SUMMARY
A canonical nucleosome architecture around promoters establishes the context in which proteins regulate gene expression. Whether gene regulatory proteins that interact with nucleosomes are selective for individual nucleosome positions across the genome is not known. Here we examine in Saccharomyces several protein-nucleosome interactions, including those that 1) bind histones (Bdf1/SWR1 and Srm1), 2) bind specific DNA sequences (Rap1 and Reb1), and 3) potentially collide with nucleosomes during transcription (RNA polymerase II). We find that the Bdf1/SWR1 complex forms a di-nucleosome complex that is selective for the +1 and +2 nucleosomes of active genes. Rap1 selectively binds to its cognate site on the rotationally exposed first and second helical turn of DNA inside either border of the −1 nucleosome, whereas Reb1 is selective for a single NFR-proximal border of the −1 nucleosome. We find that a transcribing RNA polymerase creates a delocalized state of resident nucleosomes. These findings suggest that nucleosomes around promoter regions have position-specific functions, and that some gene regulators have position-specific nucleosomal interactions.
Keywords: ChIP-seq, nucleosomes, genomics, Saccharomyces
INTRODUCTION
Genes and their promoters tend to have a canonical chromatin architecture, involving well positioned nucleosomes at precise distances from the transcriptional start site (TSS). In the budding yeast Saccharomyces, the “−1” and “+1” nucleosomes are centered ~230 bp upstream and ~60 bp downstream from the TSS, respectively (Albert et al., 2007; Yuan et al., 2005). Between the two is an intervening ~140 bp nucleosome-free region (NFR) where the general transcription machinery assembles. A similar arrangement exists in multi-cellular eukaryotes (Barski et al., 2007; Mavrich et al., 2008b; Valouev et al., 2008). Little is known about how gene regulatory proteins and the transcription machinery function in the context of this organized state of chromatin. Indeed, although histone-binding domains have been identified (Bannister et al., 2001; Lachner et al., 2001), and factor-nucleosomal DNA interactions have been defined in vitro (Carey et al., 2006; Dang and Bartholomew, 2007; Gelbart et al., 2001; Hassan et al., 2001; Hassan et al., 2002; Li et al., 1994; Prochasson et al., 2005; Rossetti et al., 2001; Saha et al., 2002; Sengupta et al., 2001), there is little direct evidence demonstrating the binding of regulatory factors to nucleosomes in vivo. Chromatin immunoprecipitation (ChIP) assays that measure in vivo occupancy do not distinguish between nucleosomal binding and direct binding to free DNA. Understanding whether and how transcription regulatory proteins interact with nucleosomes throughout a genome should provide key insights into how they function to regulate gene expression.
In principle, there are three non-mutually exclusive ways that a protein might engage a nucleosome: (i) through interactions with histones, (ii) through interactions with nucleosomal DNA, and (iii) through directed collisions having little or no intrinsic affinity. Proteins that interact with histones often have signature motifs such as bromodomains that recognize specific histone modifications (Ruthenburg et al., 2007). Proteins that interact with nucleosomal DNA might recognize the rotationally exposed DNA on the nucleosome surface or adjacent linker DNA entering and exiting the nucleosome (Polach and Widom, 1995; Rossetti et al., 2001). Proteins that potentially collide with nucleosomes in a directed manner include nucleic acid polymerases and helicases.
A number of questions arise regarding the interactions of the transcription machinery and its regulators with nucleosomes in their native context in vivo, that we examine here: 1) Given the fact that nucleosomes are evicted upon transcriptional activation and that promoters reside in nucleosome-free regions, do regulatory factors simply bind nucleosome-free DNA or do some bind to nucleosomes, perhaps during the course of activation? 2) Do regulatory factors that bind nucleosomes discriminate among nucleosome positions? That is, do factors selectively interact with nucleosomes at the −1, +1, +2, etc. positions? 3) Do factors engage single nucleosomes or arrays of nucleosomes? If so, what might be the mechanistic significance? 4) In vivo, does a factor bind to rotationally exposed DNA on the nucleosome surface, or is the cognate site rotationally buried such that nucleosome disruption is required for binding? If binding is to rotationally exposed sites, do those sites reside near nucleosome borders, as model in vitro studies suggest (Polach and Widom, 1995)? 5) Are nucleosomes re-positioned during transcription?
To address these questions we developed a genome-wide factor-nucleosome interaction assay to examine proteins that potentially make contact with nucleosomes in vivo. We examined proteins that we surmised to belong to the three broad interaction types described above, as well as a control protein that is not expected to interact with nucleosomes at all. Our goal was to identify unique as well as general principles regarding the genomic location and regulation of individual factor-nucleosome interactions. Our results suggest that regulatory proteins operate at cognate nucleosome positions at the 5′ end of genes.
RESULTS
Identification of Factor-nucleosome Interactions in vivo
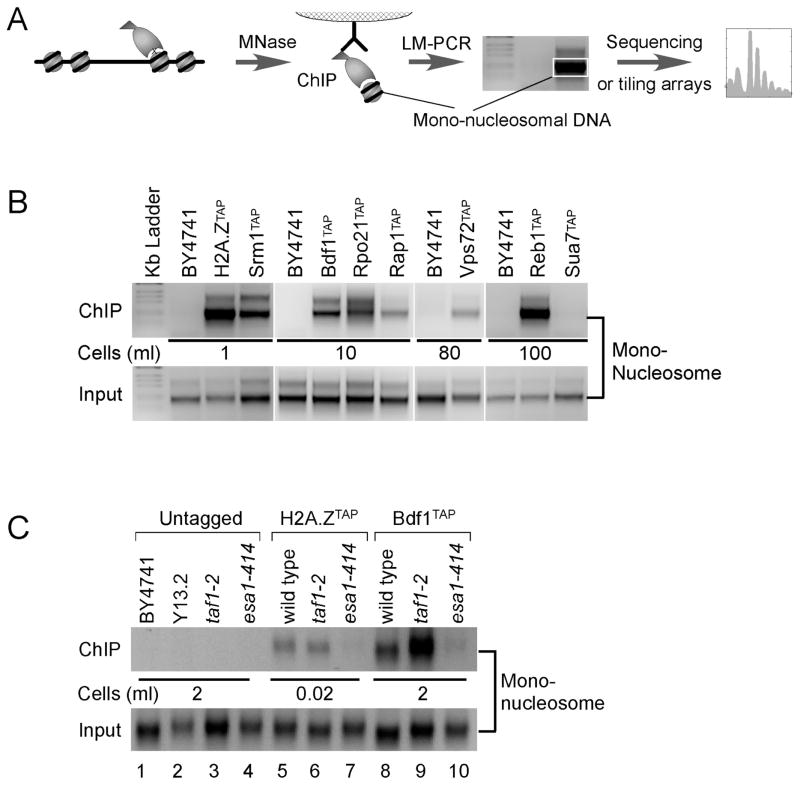
We employed an in vivo factor-nucleosome interaction assay, which is derived from the standard ChIP assay involving protein-DNA crosslinking. In this assay, the chromatin was solubilized into nucleosome core particles using high levels of MNase (Yuan et al., 2005), rather than fragmented via sonication. We also employed multiple purification steps, associated with the use of TAP-tagged proteins. The resulting immunoprecipitated factor-bound mono-nucleosomal DNA was detected by LM-PCR as a nucleosomal-sized band (Fig. 1A), and ultimately mapped across the genome using massively parallel DNA sequencing (AB SOLiD) and verified with high-density tiling microarrays (Affymetrix, 5 bp probe spacing). These genome-wide methods are expected to define a subset of all nucleosome positions in the genome that are in very close proximity (few angstroms) to the tested factor.
Figure 1. Identification of Factor-nucleosome Interactions in vivo.
(A) Diagram outlining the factor-nucleosome interaction assay. Cells encoding a TAP-tagged protein are treated with formaldehyde. Chromatin is then isolated and solubilized to mononucleosomes with MNase, then subjected to TAP purification. Adapter capture of mono-nucleosomal DNA produces an ~200 bp LM-PCR product, which can be subsequently mapped genome-wide.
(B) LM-PCR detection of the indicated factor-nucleosome interaction. BY4741 is a negative control that lacks the TAP tag; H2A.Z is a positive control. Input represents the equivalent of 10−5 ml of cell culture used in LM-PCR, and is the material used for the immunoprecipitation. The volume of cell culture (at OD600 = 0.8) used in the TAP purification is indicated.
(C) Bdf1-nucleosome interactions are dependent on Nua4 (Esa1)-directed acetylation. LM-PCR experiments were performed on the indicated strains as described in panels A and B.
