053/srao.30814. [DOI] [PubMed] [Google Scholar]
- 2.Resende AL, Mattos IE, Koifman S. [Gastric cancer mortality in the State of Para, Brazil, 1980-1997] Arq Gastroenterol. 2006;43:247–251. doi: 10.1590/s0004-28032006000300018. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization (WHO) Available from: URL: http://www.who.int/en. Accessed January 29, 2008.
- 4.Nardone G. Review article: molecular basis of gastric carcinogenesis. Aliment Pharmacol Ther. 2003;17 Suppl 2:75–81. doi: 10.1046/j.1365-2036.17.s2.10.x. [DOI] [PubMed] [Google Scholar]
- 5.Smith MG, Hold GL, Tahara E, El-Omar EM. Cellular and molecular aspects of gastric cancer. World J Gastroenterol. 2006;12:2979–2990. doi: 10.3748/wjg.v12.i19.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354–362. doi: 10.3748/wjg.v12.i3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamilton SR, Aaltonen LA. Pathology and Genetics of Tumours of the Digestive System. In: WHO Classification of Tumors., editor. Lyon: IARC Press; 2000. p. 204. [Google Scholar]
- 8.Assumpção PP, Burbano RR. Genética do Câncer Gástrico. 1st ed. In: Linhares E, Laércio L, Takeshi S, editors. Atualização em câncer gástrico. São Paulo: Tecmed Editora Ltda; 2005. pp. 95–121. [Google Scholar]
- 9.Chan AO, Luk JM, Hui WM, Lam SK. Molecular biology of gastric carcinoma: from laboratory to bedside. J Gastroenterol Hepatol. 1999;14:1150–1160. doi: 10.1046/j.1440-1746.1999.02000.x. [DOI] [PubMed] [Google Scholar]
- 10.Baffa R, Santoro R, Bullrich F, Mandes B, Ishii H, Croce CM. Definition and refinement of chromosome 8p regions of loss of heterozygosity in gastric cancer. Clin Cancer Res. 2000;6:1372–1377. [PubMed] [Google Scholar]
- 11.Oster SK, Ho CS, Soucie EL, Penn LZ. The myc oncogene: Marvelousl Y Complex. Adv Cancer Res. 2002;84:81–154. doi: 10.1016/s0065-230x(02)84004-0. [DOI] [PubMed] [Google Scholar]
- 12.Sheiness D, Fanshier L, Bishop JM. Identification of nucleotide sequences which may encode the oncogenic capacity of avian retrovirus MC29. J Virol. 1978;28:600–610. doi: 10.1128/jvi.28.2.600-610.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bishop JM. Retroviruses and cancer genes. Adv Cancer Res. 1982;37:1–32. doi: 10.1016/s0065-230x(08)60880-5. [DOI] [PubMed] [Google Scholar]
- 14.Vennstrom B, Sheiness D, Zabielski J, Bishop JM. Isolation and characterization of c-myc, a cellular homolog of the oncogene (v-myc) of avian myelocytomatosis virus strain 29. J Virol. 1982;42:773–779. doi: 10.1128/jvi.42.3.773-779.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Battey J, Moulding C, Taub R, Murphy W, Stewart T, Potter H, Lenoir G, Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983;34:779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- 17.Persson H, Leder P. Nuclear localization and DNA binding properties of a protein expressed by human c-myc oncogene. Science. 1984;225:718–721. doi: 10.1126/science.6463648. [DOI] [PubMed] [Google Scholar]
- 18.Cowling VH, Cole MD. Mechanism of transcriptional activation by the Myc oncoproteins. Semin Cancer Biol. 2006;16:242–252. doi: 10.1016/j.semcancer.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 22.McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 23.Eberhardy SR, Farnham PJ. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J Biol Chem. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- 24.O’Connell BC, Cheung AF, Simkevich CP, Tam W, Ren X, Mateyak MK, Sedivy JM. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J Biol Chem. 2003;278:12563–12573. doi: 10.1074/jbc.M210462200. [DOI] [PubMed] [Google Scholar]
- 25.Kanazawa S, Soucek L, Evan G, Okamoto T, Peterlin BM. c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene. 2003;22:5707–5711. doi: 10.1038/sj.onc.1206800. [DOI] [PubMed] [Google Scholar]
- 26.Brenner C, Deplus R, Didelot C, Loriot A, Vire E, De Smet C, Gutierrez A, Danovi D, Bernard D, Boon T, et al. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 2005;24:336–346. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayer DE, Kretzner L, Eisenman RN. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–722. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 28.Sears RC. The life cycle of C-myc: from synthesis to degradation. Cell Cycle. 2004;3:1133–1137. [PubMed] [Google Scholar]
