Abstract
A genome-scale metabolic network reconstruction for Clostridium acetobutylicum (ATCC 824) was carried out using a new semi-automated reverse engineering algorithm. The network consists of 422 intracellular metabolites involved in 552 reactions and includes 80 membrane transport reactions. The metabolic network illustrates the reliance of clostridia on the urea cycle, intracellular l-glutamate solute pools, and the acetylornithine transaminase for amino acid biosynthesis from the 2-oxoglutarate precursor. The semi-automated reverse engineering algorithm identified discrepancies in reaction network databases that are major obstacles for fully automated network-building algorithms. The proposed semi-automated approach allowed for the conservation of unique clostridial metabolic pathways, such as an incomplete TCA cycle. A thermodynamic analysis was used to determine the physiological conditions under which proposed pathways (e.g., reverse partial TCA cycle and reverse arginine biosynthesis pathway) are feasible. The reconstructed metabolic network was used to create a genome-scale model that correctly characterized the butyrate kinase knock-out and the asolventogenic M5 pSOL1 megaplasmid degenerate strains. Systematic gene knock-out simulations were performed to identify a set of genes encoding clostridial enzymes essential for growth in silico.
Keywords: Clostridium acetobutylicum, metabolic flux analysis, genome-scale model
Introduction
Genome-scale models involve the application of flux balance analysis (FBA) to the two-dimensional stoichiometric matrix of a reconstructed metabolic network (Edwards et al., 1999; Stephanopoulos et al., 1998). Maximizing the specific growth rate has become an accepted objective function of FBA (Edwards et al., 1999), but not the only one (Knorr et al., 2007). Thermodynamic (Henry et al., 2007; Kummel et al., 2006) and regulatory (Covert et al., 2001; Gianchandani et al., 2006; Thomas et al., 2004, 2007) flux constraints along with metabolite conservation relationships (Cakir et al., 2006; Nikolaev et al., 2005) have been developed to decrease the size of the steady-state flux-distribution solution space of FBA.
Solventogenic butyric-acid clostridia are of interest for industrial solvent (particularly bio-butanol) production from diverse substrates, including most hexoses and pentoses, cellulose and xylans (Demain et al., 2005; Montoya et al., 2001; Schwarz, 2001). C. acetobutylicum ATCC 824 is the first sequenced solventogenic Clostridium and can be argued that it serves as a model organism for clostridial metabolism and sporulation in general (Paredes et al., 2005; Thormann et al., 2002). It is an endospore former that displays several defined cascading sigma-factor regulated metabolic programs which impact or are driven by the extracellular environment (Husemann and Papoutsakis, 1988; Jones and Woods, 1986; Paredes et al., 2005; Zhao et al., 2005). It also has an incomplete TCA cycle that may operate in reverse to synthesize fumarate from oxaloacetate (Nolling et al., 2001). Although a genome-scale model has also been constructed for the endospore-forming Bacillus subtilis (Oh et al., 2007), clostridia differ substantially from bacilli in many different ways (Paredes et al., 2005). For example, clostridia are strict anaerobes while bacilli are facultative aerobes. Thus, a genome-scale model of C. acetobutylicum will not only serve genetic, biotechnological and physiological research needs of butyric-acid clostridia, but significantly, its genome-scale metabolic model may eventually be extrapolated to similar pathogenic and non-pathogenic clostridia with annotated genomes.