We detected nucleosomal crosslinks for representatives in each type of interaction (Fig. 1B, and quantified in Table 1 data column 1): (i) Htz1, Srm1, Vps72 and Bdf1; (ii) Rap1 and Reb1; and (iii) Rpo21 (RNA polymerase (Pol) II). No crosslinks were detected using an untagged (BY4741) control. No crosslinks were detected with the general transcription factor Sua7 (TFIIB), indicating that not all nuclear proteins are in close crosslinkable proximity to nucleosomes. TFIIB binds in the middle of the NFR (~100 bp from −1 and ~40 bp from +1) and thus is not expected to interact with nucleosomes (Venters and Pugh, 2009). These findings substantially increase the number of proteins demonstrated to crosslink with nucleosomes in vivo, rather than with DNA only, which the standard ChIP assay does not distinguish.
Table 1.
Summary of factor-nucleosome interactions
| Protein | Complex | Complex function | LM-PCRa | ChIP Effciencyb | Nucleosome Interaction Ratioc |
|---|---|---|---|---|---|
| Htz1 | H2A.Z | Nucleosome subunit | 934 ±270 | 23 | 250 |
| Srm1 | Nucleotide exchange factor | 151 ± 31 | 8 | 120 | |
| Rpo21 | Pol II | Transcription | 13.0 ± 3.2 | 11 | 7.0 |
| Bdf1 | SWR1 & TFIID | H2A.Z deposition | 10.5 ± 2.5 | 19 | 3.3 |
| Rap1 | Sequence-specific DNA binding | 5.2 ± 1.2 | 72 | 0.44 | |
| Vps72 | SWR1 | H2A.Z deposition | 0.6 ± 0.4 | 11 | 0.35 |
| Reb1 | Sequence-specific DNA binding | 2.2 ± 0.5 | 42 | 0.32 | |
| Sua7 | TFIIB | General transcription initiation factor | 0.1 ± 0.0 | 37 | 0.02 |
| No tag | BY4741 | Negative control | 0.2 ± 0.1 | 1 |
Mono-nucleosomal LM-PCR signal normalized to input and volume of cells used. The standard error of the mean is shown for at least three replicates.
ChIP Efficiency Ratio (CER) = T/B, where T = Hybridization probe value (after local background subtraction) at the top 1 percentile of occupancy (standard sonication-based ChIP), and B = Hybridization probe value at the bottom 10 percentile of occupancy. The ratios are relative to that obtained for the BY4741 negative control. Similar results were obtained using the top 5 percentile vs. bottom 20 percentile and top 10 percentile vs. bottom 30 percentile.
Nucleosome Interaction Ratio (NIR) = LM-PCR signal/CER. Values were adjusted by dividing through by the BY4741 value (0.16).
A number of addressable caveats are associated with the factor-nucleosome LM-PCR assay. First, it does not distinguish between a protein bound directly to a nucleosome vs. a protein bound to the adjacent linker/NFR regions, but close enough to be crosslinked. Below, we provide a means to distinguish these possibilities for sequence-specific DNA binding factors. Second, without demonstration that binding is actually measurable in a standard ChIP assay, a negative result is not interpretable. Moreover, any crosslinking that is detected represents a net effect of intrinsic crosslinking (i.e., ChIP efficiency) and actual nucleosomal binding.
To distinguish between ChIP efficiency and actual nucleosome binding, we measured intrinsic crosslinking by standard genome-wide ChIP-chip experiments where the chromatin is fragmented by sonication rather than by MNase over-digestion. In this assay, all binding events (nucleosomal and non-nucleosomal) are measured. To assess intrinsic ChIP efficiency, we calculated the ratio of hybridization values at the top 1% of bound sites (after probe normalization) to the bottom 10%, which we take to represent background levels of binding. ChIP efficiency is reported in data column 2 in Table 1. Factors like Rap1, Reb1, and Sua7/TFIIB have very high intrinsic ChIP efficiencies (40–70 fold over the control BY4741).
We next calculated the Nucleosome Interaction Ratio (data column 3 in Table 1), which equals the observed LM-PCR nucleosomal interaction signal normalized to ChIP efficiency (essentially data column 1 divided by data column 2). As expected, the highest Nucleosome Interaction Ratio was seen with Htz1/H2A.Z, which is a nucleosome subunit. The lowest ratio was Sua7/TFIIB, indicating that despite its strong ChIP signal, it does not crosslink to nucleosomes. Thus, despite the nucleus being crowded with nucleosomes, not all competent gene regulatory factors will crosslink with nucleosomes. We conducted further analysis to assess the physiological and mechanistic significance of such interactions.
Bdf1 interacts with NuA4-acetylated nucleosomes in vivo
Bdf1 (type I interaction) is a component of SWR-C/SWR1 (Kobor et al., 2004; Krogan et al., 2003), which is responsible for incorporating H2A.Z into nucleosomes at promoters. Bdf1 binds to acetylated lysines on isolated histone H4 tails (Jacobson et al., 2000; Matangkasombut and Buratowski, 2003), and this acetylation is catalyzed by the Esa1 subunit of the NuA4 complex (Allard et al., 1999). As further validation of Bdf1-nucleosome interactions in vivo, we found that Bdf1TAP-nucleosomal interactions were lost in a catalytically dead esa1-414 mutant (Fig. 1C, lane 8 vs 10). As expected, H2A.Z incorporation was also lost (lane 7). Bdf1 also interacts with TFIID (Matangkasombut et al., 2000; Sanders et al., 2002), which is responsible for assembling the pre-initiation complex. However, loss of the main TFIID subunit in a taf1-2 strain failed to eliminate Bdf1-nucleosomal interactions (lane 9). Together, the results indicate that Bdf1-nucleosomal interactions are mediated through NuA4-directed histone acetylation rather than TFIID. Thus, the factor-nucleosome interaction assay is further validated by the demonstration that the expected NuA4-dependent Bdf1-histone interactions that have been largely defined in vitro, produce the expected dependencies in vivo.
Bdf1 interacts with the +1 and +2 nucleosomes
The genomic locations of Bdf1-crosslinked nucleosomes were determined by sequencing 1,202,352 of these nucleosomes (examples of mapped positions are shown in Fig. 2A), and were verified by hybridization to high-density tiling arrays. Approximately 3% (1,853) of all 54,753 nucleosomes in the yeast genome were significantly crosslinked to Bdf1 (P <0.05, Supplementary Fig. 1A; and listed in Supplementary Table 1), many of which may represent low levels of binding. We selected the genes having the strongest 150 Bdf1-bound nucleosomes as a robust subset for further analysis (listed in Supplementary Table 2; cutoffs of 50, 450, and 1,853 produced essentially the same results, as shown in Supplementary Fig. 1).
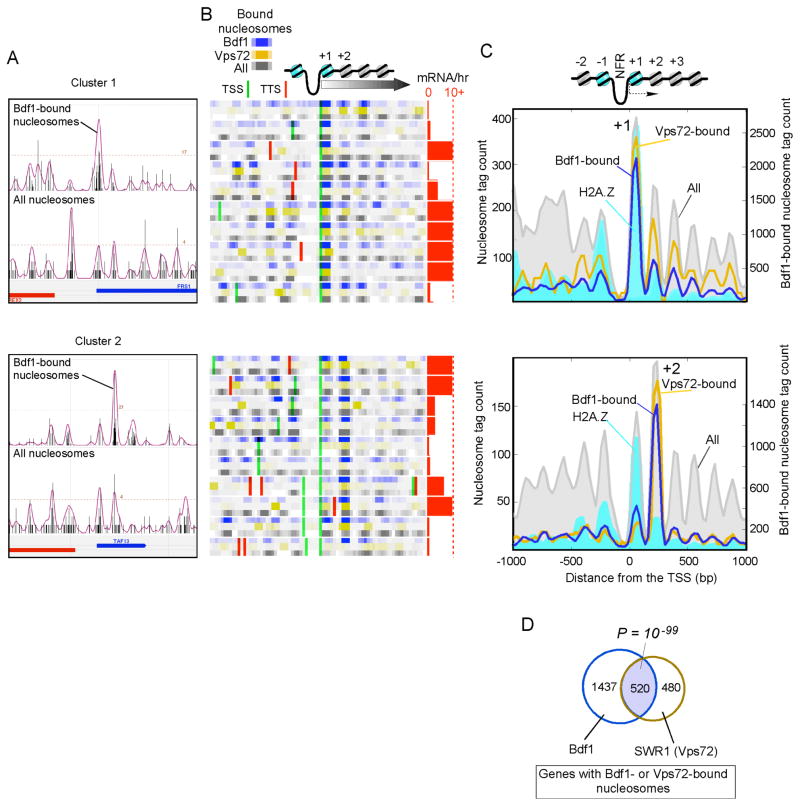
Figure 2. SWR1/Bdf1 are Enriched at the +1 and +2 Nucleosomes.