- 29.Meyer N, Kim SS, Penn LZ. The Oscar-worthy role of Myc in apoptosis. Semin Cancer Biol. 2006;16:275–287. doi: 10.1016/j.semcancer.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16:318–330. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 31.Chung HJ, Levens D. c-myc expression: keep the noise down! Mol Cells. 2005;20:157–166. [PubMed] [Google Scholar]
- 32.Prochownik EV, Li Y. The ever expanding role for c-Myc in promoting genomic instability. Cell Cycle. 2007;6:1024–1029. doi: 10.4161/cc.6.9.4161. [DOI] [PubMed] [Google Scholar]
- 33.Lima EM, Rissino JD, Harada ML, Assumpcao PP, Demachki S, Guimaraes AC, Casartelli C, Smith MA, Burbano RR. Conventional cytogenetic characterization of a new cell line, ACP01, established from a primary human gastric tumor. Braz J Med Biol Res. 2004;37:1831–1838. doi: 10.1590/s0100-879x2004001200008. [DOI] [PubMed] [Google Scholar]
- 34.Costa Guimarães A, Gonçalves Quintana L, Ferreira Leal M, Satomi Takeno S, Pimentel Assumpção P, Moura Lima E, Salim Khayat A, Suchi Chen E, de Arruda Cardoso Smith M, Rodríguez Burbano R. Aneuploidy of chromosome 8 detected by fluorescence in situ hybridisation in ACP01 cell line gastric adenocarcinoma. Clin Exp Med. 2006;6:129–133. doi: 10.1007/s10238-006-0108-5. [DOI] [PubMed] [Google Scholar]
- 35.Assumpcao PP, Seabra AD, Leal MF, Guimaraes AC, Calcagno DQ, Khayat AS, Smith MC, Burbano RR. Chromosome instability in carcinomas. Int J Morphol. 2006;24:335–338. [Google Scholar]
- 36.Milne AN, Sitarz R, Carvalho R, Carneiro F, Offerhaus GJ. Early onset gastric cancer: on the road to unraveling gastric carcinogenesis. Curr Mol Med. 2007;7:15–28. doi: 10.2174/156652407779940503. [DOI] [PubMed] [Google Scholar]
- 37.Calcagno DQ, Leal MF, Seabra AD, Khayat AS, Chen ES, Demachki S, Assumpcao PP, Faria MH, Rabenhorst SH, Ferreira MV, et al. Interrelationship between chromosome 8 aneuploidy, C-MYC amplification and increased expression in individuals from northern Brazil with gastric adenocarcinoma. World J Gastroenterol. 2006;12:6207–6211. doi: 10.3748/wjg.v12.i38.6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spandidos DA, Karayiannis M, Yiagnisis M, Papadimitriou K, Field JK. Immunohistochemical analysis of the expression of the c-myc oncoprotein in human stomach cancers. Digestion. 1991;50:127–134. doi: 10.1159/000200752. [DOI] [PubMed] [Google Scholar]
- 39.Ninomiya I, Yonemura Y, Matsumoto H, Sugiyama K, Kamata T, Miwa K, Miyazaki I, Shiku H. Expression of c-myc gene product in gastric carcinoma. Oncology. 1991;48:149–153. doi: 10.1159/000226915. [DOI] [PubMed] [Google Scholar]
- 40.Nakata B, Onoda N, Chung YS, Maeda K, Nishimura S, Yashiro M, Nitta A, Kubo T, Kato Y, Sowa M. [Correlation between malignancy of gastric cancer and c-myc DNA amplification or overexpression of c-myc protein] Gan To Kagaku Ryoho. 1995;22 Suppl 2:176–179. [PubMed] [Google Scholar]
- 41.Onoda N, Maeda K, Chung YS, Yano Y, Matsui-Yuasa I, Otani S, Sowa M. Overexpression of c-myc messenger RNA in primary and metastatic lesions of carcinoma of the stomach. J Am Coll Surg. 1996;182:55–59. [PubMed] [Google Scholar]
- 42.Hajdu J, Kozma L, Kiss I, Szentkereszty Z, Szakall S, Ember I. Is the presence of distant metastasis associated with c-myc amplification in gastric cancer? Acta Chir Hung. 1997;36:119–121. [PubMed] [Google Scholar]
- 43.Suzuki S, Tenjin T, Watanabe H, Matsushima S, Shibuya T, Tanaka S. Low level c-myc gene amplification in gastric cancer detected by dual color fluorescence in situ hybridization analysis. J Surg Oncol. 1997;66:173–178. doi: 10.1002/(sici)1096-9098(199711)66:3<173::aid-jso4>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 44.Hara T, Ooi A, Kobayashi M, Mai M, Yanagihara K, Nakanishi I. Amplification of c-myc, K-sam, and c-met in gastric cancers: detection by fluorescence in situ hybridization. Lab Invest. 1998;78:1143–1153. [PubMed] [Google Scholar]
- 45.Han S, Kim HY, Park K, Cho HJ, Lee MS, Kim HJ, Kim YD. c-Myc expression is related with cell proliferation and associated with poor clinical outcome in human gastric cancer. J Korean Med Sci. 1999;14:526–530. doi: 10.3346/jkms.1999.14.5.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanz-Ortega J, Steinberg SM, Moro E, Saez M, Lopez JA, Sierra E, Sanz-Esponera J, Merino MJ. Comparative study of tumor angiogenesis and immunohistochemistry for p53, c-ErbB2, c-myc and EGFr as prognostic factors in gastric cancer. Histol Histopathol. 2000;15:455–462. doi: 10.14670/HH-15.455. [DOI] [PubMed] [Google Scholar]