The development of a genome-scale metabolic network reconstruction and associated stoichiometric matrix requires the piece-wise integration of: (i) enzymes with annotated Enzyme Commission (EC) numbers and associated biological reactions; (ii) metabolic pathway blueprints from biochemical reaction, enzymatic, and membrane transport databases; and (iii) physiological knowledge of the organism transcriptome, proteome and metabolome, including high-throughput data when available. The traditional model-building methodology involves iterative organization of these data into a functional flux network (Becker and Palsson, 2005; Forster et al., 2003; Heinemann et al., 2005). Automation of a metabolic network reconstruction, based on enzyme homology, requires the use of a generalized metabolic network topology readily available from reaction network databases such as Kyoto Encyclopedia of Genes and Genomes (KEGG) and MetaCyc (Caspi et al., 2006; Francke et al., 2005; Kanehisa and Goto, 2000). Due to incomplete genome annotation, these methods commonly result in a non-functional metabolic network due to missing enzymes and other gaps in the network. Thus, algorithms have been developed to automate the processes needed to rectify these discrepancies in metabolic network drafts.
From initial drafts of the genome-scale metabolic network for C. acetobutylicum presented here, two categories of network gaps were identified: (i) gaps resulting from missing enzymes or unknown biological reactions and (ii) gaps resulting from discrepancies in biological reaction databases due to incorrect and mislabeling of compounds and reactions. The first category of network gaps have been addressed by many recently developed algorithms. Techniques used by these algorithms include: genome context analysis (advances of comparative genomics), metabolic pathway homology, enzymatic databases, and high-throughput-omics data (Francke et al., 2005; Kharchenko et al., 2006; Kumar et al., 2007; Notebaart et al., 2006; Osterman and Overbeek, 2003). Other useful algorithms make use of growth phenotyping data (Reed et al., 2006) and genetic perturbations (MacCarthy et al., 2005; Tegner et al., 2003), but these data exist only for a very small percentage of organisms with sequenced and annotated genomes. To address both types of network gaps, analysis of the stoichiometric matrix can be used to identify compounds without both an origin of biosynthesis and degradation (or transport in/out of the network) (Kumar et al., 2007; Reed et al., 2003). From our experience, many discrepancies of the reconstructed metabolic network are not evident from direct analysis of the stoichiometric matrix itself. We found that some discrepancies result in internal cycling of isolated pathways within the metabolic network. Common fixes to metabolic network discrepancies allow transport of inadequately synthesized (or degraded) biological macromolecules into (or out of) the network. This methodology may result in a miscalculation of the metabolic flux profile. Here, we propose a new semi-automated algorithm, based on reverse engineering, to quickly identify both categories of discrepancies in the stoichiometric matrix and illustrate a few examples encountered in metabolic network reconstruction for C. acetobutylicum. Our method allows for the conservation of pathways unique to each bacterial genome. We also demonstrate the usefulness of thermodynamic analysis of proposed pathways here.
Methods
Genome-Scale Metabolic Network Reconstruction for C. acetobutylicum
The genome-scale metabolic model for C. acetobutylicum was derived from mass balances given all known or predicted intracellular metabolic and membrane transport reactions as well as empirical relations for biomass composition. The pseudo-steady state assumption was assumed for all mass balances, resulting in a system of linear equations (Edwards et al., 1999; Papoutsakis, 1984). Prediction of metabolic reactions or transport processes were based on the annotated genome (Nolling et al., 2001) in conjunction with accumulated physiological data. Reactions of the genome-scale model consist of biochemical reactions given constant physiological pH 7 and concentration of free metal ions, as opposed to charge-balanced chemical reactions, according to a previously developed formalism (Alberty, 1993, 1994, 2002). The reconstruction of the metabolic network and integration of these pathways to simulate cell growth in silico was divided into the following separate processes: (i) building metabolic pathways and membrane transport reactions based on genomic annotation, enzyme homology and experimental observations; (ii) developing biomass constituting equations based on physiological data; and (iii) identifying incomplete metabolic pathways and missing metabolite membrane transport reactions through semiautomated reverse engineering of the metabolic network. These three model-building processes are discussed in detail below and were used iteratively to generate a genome-scale model of C. acetobutylicum capable of cell growth in silico.