(A) Distribution of sequence tags for Bdf1-bound nucleosomes and all nucleosomes in a representative section of the genome. The upper panel (cluster 1) displays the enrichment at the +1 nucleosome, and the bottom panel (cluster 2) displays the enrichment at the +2 nucleosome.
(B) Same as panel A, for a collection of loci in which the tag counts have been binned (25 bp bins, and smoothed via a 3-bin moving average) and converted to color intensities. Each gene is represented by three tracks (Bdf1-bound nucleosomes in blue, Vps72-bound nucleosomes in gold, and all nucleosomes in gray). Genes are aligned by their transcriptional start site (TSS) (David et al., 2006). TSS (green lines) and transcript termination sites (TTS, red lines) in the region are shown. Transcription frequency (mRNA/hr) (Holstege et al., 1998) is shown as horizontal red bars, with 10 mRNA/hr indicated by the dashed line.
(C) A composite distribution of tags around the TSS for those genes having the top 150 Bdf1-bound nucleosomes (blue trace). Also shown for the same set of genes are tag distributions for nucleosomes bound by Vps72 (gold trace), H2A.Z (cyan fill), and H3/H4 (“All”, in gray fill).
(D) Venn diagram of the overlap of genes having significant (P < 0.05) Bdf1- or Vps72-interacting nucleosomes.
Surprisingly, at individual genes, Bdf1 bound predominantly to either the +1 or the +2 nucleosome (upper vs lower panels in Fig. 2B, C and Supplementary Figs. 1B, C). This was not a consequence of mis-identifying the +2 nucleosome, because the hallmark of the +1 nucleosome, H2A.Z, was enriched at the +1 nucleosome in both cases (cyan filled plot in Fig. 2C). Moreover, the NFR that is adjacent to the +1 nucleosome is evident in both cases. We also found many cases where Bdf1 bound to the −1 and −2 positions, but these turned out to also be the +1 and +2 nucleosomes of divergently transcribed genes (Supplementary Fig. 2B). Approximately 63% of the top 150 bound nucleosomes were found at the +1/+2 positions compared to 15% expected by chance (P =10−59); 51% of all 1,853 significantly bound nucleosomes were at this position (P = 0). Therefore, Bdf1 is selective for the +1 and +2 nucleosomes. Those not at +1/+2 positions may represent a combination of false positives, occupancy at non-protein encoding genes, and/or additional functionalities associated with Bdf1.
The selectivity of Bdf1 for the +1/+2 nucleosomes was not due to any intrinsically strong positioning of these nucleosomes, making them more detectable, because Bdf1-bound nucleosomes were about average for positioning strength when compared to all nucleosomes (Supplementary Fig. 1D). Furthermore, the distribution of Bdf1-bound nucleosomes from −1 kb to +1 kb of the TSS did not follow the canonical distribution of all nucleosomes at the same set of genes (Fig. 2C), which would be expected if the interactions were simply selecting the best-phased nucleosomes.
Since Bdf1 is part of the SWR1 complex, we examined the genome-wide distribution of SWR1-nucleosomal interactions (via its Vps72 subunit). Genes having Bdf1- and Vps72-bound nucleosomes were statistically co-incident (P ~10−99, Fig. 2D). Moreover, when Bdf1 was enriched at the +1 nucleosome so was SWR1 (Vps72), and when enriched at +2 so was SWR1 (Vps72) (Fig. 2C, blue vs gold traces). This further supports the notion that the SWR1(Vps72)/Bdf1 complex together segregates between either the +1 or the +2 nucleosome, depending on the gene. Both clusters of genes tended to be transcriptionally active (red bar graph in Fig. 2C), indicating that the +1/+2 Bdf1 interactions are associated with transcription. However, neither group was differentially enriched with any Gene Ontology function, which is consistent with such interactions being associated with the transcription process rather than any gene-specific control mechanism. We also examined over two thousand genomic datasets in the public domain for differential properties between the two clusters. We found that cluster 1 tended to have higher levels of intergenic H4 acetylation (largely probing the status of the −1 and +1 nucleosomes) compared to cluster 2 (not shown), which is consistent with cluster 2 being relatively depleted of crosslinkable +1 nucleosomes, and cluster 1 having relatively high levels of acetylated +1 nucleosomes for Bdf1 binding.
Bdf1 Forms a Di-nucleosome Complex Specifically with the +1 and +2 Nucleosomes
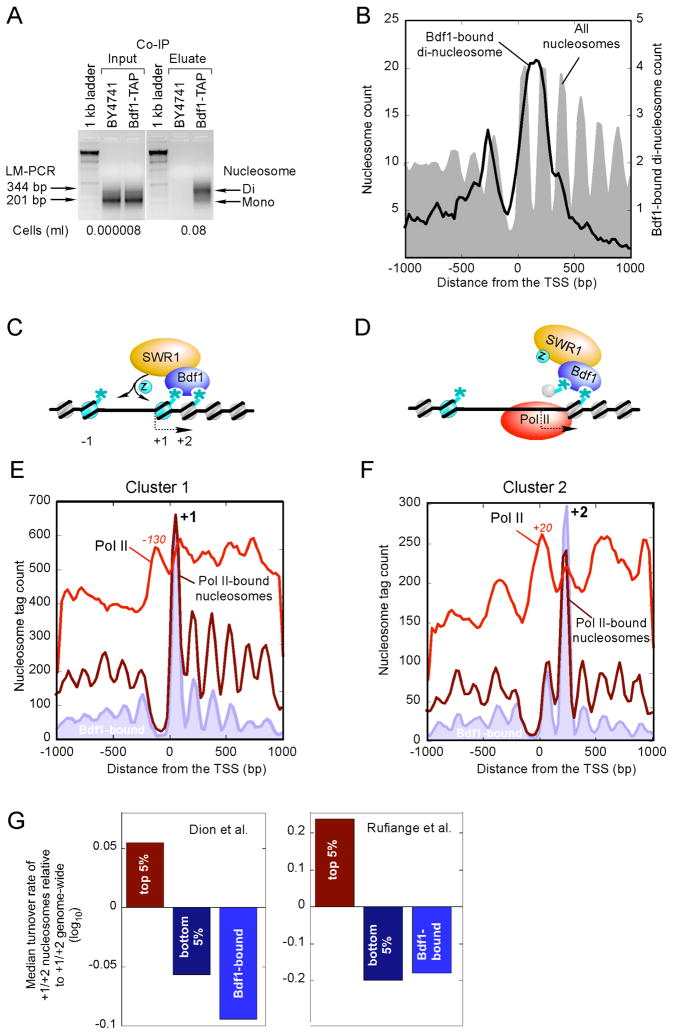
We next sought to understand the relationship between Bdf1 binding to the +1 vs +2 nucleosome by biochemically isolating native Bdf1-nucleosomal complexes (i.e., no formaldehyde and use of a less chaotropic buffer). Surprisingly, these complexes were resistant to MNase (unlike other immunoprecipitated nucleosomal complexes), yielding predominantly di-nucleosomes rather than mononucleosomes (Fig. 3A). This observation suggests that a native Bdf1-containing complex simultaneously binds to two nucleosomes and protects the intervening linker DNA from MNase digestion.
Figure 3. Bdf1 Forms a Di-nucleosome Complex with the +1 and +2 Nucleosomes.
(A) LM-PCR of Bdf1-bound di-nucleosomes. The assay was performed as in Fig. 1A, except that formaldehyde crosslinking was omitted and less chaotropic extraction buffers were used (termed “CoIP”).
(B) Bdf1 di-nucleosome material from panel A was analyzed by Affymetrix high density tiling arrays. The distribution of the highest 4,722 (P <0.05) peaks, representing the di-nucleosome midpoint, around the nearest TSS is shown.
(C) Model of how binding of Bdf1 to acetylated +1/+2 nucleosomes might promote H2A.Z incorporation via the SWR1 complex. The asterisks represent histone acetylation marks. “Z” denotes H2A.Z.
(D) Illustration of how the model in Fig. 3C might allow Pol II to traverse the region and maintain histone modification states.