- 47.Kitayama Y, Igarashi H, Sugimura H. Different vulnerability among chromosomes to numerical instability in gastric carcinogenesis: stage-dependent analysis by FISH with the use of microwave irradiation. Clin Cancer Res. 2000;6:3139–3146. [PubMed] [Google Scholar]
- 48.Koo SH, Kwon KC, Shin SY, Jeon YM, Park JW, Kim SH, Noh SM. Genetic alterations of gastric cancer: comparative genomic hybridization and fluorescence In situ hybridization studies. Cancer Genet Cytogenet. 2000;117:97–103. doi: 10.1016/s0165-4608(99)00152-1. [DOI] [PubMed] [Google Scholar]
- 49.Ishii H, Gobe G, Kawakubo Y, Sato Y, Ebihara Y. Interrelationship between Epstein-Barr virus infection in gastric carcinomas and the expression of apoptosis-associated proteins. Histopathology. 2001;38:111–119. doi: 10.1046/j.1365-2559.2001.01037.x. [DOI] [PubMed] [Google Scholar]
- 50.Kozma L, Kiss I, Hajdu J, Szentkereszty Z, Szakall S, Ember I. C-myc amplification and cluster analysis in human gastric carcinoma. Anticancer Res. 2001;21:707–710. [PubMed] [Google Scholar]
- 51.Xu AG, Li SG, Liu JH, Gan AH. Function of apoptosis and expression of the proteins Bcl-2, p53 and C-myc in the development of gastric cancer. World J Gastroenterol. 2001;7:403–406. doi: 10.3748/wjg.v7.i3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishii HH, Gobe GC, Pan W, Yoneyama J, Ebihara Y. Apoptosis and cell proliferation in the development of gastric carcinomas: associations with c-myc and p53 protein expression. J Gastroenterol Hepatol. 2002;17:966–972. doi: 10.1046/j.1440-1746.2002.02805.x. [DOI] [PubMed] [Google Scholar]
- 53.Lan J, Xiong YY, Lin YX, Wang BC, Gong LL, Xu HS, Guo GS. Helicobacter pylori infection generated gastric cancer through p53-Rb tumor-suppressor system mutation and telomerase reactivation. World J Gastroenterol. 2003;9:54–58. doi: 10.3748/wjg.v9.i1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang GF, Deng CS, Xiong YY, Gong LL, Wang BC, Luo J. Expression of nuclear factor-kappa B and target genes in gastric precancerous lesions and adenocarcinoma: association with Helicobactor pylori cagA (+) infection. World J Gastroenterol. 2004;10:491–496. doi: 10.3748/wjg.v10.i4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang GX, Gu YH, Zhao ZQ, Xu SF, Zhang HJ, Wang HD, Hao B. Coordinate increase of telomerase activity and c-Myc expression in Helicobacter pylori-associated gastric diseases. World J Gastroenterol. 2004;10:1759–1762. doi: 10.3748/wjg.v10.i12.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Calcagno DQ, Leal MF, Taken SS, Assumpcao PP, Demachki S, Smith Mde A, Burbano RR. Aneuploidy of chromosome 8 and C-MYC amplification in individuals from northern Brazil with gastric adenocarcinoma. Anticancer Res. 2005;25:4069–4074. [PubMed] [Google Scholar]
- 57.Milne AN, Carvalho R, Morsink FM, Musler AR, de Leng WW, Ristimaki A, Offerhaus GJ. Early-onset gastric cancers have a different molecular expression profile than conventional gastric cancers. Mod Pathol. 2006;19:564–572. doi: 10.1038/modpathol.3800563. [DOI] [PubMed] [Google Scholar]
- 58.Lima VP, de Lima MA, Andre AR, Ferreira MV, Barros MA, Rabenhorst SH. H pylori (CagA) and Epstein-Barr virus infection in gastric carcinomas: correlation with p53 mutation and c-Myc, Bcl-2 and Bax expression. World J Gastroenterol. 2008;14:884–891. doi: 10.3748/wjg.14.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Costa Raiol LC, Figueira Silva EC, Mendes da Fonseca D, Leal MF, Guimaraes AC, Calcagno DQ, Khayat AS, Assumpcao PP, de Arruda Cardoso Smith M, Burbano RR. Interrelationship between MYC gene numerical aberrations and protein expression in individuals from northern Brazil with early gastric adenocarcinoma. Cancer Genet Cytogenet. 2008;181:31–35. doi: 10.1016/j.cancergencyto.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 60.Cancer Databases and Other Resourcess. International Agency for Research on Cancer (IARC) page. Available from: URL: http://www.iarc.fr.