The Stoichiometric Matrix and Constraints
The resulting composite equation, S · ν=0, consisted of a two-dimensional matrix, S, and a vector, ν, of all intracellular and membrane transport fluxes. Integration of transport reaction fluxes into the stoichiometric matrix of a metabolic model was published (Edwards et al., 2001). Constraints, in the form αi≤νi≤βi were applied to all components of the flux vector. A constraint for irreversibility consisted of setting αi or βi to zero (depending on the reaction-flux direction) while setting the opposite constraint near infinity. The flux vector was optimized through linear programming, a technique commonly referred to as FBA (Edwards et al., 1999; Papoutsakis, 1984). The objective function used in the optimization algorithm was to maximize the specific growth rate. The stoichiometric matrix was constructed in MATLAB (The Mathworks, Inc., Natick, MA). Constrained optimization by linear programming was performed with LINDO API (Lindo Systems, Chicago, IL), within the MATLAB environment. A list of all biochemical reactions, biomass constituting equations, exchange reactions, and associated ranges of applied constraints for FBA is given as Supplementary Appendix 1.
Identification of Metabolic Pathways and Transporters
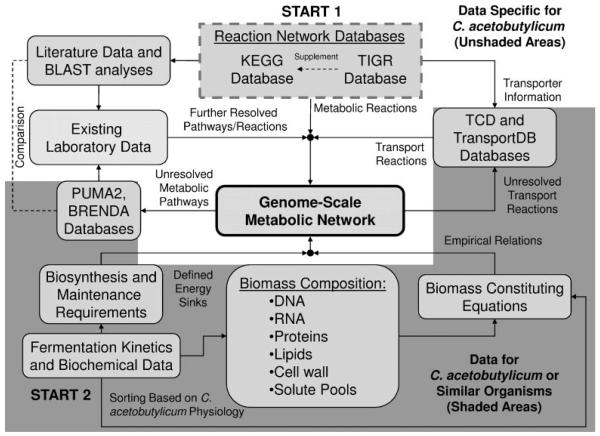
The iterative metabolic pathway construction procedure is summarized in Figure 1. The procedure was initiated with data mining of metabolic pathways specific to C. acetobutylicum contained in the KEGG (Kanehisa and Goto, 2000), the GenomeNet (Kanehisa et al., 2002), MetaCyc (Caspi et al., 2006) and the Comprehensive Microbial Resource (CMR) (Peterson et al., 2001) at The Institute for Genomic Research (TIGR; http://www.tigr.org/). This set of metabolic reactions was further supplemented with metabolite transport reactions obtained from the Transport Classification Database (TCD) (Busch and Saier, 2002; Saier et al., 2006) and TransportDB (Ren et al., 2007). Unresolved metabolic pathways were identified through reverse engineering of metabolic network reconstruction (discussed below). Additional metabolic and transport reactions were identified through the PUMA2 database (Maltsev et al., 2006) and literature specific to the C. acetobutylicum physiology. Furthermore, BLASTP analyses of C. acetobutylicum proteins of unknown function to other annotated clostridial genomes were used to identify additional enzymes contained in KEGG and CMR that were required by the metabolic network. In the absence of clostridial data, genomes of the well-studied bacteria (in order) B. subtilis (Kunst et al., 1997), Staphylococcus aureus N315 (Kuroda et al., 2001), and Escherichia coli K-12 MG1655 (Blattner et al., 1997) were used. The BRENDA enzymatic database (Schomburg et al., 2004) and ExPASy ENZYME database (Bairoch, 2000) were used to further identify substrates/products and stoichiometry of reactions catalyzed by individual enzymes and characterize unresolved pathways. The BRENDA database was also parsed to obtain a list of all enzymes catalyzing irreversible reactions under physiological conditions, and this list was used to identify enzymes in the C. acetobutylicum metabolic network catalyzing irreversible reactions.
Figure 1.
Flow diagram of iterative construction of the genome-scale metabolic network. The un-shaded (white) background corresponds to data obtained from resources specific to C. acetobutylicum. Elements of the flow diagram located in shaded (dark gray) background regions represent data compiled from resources specific to C. acetobutylicum and supplemented with information obtained from organisms similar to C. acetobutylicum, namely: other clostridia, B. subtilis, S. aureus, and E. coli.