(E,F) Distribution of Pol II (produced by standard sonication-based ChIP) (Venters and Pugh, 2009) and Pol II-bound nucleosomes around the TSS. The Bdf1-bound filled trace is from Fig. 2C. Panels B and C are for the same genes as in Fig. 2C.
(G) Nucleosome exchange rate of Bdf1-bound nucleosomes. Nucleosome exchange rates were from (Dion et al., 2007; Rufiange et al., 2007). Exchange rates for the top and bottom 5 percentile at the +1/+2 nucleosomal positions, along with the median value for the top 150 Bdf1-bound genes +1/+2 nucleosomes, were divided by the genome-wide median for +1/+2, then log10 transformed and plotted.
To verify that the di-nucleosomal complex represents interactions at the +1 and +2 positions, as opposed to minor or nonspecific complexes at other locations, the di-nucleosomal DNA was mapped at high resolution to the yeast genome. The di-nucleosomal DNA mapped to a region spanning the +1 and +2 nucleosomes (Fig. 3B, note that occupancy between −1 and −2 is due to +1/+2 occupancy of divergent genes), which demonstrates that the Bdf1-bound di-nucleosomal complex is indeed specific to the +1/+2 nucleosomes. Taken together, our findings suggest that the SWR1/Bdf1 complex binds to a NuA4-acetylated di-nucleosomal complex that resides at the +1 and +2 positions of active genes (Fig. 3C). The SWR1 complex then inserts H2A.Z preferentially at the −1 and +1 nucleosomes.
The strong bias of Bdf1 binding towards the +1 vs. the +2 nucleosome position (or visa versa) at individual genes might be a consequence of greater intrinsic nucleosome occupancy levels at the biased position, as shown in Fig. 2C (gray filled plot). To identify a possible source of this bias we hypothesized that as Pol II transcription moves through this region, the +1 acetylated histones are ejected but perhaps retained locally by the SWR1/Bdf1 complex bound at +2 (Fig. 3D). These histones are returned to +1 and a reciprocal process happens at +2 as Pol II moves through the +2 region.
Because Bdf1-bound nucleosomes might present a stronger barrier to Pol II movement, such a model predicts that Pol II occupancy (measured by standard sonication-based ChIP) would be enriched just before the nucleosome that SWR1/Bdf1 is bound to. In addition, the same SWR1/Bdf1-bound nucleosome might be preferentially crosslinked to Pol II due to their close proximity. Indeed, we find evidence to support these predictions at both the +1 (Fig. 3E) and +2 (Fig. 3F) nucleosome positions, where a local enrichment of Pol II (red trace) is found at a fixed distance just upstream of a SWR1/Bdf1-bound (blue-filled plot) and Pol II-crosslinked nucleosome (dark red trace). Additional Pol II is found in the body of the genes, as expected of their transcriptionally active state. Interestingly, in examining over two thousand public genomic datasets for distinguishing features between cluster 1 and 2, one of the strongest distinctions was the enrichment of the Bye1 negative regulator of transcription at some cluster 1 genes (not shown), which might indicate that the hold-up of Pol II before the +1 nucleosome might be regulated at least in part through pol II at these genes.
The notion that Bdf1 might help retain nucleosomes at some promoters is in apparent conflict with the findings that nucleosomes are highly dynamic at promoter regions (Dion et al., 2007; Rufiange et al., 2007). We addressed this by comparing the dynamic state of Bdf1-bound nucleosomes to all other nucleosomes at the +1/+2 position. Strikingly, Bdf1-bound nucleosomes were as cold or even colder (i.e. slower exchange dynamics) than the coldest 5% of +1/+2 nucleosomes (Fig. 3G). This finding lends further credence, from two independent data sets, to the idea that Bdf1 promotes retention of nucleosomes at promoters during the passage of Pol II.
Rap1 Selectively Binds to the −1 Nucleosome that is Shared Between Two Divergent Genes
As a representative of type ii nucleosome-interacting proteins, the sequence-specific DNA binding transcription factor Rap1 is both an activator and repressor of some of the most highly and lowly expressed genes in the cell (Kurtz and Shore, 1991; Shore, 1994). Rap1’s positive role in transcription might be to direct nucleosome disruption and/or recruit TFIID to promoters (Garbett et al., 2007; Yu and Morse, 1999), whereas its negative role, paradoxically, may be to promote nucleosome formation (Gartenberg, 2000; Shore, 1994). These apparent opposing functions remain enigmatic, but could be linked to the location of Rap1 and nucleosomes in promoter regions.
The genomic locations of Rap1-crosslinked nucleosomes were determined by sequencing 383,892 of these nucleosomes (Fig. 4A), and were verified by hybridization to high-density tiling arrays. Approximately 0.4% (229) of all 54,753 nucleosomes in the yeast genome were significantly crosslinked to Rap1 (P <0.05, Supplementary Fig. 2A and listed in Supplementary Table 1). Thirty percent of the previously determined Rap1-bound loci (Lieb et al., 2001) overlapped with these nucleosomes (the remainder being nucleosome-free sites). The genes associated with the top 150 Rap1-crosslinked nucleosomes were selected for further study (listed in Supplementary Table 2).
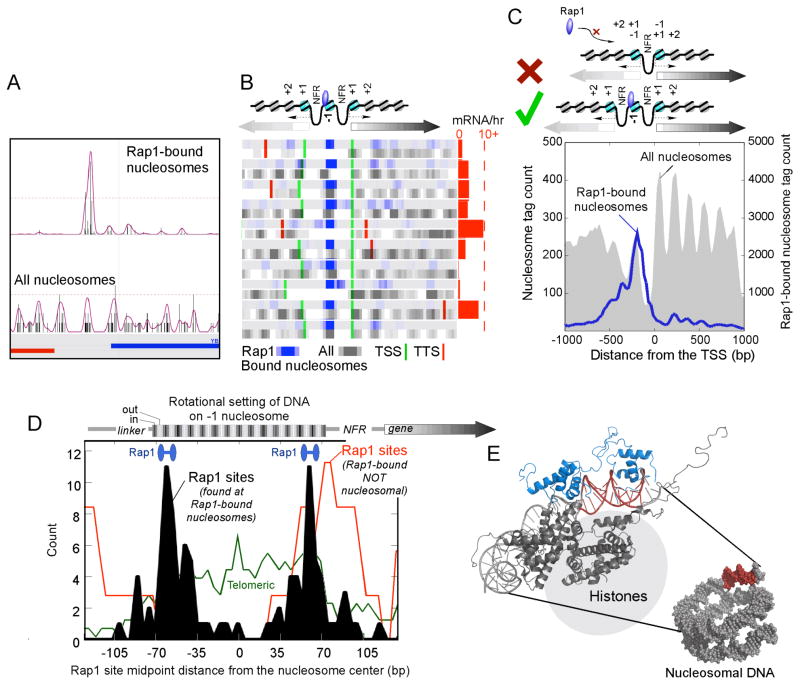
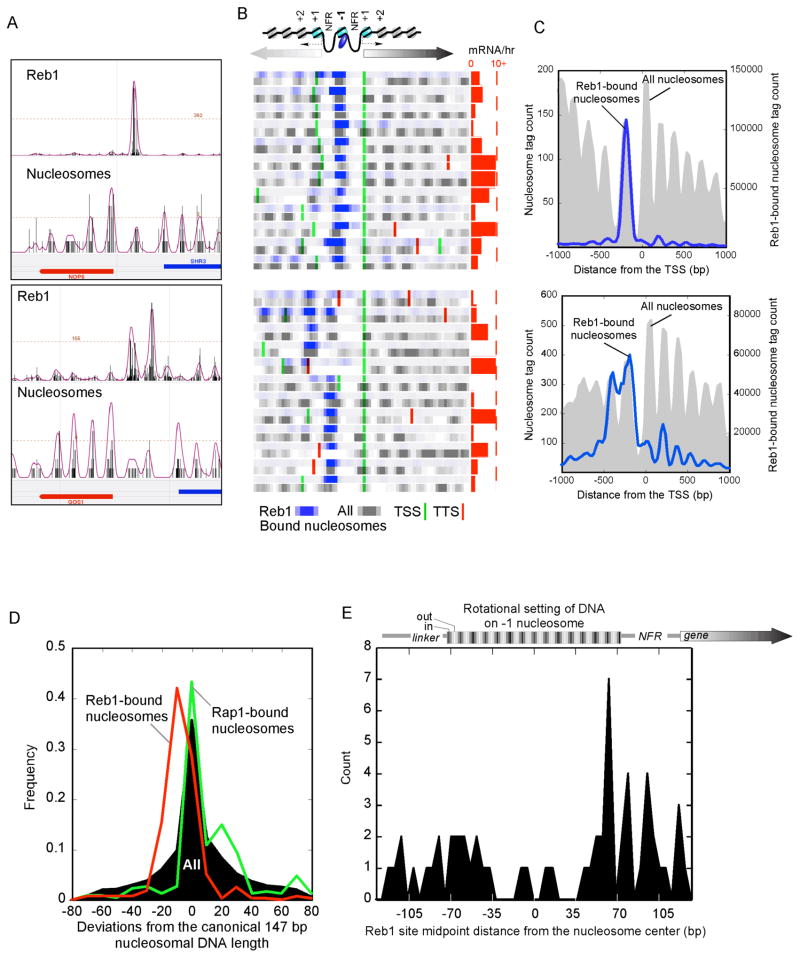
Figure 4. Rap1 Associates with a Specific Rotational Setting on the −1 Nucleosome.