- 61.Wu MS, Chen SY, Shun CT, Lee WJ, Wang HP, Wang TH, Chen CJ, Lin JT. Increased prevalence of Helicobacter pylori infection among patients affected with intestinal-type gastric cancer at non-cardiac locations. J Gastroenterol Hepatol. 1997;12:425–428. doi: 10.1111/j.1440-1746.1997.tb00460.x. [DOI] [PubMed] [Google Scholar]
- 62.Wu MS, Shun CT, Wu CC, Hsu TY, Lin MT, Chang MC, Wang HP, Lin JT. Epstein-Barr virus-associated gastric carcinomas: relation to H. pylori infection and genetic alterations. Gastroenterology. 2000;118:1031–1038. doi: 10.1016/s0016-5085(00)70355-6. [DOI] [PubMed] [Google Scholar]
- 63.Tatsuta M, Iishi H, Baba M, Nakaizumi A, Uehara H, Taniguchi H. Expression of c-myc mRNA as an aid in histologic differentiation of adenoma from well differentiated adenocarcinoma in the stomach. Cancer. 1994;73:1795–1799. doi: 10.1002/1097-0142(19940401)73:7<1795::aid-cncr2820730704>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 64.Nardone G, Staibano S, Rocco A, Mezza E, D’armiento FP, Insabato L, Coppola A, Salvatore G, Lucariello A, Figura N, et al. Effect of Helicobacter pylori infection and its eradication on cell proliferation, DNA status, and oncogene expression in patients with chronic gastritis. Gut. 1999;44:789–799. doi: 10.1136/gut.44.6.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim SS, Meitner P, Konkin TA, Cho YS, Resnick MB, Moss SF. Altered expression of Skp2, c-Myc and p27 proteins but not mRNA after H. pylori eradication in chronic gastritis. Mod Pathol. 2006;19:49–58. doi: 10.1038/modpathol.3800476. [DOI] [PubMed] [Google Scholar]
- 66.Yang Y, Deng CS, Peng JZ, Wong BC, Lam SK, Xia HH. Effect of Helicobacter pylori on apoptosis and apoptosis related genes in gastric cancer cells. Mol Pathol. 2003;56:19–24. doi: 10.1136/mp.56.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luo B, Wang Y, Wang XF, Gao Y, Huang BH, Zhao P. Correlation of Epstein-Barr virus and its encoded proteins with Helicobacter pylori and expression of c-met and c-myc in gastric carcinoma. World J Gastroenterol. 2006;12:1842–1848. doi: 10.3748/wjg.v12.i12.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sakakura C, Mori T, Sakabe T, Ariyama Y, Shinomiya T, Date K, Hagiwara A, Yamaguchi T, Takahashi T, Nakamura Y, et al. Gains, losses, and amplifications of genomic materials in primary gastric cancers analyzed by comparative genomic hybridization. Genes Chromosomes Cancer. 1999;24:299–305. doi: 10.1002/(sici)1098-2264(199904)24:4<299::aid-gcc2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 69.Panani AD, Ferti AD, Avgerinos A, Raptis SA. Numerical aberrations of chromosome 8 in gastric cancer detected by fluorescence in situ hybridization. Anticancer Res. 2004;24:155–159. [PubMed] [Google Scholar]
- 70.Kitayama Y, Sugimura H. Nonrandom chromosomal numerical abnormality as a new molecular cytogenetic tumor marker--a retrospective study of 60 gastric cancer cases. Rinsho Byori. 2005;53:881–886. [PubMed] [Google Scholar]
- 71.Assumpcao PP, Ishak G, Chen ES, Takeno SS, Leal MF, Guimaraes AC, Calcagno DQ, Khayat AS, Demachki S, Smith Mde A, et al. Numerical aberrations of chromosome 8 detected by conventional cytogenetics and fluorescence in situ hybridization in individuals from northern Brazil with gastric adenocarcinoma. Cancer Genet Cytogenet. 2006;169:45–49. doi: 10.1016/j.cancergencyto.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 72.Burbano RR, Assumpcao PP, Leal MF, Calcagno DQ, Guimaraes AC, Khayat AS, Takeno SS, Chen ES, De Arruda Cardoso Smith M. C-MYC locus amplification as metastasis predictor in intestinal-type gastric adenocarcinomas: CGH study in Brazil. Anticancer Res. 2006;26:2909–2914. [PubMed] [Google Scholar]