(A–C), Distribution of sequence tags for Rap1-bound nucleosomes. The panels show A) individual loci, B) a collection of genes, C) and a composite profile of tags, as described in Fig. 2. Also illustrated in C is a Rap1-unallowable (X) and –allowable (√) promoter nucleosome configuration.
(D) Distribution of Rap1 binding motifs on the −1 nucleosome. Plotted is the midpoint of the 13-bp motif (ACACCCRYACAYM for the top 150 Rap1-bound nucleosomes), which is composed of two separate half-sites as illustrated by the blue barbells. The rotational setting of DNA on the histone surface is indicated above the plot, where black indicates that the major groove faces inward. Motifs that existed as tandem repeats (found near telomeres) were removed and plotted separately (green trace). Also shown is the distribution of Rap1 sites that have previously been shown to be bound by Rap1 (Buck and Lieb, 2006), but were detected here as having insignificant (P >0.3) Rap1-nucleosomal interactions (red trace).
(E) Model depicting the interaction of Rap1 with the nucleosome core particle in the rotational and translational setting defined in vivo. The model represents a merge of the Rap1/DNA structure (blue/red, 1IGN) with a portion of the nucleosome core particle structure (gray, 1KX5).
Approximately 43% of the Rap1-bound nucleosomes were at the −1 position (P <10−58) (Fig. 4B,C and Supplementary Fig. 2B,C). For the same reasons presented above for Bdf1, detection of the Rap1-crosslinked nucleosomes was not a consequence of biased selection of nucleosomes that are intrinsically the most detectable (Fig. 4B,C and Supplementary Fig. 2B–D).
Rap1-nucleosome crosslinking was not a consequence of Rap1 binding to adjacent linker DNA and fortuitously crosslinking to a neighboring nucleosome because when crosslinking was omitted, Rap1-nucleosomal binding was still detected on fully digested nucleosome core particles (presumably eliminating linker sites) (Supplementary Fig. 2E). Moreover, the MNase resistant DNA present in Rap1-bound nucleosomes was not longer than that found in other nucleosomes (see Fig. 5D), indicating that Rap1 was not protecting additional flanking sequence as a potential consequence of adjacent binding. More importantly, 80% of Rap1-bound nucleosomal DNA possessed a Rap1 binding site within it borders, and very few had sites in adjacent linker regions (Fig. 4D, black filled plot). Rap1-bound sites (Buck and Lieb, 2006) that were not detected as Rap1-bound nucleosomes in our study, were found adjacent to nearby nucleosomes (red trace). This further confirms that Rap1 in linker/NFR regions does not fortuitously crosslink to adjacent nucleosomes. Interestingly, telomeric Rap1 sites tended to be internal to nucleosomes (green trace), suggesting that nucleosomal Rap1 interactions may be different in telomeric regions compared to promoter regions.
Figure 5. Reb1 Associates Specifically with the −1 Nucleosome.
Panel descriptions are as indicated in Fig. 4, but for Reb1. Panel D shows the frequency distribution of nucleosomal DNA lengths as deviations from the canonical 147 bp, for Reb1-bound, Rap1-bound, and all nucleosomes. In panel E, −1 nucleosomes shared between divergently transcribed genes were removed, so as to clearly assess any asymmetry in Reb1 binding to nucleosome borders.
Strikingly, 23% of Rap1-bound “−1” nucleosomes were shared between two divergently-transcribed genes (i.e., the same nucleosome serving the −1 role for both genes), compared to <5% expected by chance (P <10−12) (Fig. 4B and Supplementary Fig. 2B, and illustrated as the “√” configuration in Fig. 4C). In contrast, for 27% of all divergently transcribed genes, the +1 nucleosome of one gene is the −1 nucleosome of the other gene (illustrated as the “X” configuration in Fig. 5C). None of these genes harbored a Rap1-bound nucleosome (P <10−6). Thus, Rap1 may place an evolutionary constraint on the spacing between two divergent Rap1-regulated promoters, such that promoter Rap1-nucleosomal interactions are restricted to configurations where the bound −1 nucleosome does not also serve as a +1 nucleosome.
Rap1 Binds to the First and Second Rotationally Exposed Major Groove inside either Nucleosome Border
We further examined the distribution of the 13-bp bipartite directionally-oriented Rap1 binding site (ACACCCRYACAYM) on the mapped Rap1-nucleosome positions at −1. The midpoint of the Rap1 sites peaked 14 bp from either nucleosome border (Fig. 4D, black filled plot), and was independent of site orientation (not shown). This places the bipartite Rap1 DNA binding domain and the bipartite DNA recognition site on the first and second turn from the nucleosome border of the rotationally exposed major groove (Fig. 4E), which biochemical studies have shown to be the preferred location for Rap1 binding (Rossetti et al., 2001). Together these findings provide near base-pair resolution for the placement of Rap1-nucleosomal interactions in the yeast genome.
Reb1 Selectively Binds to the NFR-proximal Border of the −1 Nucleosomal DNA
As a second representative of type ii nucleosome-interacting proteins, the sequence-specific DNA binding transcription factor Reb1 is thought to bind promoter regions and promote NFR formation (Angermayr and Bandlow, 1997; Hartley and Madhani, 2009; Raisner et al., 2005), although NFR formation may be Reb1-independent at some sites (Erkine et al., 1996; Moreira et al., 2002; Reagan and Majors, 1998). Conceivably, Reb1 might promote NFR formation in part by creating a boundary to which a nucleosome may not encroach. In such situations Reb1 might reside at or near the NFR-proximal nucleosome border. Alternatively, instead of a boundary, Reb1 might position a nucleosome by engaging in specific contacts with histones at some position along the nucleosomal DNA.
The genomic locations of Reb1-crosslinked nucleosomes were determined by sequencing 7,004,145 of these nucleosomes (Fig. 5A). Approximately 0.5% (281) of all detectable 54,753 nucleosomes in the yeast genome were significantly crosslinked to Reb1 (P <0.05, Supplementary Fig. 3A and listed in Supplementary Table 1). The genes associated with the top 150 Reb1-crosslinked nucleosomes were selected for further study (listed in Supplementary Table 2).
Remarkably, 82% of the Reb1-bound nucleosomes were at the −1 position (P <10−257) and 94% of the associated genes were divergently transcribed (upper panels in Fig. 5B,C and Supplementary Fig. 3B,C). Thus, like Rap1, Reb1 strongly favors the −1 nucleosome of divergently transcribed genes. However, unlike Rap1, Reb1-bound nucleosomal DNAs were ~12 bp shorter than the expected length (Fig. 5D), suggesting that Reb1 binding might promote MNase invasion by enhancing the breathing of DNA at the nucleosome border in accordance with the site exposure model (Polach and Widom, 1995).
When the distribution of Reb1 binding sites were examined around Reb1-bound nucleosomes at the −1 position, the Reb1 sites were found to be enriched at the border (Fig. 5E), and were independent of recognition motif orientation (not shown). Strikingly, they were particularly enriched at the NFR-proximal border. The increased nuclease accessibility of the borders of Reb1-bound nucleosomes, which could be particular to the Reb1-bound border, precluded an accurate determination of their position, and so we were less certain as to the rotational setting of the Reb1 binding site. Nonetheless, the NFR-proximal location of Reb1 binding is in accord with the notion of Reb1 setting a boundary for nucleosome positioning adjacent to an NFR (Hartley and Madhani, 2009; Raisner et al., 2005). Since we do not see enrichment of Reb1 at the +1 nucleosome, some other factor may be responsible for establishing the downstream border of the NFR.
Srm1 Abundantly but Non-selectively Occupies Nucleosomes Genome-wide
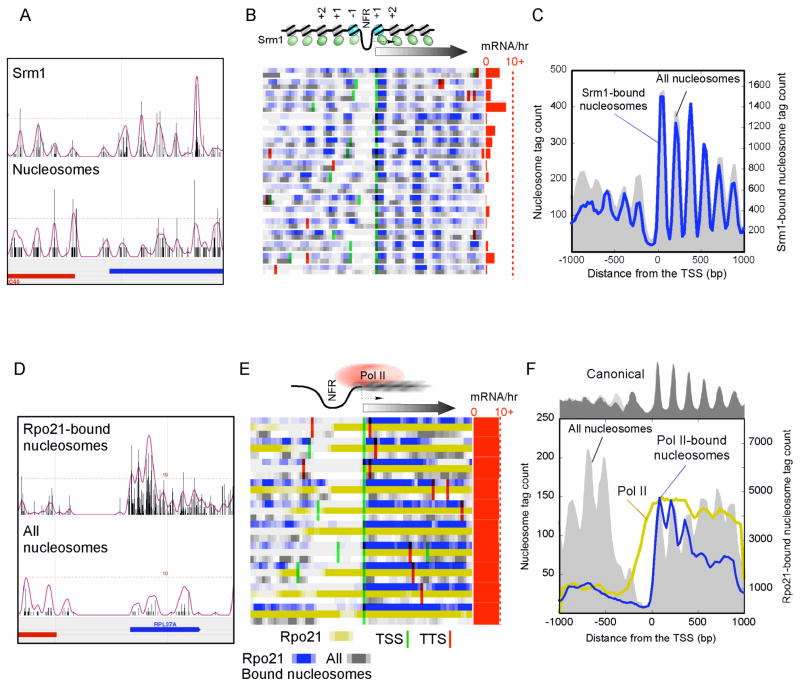
Srm1 (RCC1 in human) is a guanine nucleotide exchange factor that is thought to regulate chromatin condensation and nucleocytoplasmic shuffling (Aebi et al., 1990; Hadjebi et al., 2008). Importantly, Srm1 is nuclear and binds nucleosomes (Nemergut et al., 2001). In our in vivo factor-nucleosome interaction assay, Srm1 generated the strongest interaction ratio (Table 1).
Genome mapping of Srm1-nucleosome interactions revealed a distribution pattern around genes that was essentially indistinguishable from bulk nucleosomes (Fig. 6A, B, C and Supplementary Fig. 4). Thus, while Srm1 binds abundantly to nucleosomes it does not appear to bind specifically. This is in accord with a general role of Srm1/RCC1 in maintaining chromatin structure, particularly in light of the fact that an srm1-1 mutant displays gross chromosomal structural abnormalities (Aebi et al., 1990).
Figure 6. Srm1 Binds Nucleosomes Broadly and Pol II-bound Nucleosomes are Delocalized.
The panels show the distribution of sequence tags for Srm1-bound (A–C) and Pol II-bound (D–F) nucleosomes for individual loci (A, D), a collection of genes (B, E in blue), and a composite profile (C, F blue trace), as described in Fig. 2. Rpo21 is the largest subunit of Pol II. The data shown in gold (E, F) represent the distribution of Pol II from standard sonication-based ChIP-chip.
Pol II-bound Nucleosomes are Delocalized
As a representative of type iii nucleosome-interacting proteins, Pol II is not expected to stably bind to an intact nucleosome. However, due to the fact that it must translocate along DNA, Pol II might collide with nucleosomes, and this could present a barrier to elongation (Bondarenko et al., 2006). Indeed, in Drosophila, Pol II initiates transcription and then pauses as it contacts the +1 nucleosome (Mavrich et al., 2008b; Muse et al., 2007; Zeitlinger et al., 2007). Continued transcription elongation requires that Pol II either eject a nucleosome barrier or traverse some remodeled state of the nucleosome.
The genomic locations of Pol II-crosslinked nucleosomes were determined by sequencing 5,097,371 of these nucleosomes (Fig. 6D), and were verified by hybridization to high-density tiling arrays. Size selection after MNase digestion (Fig. 1B) ensured that intact nucleosomes were being examined.
The genes associated with the top 150 peaks were analyzed further (Supplementary Fig. 5A and listed in Supplementary Tables 1 and 2). The positions of the pol II-crosslinked nucleosomes lacked phasing (Fig. 6D, E and Supplementary Fig. 5B), and so making consensus calls of their positions was not informative. Individual nucleosomal tags were not enriched at canonical locations, as evidenced by a lack of well-defined peaks and valleys of tags around the TSS (Fig. 6F and Supplementary Fig. 5C). Genes that contained relatively high levels of Pol II-crosslinked nucleosomes were generally highly transcribed (red bars in Fig. 6E) and depleted of nucleosomes. We interpret these findings to suggest that during transcription, Pol II collides with nucleosomes (detected as Pol II-crosslinked nucleosomes), and this results in their random repositioning and ultimately their eviction or partial dismantling to allow passage of Pol II (hence nucleosome depletion). The enrichment of Pol II-nucleosomal interactions towards the 5′ end of genes might reflect slower release or a quicker return of nucleosomes at the 5′ end of genes upon transcription.
DISCUSSION
The results presented here advance our understanding of the interplay between the transcription machinery and the highly organized chromatin structure of the yeast genome. The conservation of chromatin architecture across eukaryotes indicates that these findings are likely to be applicable to higher eukaryotes. The −1, NFR, +1, +2 canonical positions can be thought of as providing a fixed scaffold upon which the transcription machinery assembles, and where individual nucleosome positions take on specific functions. The fact that the transcription machinery and its regulators occupy nucleosomes that span from the −1 to the +2 position, a range of nearly 600 bp of DNA sequence, indicates that transcription complex assembly may encompass a much larger stretch of DNA than previously recognized.
Rap1 binds Stereoselectively to the −1 Nucleosome
If a nucleosome resides over the core promoter, then the nucleosome must be removed prior to assembly of the transcription machinery. However, most genes have constitutively nucleosome-free core promoters (NFRs) and thus should be intrinsically accessible. Nevertheless, assembly of the transcription machinery (general transcription factors or GTFs) at the nucleosome-free core promoter requires sequence-specific DNA binding proteins, such as Rap1. If GTF recruitment requires other sequence-specific activators located further upstream, and those binding sites are occluded by nucleosomes (specifically, the −1 nucleosome), then nucleosome disruption or displacement is necessary. In this way, the −1 nucleosome serves a regulatory function.
Rap1 appears to bind to both nucleosomal and non-nucleosomal DNA. When binding to nucleosomal DNA, it is selective for the −1 nucleosome, and is enriched at divergent genes that share the same −1 nucleosomes. Rap1 regulates ribosomal protein genes. However, these genes are highly expressed and tend to lack a −1 nucleosome. Indeed, we find that genes associated with Rap1-nucleosomal interactions tended to be devoid of ribosomal protein genes when compared to the set of genes having non-nucleosomal Rap1 interactions (not shown).
Our results provide the first demonstration on a genomic scale and in vivo that Rap1 binds to the rotationally exposed first and second major groove of DNA inside the nucleosome border, essentially as previously determined in an in vitro reconstitution experiment (Rossetti et al., 2001). Such locations are the least curved and most “breathable” of the nucleosomal DNA (Polach and Widom, 1995; Rossetti et al., 2001), and thus may be as suitable of a binding site as a nucleosome-free site. The “locking in” of Rap1 into the first and second major groove of the −1 nucleosome might impose phasing onto this nucleosome. Indeed, we find that unlike the situation with Reb1, Rap1-bound nucleosomes have a substantially higher degree of phasing compared to all other −1 nucleosomes (not shown).
Interestingly, many nucleosome-free Rap1-bound sites are located immediately adjacent to the −1 nucleosome border. Thus, translational repositioning of the −1 nucleosome over a very short distance could convert nucleosomal-bound sites to non-nucleosomal and vise versa. At some genes Rap1 might promote nucleosome displacement or eviction, with the detected Rap1-nucleosomal interactions being a consequence of a transient interaction. Consistent with this, the −1 nucleosome is relatively depleted (when all nucleosomes are examined) at sites where Rap1-nucleosome interactions are detected.