- 73.Yang S, Jeung HC, Jeong HJ, Choi YH, Kim JE, Jung JJ, Rha SY, Yang WI, Chung HC. Identification of genes with correlated patterns of variations in DNA copy number and gene expression level in gastric cancer. Genomics. 2007;89:451–459. doi: 10.1016/j.ygeno.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 74.Yamashita Y, Nishida K, Okuda T, Nomura K, Matsumoto Y, Mitsufuji S, Horiike S, Hata H, Sakakura C, Hagiwara A, et al. Recurrent chromosomal rearrangements at bands 8q24 and 11q13 in gastric cancer as detected by multicolor spectral karyotyping. World J Gastroenterol. 2005;11:5129–5135. doi: 10.3748/wjg.v11.i33.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fang JY, Zhu SS, Xiao SD, Jiang SJ, Shi Y, Chen XY, Zhou XM, Qian LF. Studies on the hypomethylation of c-myc, c-Ha-ras oncogenes and histopathological changes in human gastric carcinoma. J Gastroenterol Hepatol. 1996;11:1079–1082. doi: 10.1111/j.1440-1746.1996.tb00040.x. [DOI] [PubMed] [Google Scholar]
- 76.Fang JY, Cheng ZH, Chen YX, Lu R, Yang L, Zhu HY, Lu LG. Expression of Dnmt1, demethylase, MeCP2 and methylation of tumor-related genes in human gastric cancer. World J Gastroenterol. 2004;10:3394–3398. doi: 10.3748/wjg.v10.i23.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fang JY, Xiao SD, Zhu SS, Yuan JM, Qiu DK, Jiang SJ. Relationship of plasma folic acid and status of DNA methylation in human gastric cancer. J Gastroenterol. 1997;32:171–175. doi: 10.1007/BF02936363. [DOI] [PubMed] [Google Scholar]
- 78.Weng YR, Sun DF, Fang JY, Gu WQ, Zhu HY. Folate levels in mucosal tissue but not methylenetetrahydrofolate reductase polymorphisms are associated with gastric carcinogenesis. World J Gastroenterol. 2006;12:7591–7597. doi: 10.3748/wjg.v12.i47.7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer. 2004;4:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- 80.Boxer RB, Jang JW, Sintasath L, Chodosh LA. Lack of sustained regression of c-MYC-induced mammary adenocarcinomas following brief or prolonged MYC inactivation. Cancer Cell. 2004;6:577–586. doi: 10.1016/j.ccr.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 81.Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 82.Shachaf CM, Felsher DW. Tumor dormancy and MYC inactivation: pushing cancer to the brink of normalcy. Cancer Res. 2005;65:4471–4474. doi: 10.1158/0008-5472.CAN-05-1172. [DOI] [PubMed] [Google Scholar]
- 83.Yu D, Dews M, Park A, Tobias JW, Thomas-Tikhonenko A. Inactivation of Myc in murine two-hit B lymphomas causes dormancy with elevated levels of interleukin 10 receptor and CD20: implications for adjuvant therapies. Cancer Res. 2005;65:5454–5461. doi: 10.1158/0008-5472.CAN-04-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen JP, Lin C, Xu CP, Zhang XY, Fu M, Deng YP, Wei Y, Wu M. Molecular therapy with recombinant antisense c-myc adenovirus for human gastric carcinoma cells in vitro and in vivo. J Gastroenterol Hepatol. 2001;16:22–28. doi: 10.1046/j.1440-1746.2001.02361.x. [DOI] [PubMed] [Google Scholar]