Whereas Rap1-nucleosomal interactions appear to be confined to the 1st and 2nd rotationally exposed major groove from either nucleosome border, Reb1-nucleosomal interactions appear to be more limited to the NFR-proximal border. Consequently, Reb1 appears to be in position to create a boundary for positioning of the −1 nucleosome, thereby creating the upstream NFR border, in agreement with a recent study (Hartley and Madhani, 2009). Consistent with this, genes that have Reb1-bound nucleosomes have smaller NFRs in a Reb1-depleted strain (determined from analysis of Badis et al., 2008). However, Reb1-bound nucleosomes are no more phased than other −1 nucleosomes (not shown), indicating that phasing and boundary formation may not be entirely linked at the −1 position. Within the NFR, poly dA:dT tracks appear to be responsible for actual nucleosome exclusion. Thus, both Rap1 and Reb1 interact predominantly with −1 nucleosomes near the nucleosome border, but may do so in different ways with distinct functional outcomes.
In contrast to the −1 nucleosome, we see no direct evidence for either Rap1 or Reb1 involved in establishing the position of the +1 nucleosome. While the Bdf1/SWR1 complex binds to the +1/+2 nucleosomes and thus could stabilize binding of these nucleosomes, they are not sequence-specific DNA binding proteins, and thus cannot be directly responsible for positioning the +1/+2 nucleosomes. Instead other mechanisms may be involved, as discussed elsewhere (Albert et al., 2007; Hartley and Madhani, 2009; Zhang et al., 2009).
Repositioning of Nucleosomes in the Wake of Pol II
While the assembly of the GTFs at the NFR does not require eviction of the −1 nucleosome, it is ultimately evicted upon subsequent recruitment of Pol II (Venters and Pugh, 2009). Consistent with this, we see negligible interactions of Pol II with the −1 nucleosome, despite an abundance of Pol II in this region. As Pol II transcribes a gene, it appears to collide with nucleosomes, displacing them from their canonical positions. Those nucleosomes do not adopt new phased positions, but instead are randomly positioned. This may reflect the continuity of Pol II positions along a transcribed gene, and indicates that nucleosome phasing can be disrupted by a transcribing polymerase. Importantly, any mechanism to account for the traversal of Pol II through chromatin must account for nucleosome repositioning as an intermediate stage, as opposed to simple nucleosome ejection or traversal of a fixed-position nucleosome.
How Pol II might traverse nucleosomes while maintaining histone modification states
Given that Bdf1 is homologous to the bromodomain region of TAF1 in higher eukaryotes, we initially expected Bdf1 to bind to the −1 nucleosome, where it could facilitate GTF assembly in the NFR (via its interaction with TFIID). Bdf1 is also part of the SWR1 complex (Kobor et al., 2004; Krogan et al., 2003), and we envisioned that an interaction with the −1 nucleosome could also position SWR1 to load H2A.Z into the −1 and +1 nucleosomes, where it is found. We were therefore surprised to find Bdf1 bound to the +1 and +2 nucleosomes, and that its function seemed more linked to SWR1-directed deposition of H2A.Z than with TFIID.
Given that the −1 nucleosome is evicted upon Pol II recruitment to promoters, and that the +1 and the +2 nucleosomes appear to be evicted during transcription, no nucleosomal location for Bdf1 seemed suitable. However, the data presented here provides a potential explanation in that simultaneous binding of Bdf1 to both +1 and +2 nucleosomes allows one or the other of these nucleosome pairs to be evicted by Pol II and yet retained in the local region for reassembly after Pol II has passed. Consistent with this, Bdf1-bound nucleosomes appear to have less dynamic histone exchange than other +1/+2 nucleosomes. The observed enrichment of Pol II immediately upstream to the Bdf1-bound nucleosome and the crosslinking of Pol II to the Bdf1-bound nucleosome support the notion of stable Bdf1-bound +1/+2 nucleosomes. The broader significance of a SWR1/Bdf1 complex engaged with a +1/+2 di-nucleosome is that it provides a possible model for an epigenetic mechanism to maintain histone modification states at specific nucleosomal positions as Pol II transcription (and in principle, DNA replication) passes through the region.
EXPERIMENTAL PROCEDURES
Factor-nucleosome Interaction Assay and Genome-wide Mapping of Interactions
C-terminally TAP-tagged strains were obtained from Open Biosystems, and grown in 0.5 L YPD at 25°C until OD=0.8. For Bdf1-nuclesome ChIP in temperature-sensitive mutant strains (Y13.2), yeast strains yMD26 (untagged taf1-2), yMD34 (untagged esa1-414), yMD59 (taf1-2 Bdf1-TAP), yMD65 (taf1-2 Htz1-TAP), yMD67 (esa1-414 Bdf1-TAP), & yMD73 (esa1-414 Htz1-TAP) were used (Durant and Pugh, 2007), and crosslinking was performed after a 45 min. temperature shift to 37°C. All crosslinking was performed with 1% formaldehyde at 25°C for 15 min.
Cells were harvested, disrupted, and chromatin pellets washed extensively with FA lysis buffer (50 mM Hepes pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, and 0.1% Sodium Deoxycholate), as previously described (Albert et al., 2007). Mononucleosomes were solubilized via digestion with 160 units of MNase in 600 ul of NP-S Buffer (Yuan et al., 2005) (0.5 mM Spermidine, 0.075% IGEPAL, 50 mM NaCl, 10 mM Tris-Cl (pH 7.5), 5 mM MgCl2, 1 mM CaCl2) at 37°C for 30 min. Mononucleosomes crosslinked to TAP-tagged factors were immunoprecipitated with IgG sepharose, washed with FA lysis buffer and TEV eluted. Stringent washes were used so that nucleosome isolation depended upon the use of formaldehyde and TAP tags. Details of this procedure can be found elsewhere (Albert et al., 2007). Mononucleosomes bound to TAP-tagged factors were further purified via calmodulin sepharose. Eluate DNA was subjected to ligation-mediated PCR (LM-PCR) and electrophoresed on a 2% agarose gel. The data shown are representative of at least three biological replicates. Adaptor sequences are as follows: 5′-GCGGTGACCCGGGAGATCTGAATTC-3′ and 5′-GAATTCAGATC-3′. Several technical factors that affect the yield of factor-bound nucleosomal DNA include, the number of nucleosomes bound by the factor, crosslinking efficiency of the factor, and the use of multiple purification steps.
Following gel extraction of the mono-nucleosomal band, samples were prepared for either DNA sequencing (Bdf1, Vps72, Rap1, Reb1, Srm1, and Rpo21) using Applied Biosystems SOLiD genome sequencer or hybridization with Affymetrix 1.0 GeneChips. Two biological replicates were used in each platform and found to be highly correlated. When samples were to be sequenced, the adaptors were replaced with SOLiD-specific adaptors.
For measuring intrinsic ChIP efficiency of factors, the location of each of the factors listed in Table 1 was measured by ChIP-Chip using customized microarrays containing 20,000 probes (two probes per promoter and one probe internal to genes), using published and unpublished data (Venters and Pugh, 2009). The two promoter probes (~12,000 in total) were used in the rank ordering of hybridization signals (after local background subtraction and normalization to the corresponding probe intensities of the null data set).
Bdf1 and Rap1 Co-immunoprecipitation of Nucleosomes
Bdf1-TAP and Rap1-TAP tagged cultures were grown and harvested as described above but without use of formaldehyde, and lysis was performed in NP-S Buffer (Yuan et al., 2005). Chromatin pellets were isolated and washed in NP-S buffer, and native nucleosomes were released using 15 units of MNase in a volume of 300 ul for 20 minutes. Solubilization of the MNase digested chromatin was accomplished by washing the spun pellet with FA lysis buffer and combining the NP-S and FA lysis buffer supernatant after MNase digestion. Bdf1-bound native nucleosomes were isolated by conventional TAP tag isolation (IgG immunoprecipitation followed by TEV elution), and detected by LM-PCR and whole genome tiling arrays as described above.
SOLiD Sequence Analysis
Sequence tags have been deposited at NCBI Trace Archives under accession number xxxxx. Sequence tags were mapped to the yeast genome using software provide by the SOLiD system, and nucleosome calls were made using GeneTrack software (Albert et al., 2008) (Supplementary Table 1). The tags and resulting nucleosome calls are displayed in a browsable and searchable form at the Penn State Genome Cartography website at http://atlas.bx.psu.edu/.
For the analysis conducted here, significant nucleosome calls were determined to be any call with a peak height value above the mean plus two standard deviations (P <0.05). For all genome-wide analysis, the top 50, 150, & 450 nucleosome calls were analyzed (just top 50 & 150 for Rap1 and Reb1), which showed similar results in all cases; therefore, the top 150 nucleosome calls were chosen for further analysis (Supplementary Table 1).
Gene cluster graphs represent the tag count per bin relative to the TSS (binned every 25 bp), which were smoothed on a 3 bin moving average. K-means analysis and hierarchical cluster analysis was performed on the dataset for each factor. The H3/H4 nucleosome tag counts (Mavrich et al., 2008a) were generated and ordered based on the gene list for each factor. Transcription frequency data was then attached to the gene cluster data, based on the previously described transcription frequency (mRNA/hr) (Holstege et al., 1998). Treeview (Eisen et al., 1998) was used to visualize the cluster plots, and to generate the cluster images.
Composite graphs were generated by binning nucleosome distances to the TSS for the genes to which the top 150 nucleosomes mapped (binned every 25 bp, smoothed every 3 bins) or for the genes in a particular cluster.
Statistical Analysis
P-values reported in the text were calculated via a chi-squared tests in EXCEL assuming a Gaussian distribution of the population. The null hypothesis posits that the bound nucleosomes are distributed randomly among the 54,000 total nucleosomes.
Distribution of Rap1- and Reb1-bound nucleosomal sites
The Rap1 and Reb1 consensus sequence was used to scan top 150 bound nucleosomal DNA sequence along with 100 bp upstream and downstream of the nucleosome borders using FIMO (http://meme.sdsc.edu). A p-value output threshold of 1e-4 was used for the FIMO program.
Distribution of Rap1 Sites that are Rap1-bound but were not Detected as having Significant Rap1-nucleosomal Interactions
The consensus sequence of the Rap1 binding site was used to scan promoter regions (from −600 to +200 bp of TSS) of all yeast genes. 435 sites were identified. 892 Rap1-bound nucleosomes that have a peak height great than or equal to the mean plus the standard deviation of all peak heights were used as a filter. 73 binding sites on these nucleosomes were removed from the set of 435 sites. Finally, 37 of the remaining 362 sites were detected in the 262 static target genes to which Rap1 bound throughout the time course in the previous study (Buck and Lieb, 2006). These 37 sites were defined as Rap1-bound sites that were not detected as Rap1-bound nucleosomes. Their distance from the −1 H3/H4 nucleosome (Mavrich et al., 2008a) was plotted in Fig. 4D (red trace).
Affymetrix Array Analysis
Model-based Analysis of Tiling Arrays (MAT) software was used to determine enrichment regions compared to background of the Affymetrix arrays, as well as cutoff parameters (Johnson et al., 2006). Significance values (P <0.05) were used for Bdf1 (ChIP and CoIP), Rpo21, & Vps72; a more stringent value (P <0.005) was used for the sequence-specific factor, Rap1. After cutoff parameters (derived from MAT interval analysis using significance threshold) were determined, nucleosome calls were made using GeneTrack software (Albert et al., 2008). Composite genome-wide nucleosome interaction distributions relative to the TSS were generated as described elsewhere (Mavrich et al., 2008a).
Supplementary Material
Acknowledgments
This work was supported by NIH grant HG004160. We thank Song Tan and Joe Reese for valuable advice, and Bryan Venters for supplying unpublished ChIP-chip data. The SOLiD sequencing described here was performed at the Penn State Nucleic Acid Facility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aebi M, Clark MW, Vijayraghavan U, Abelson J. A yeast mutant, PRP20, altered in mRNA metabolism and maintenance of the nuclear structure, is defective in a gene homologous to the human gene RCC1 which is involved in the control of chromosome condensation. Mol Gen Genet. 1990;224:72–80. doi: 10.1007/BF00259453. [DOI] [PubMed] [Google Scholar]
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Albert I, Wachi S, Jiang C, Pugh BF. GeneTrack--a genomic data processing and visualization framework. Bioinformatics. 2008;24:1305–1306. doi: 10.1093/bioinformatics/btn119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. Embo J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermayr M, Bandlow W. The general regulatory factor Reb1p controls basal, but not Gal4p-mediated, transcription of the GCY1 gene in yeast. Mol Gen Genet. 1997;256:682–689. doi: 10.1007/s004380050616. [DOI] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24:469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Buck MJ, Lieb JD. A chromatin-mediated mechanism for specification of conditional transcription factor targets. Nat Genet. 2006;38:1446–1451. doi: 10.1038/ng1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Bartholomew B. Domain architecture of the catalytic subunit in the ISW2-nucleosome complex. Molecular and cellular biology. 2007;27:8306–8317. doi: 10.1128/MCB.01351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- Durant M, Pugh BF. NuA4-directed chromatin transactions throughout the Saccharomyces cerevisiae genome. Mol Cell Biol. 2007;27:5327–5335. doi: 10.1128/MCB.00468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkine AM, Adams CC, Diken T, Gross DS. Heat shock factor gains access to the yeast HSC82 promoter independently of other sequence-specific factors and antagonizes nucleosomal repression of basal and induced transcription. Mol Cell Biol. 1996;16:7004–7017. doi: 10.1128/mcb.16.12.7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett KA, Tripathi MK, Cencki B, Layer JH, Weil PA. Yeast TFIID serves as a coactivator for Rap1p by direct protein-protein interaction. Mol Cell Biol. 2007;27:297–311. doi: 10.1128/MCB.01558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg MR. The Sir proteins of Saccharomyces cerevisiae: mediators of transcriptional silencing and much more. Curr Opin Microbiol. 2000;3:132–137. doi: 10.1016/s1369-5274(00)00064-3. [DOI] [PubMed] [Google Scholar]
- Gelbart ME, Rechsteiner T, Richmond TJ, Tsukiyama T. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Molecular and cellular biology. 2001;21:2098–2106. doi: 10.1128/MCB.21.6.2098-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjebi O, Casas-Terradellas E, Garcia-Gonzalo FR, Rosa JL. The RCC1 superfamily: from genes, to function, to disease. Biochim Biophys Acta. 2008;1783:1467–1479. doi: 10.1016/j.bbamcr.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAF(II)250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci U S A. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Shore D. RAP1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev. 1991;5:616–628. doi: 10.1101/gad.5.4.616. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Li B, Adams CC, Workman JL. Nucleosome binding by the constitutive transcription factor Sp1. The Journal of biological chemistry. 1994;269:7756–7763. [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Matangkasombut O, Buratowski RM, Swilling NW, Buratowski S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 2000;14:951–962. [PMC free article] [PubMed] [Google Scholar]
- Matangkasombut O, Buratowski S. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol Cell. 2003;11:353–363. doi: 10.1016/s1097-2765(03)00033-9. [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008a;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008b;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira JM, Horz W, Holmberg S. Neither Reb1p nor poly(dA*T) elements are responsible for the highly specific chromatin organization at the ILV1 promoter. J Biol Chem. 2002;277:3202–3209. doi: 10.1074/jbc.M108962200. [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut ME, Mizzen CA, Stukenberg T, Allis CD, Macara IG. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science. 2001;292:1540–1543. doi: 10.1126/science.292.5521.1540. [DOI] [PubMed] [Google Scholar]
- Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. Journal of molecular biology. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- Prochasson P, Florens L, Swanson SK, Washburn MP, Workman JL. The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes Dev. 2005;19:2534–2539. doi: 10.1101/gad.1341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagan MS, Majors JE. The chromatin structure of the GAL1 promoter forms independently of Reb1p in Saccharomyces cerevisiae. Mol Gen Genet. 1998;259:142–149. doi: 10.1007/s004380050799. [DOI] [PubMed] [Google Scholar]
- Rossetti L, Cacchione S, De Menna A, Chapman L, Rhodes D, Savino M. Specific interactions of the telomeric protein Rap1p with nucleosomal binding sites. Journal of molecular biology. 2001;306:903–913. doi: 10.1006/jmbi.2001.4458. [DOI] [PubMed] [Google Scholar]
- Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nature reviews. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes & development. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol Cell Biol. 2002;22:4723–4738. doi: 10.1128/MCB.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta SM, VanKanegan M, Persinger J, Logie C, Cairns BR, Peterson CL, Bartholomew B. The interactions of yeast SWI/SNF and RSC with the nucleosome before and after chromatin remodeling. The Journal of biological chemistry. 2001;276:12636–12644. doi: 10.1074/jbc.m010470200. [DOI] [PubMed] [Google Scholar]
- Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Morse RH. Chromatin opening and transactivator potentiation by RAP1 in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5279–5288. doi: 10.1128/mcb.19.8.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner B, Euskirchen G, Snyder M, Kadonaga J, Liu X, Struhl K. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo: evidence against a nucleosome code. Nat Struct Mol Biol . 2009 doi: 10.1038/nsmb.1636. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.