Overview of Biomass Constituting Equations
The contribution of the metabolic network to the production of biomass was calculated based on genomic and physiological data available for C. acetobutylicum. The components of the biomass constituting equation were adapted from a platform initially created for S. aureus N315 (Heinemann et al., 2005) and recently used for Methanosarcina barkeri (Feist et al., 2006). Specifically, biomass was defined as a sum of: RNA, DNA, protein, lipids, cell wall, and solute pools of the cytoplasm. The specific definition of each of these broad terms was constructed according to genomic information obtained from NCBI and from literature data. The total list of biomass constituting equations and energetic requirements are shown in Supplementary Appendix 1. The average DNA composition was based on the nucleotide content of the entire genome and the pSOL1 megaplasmid. The average protein and RNA compositions were calculated from an analysis of known ORFs. The calculation of the average RNA sequence included ribosomal and tRNA sequences in addition to ORFs. Previously published data, specific to C. acetobutylicum and B. subtilis, enabled specifically tailored constituting equations for lipids, teichoic acids, and peptidoglycan biosyntheses. These equations are also shown in Supplementary Appendix 1. Due to the unavailability of specific data, the composition of the intracellular solute pool (shown in Supplementary Appendix 1) was assumed similar to those published for S. aureus N315 (Heinemann et al., 2005) with some notable exceptions (discussed later). Also consistent with the model for S. aureus (Heinemann et al., 2005), a growth maintenance value of 40 mmol ATP/(g cell dry weight per hour) was assumed (Stephanopoulos et al., 1998).
Pathway Resolution Through Reverse Engineering of the Metabolic Network
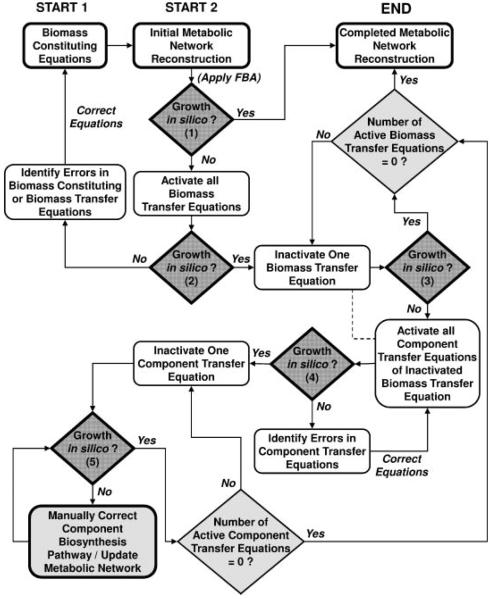
Data mining of biochemical pathway databases (KEGG, in particular) were used in compiling initial drafts of the metabolic network for C. acetobutylicum. However, as is currently the case for most genomes, incomplete gene annotation leads to several incomplete metabolic pathways within such biochemical pathway databases. In addition, other inconsistencies were observed in data obtained directly from these biochemical pathway databases. These included: (i) multiple identity markers for the same compound; (ii) compounds that lacked an origin of synthesis/degradation within the biochemical database; (iii) incorrect stoichiometry of metabolic reactions; and (iv) misappropriated enzymes to a particular cell type. Identification of the source of a broken metabolic pathway (gaps) of the network is a laborious task, especially in the case where multiple sources of inconsistencies may exist (Kumar et al., 2007; Reed et al., 2003). Thus, a reverse engineering approach was developed to identify such inconsistencies within the metabolic network. The approach was designed to be used in conjunction with or after the identification of dead-ends through stoichiometric matrix analysis (Reed et al., 2003). The proposed reverse engineering approach includes optimizing the reaction flux network with an objective function of maximizing the specific growth rate. In general, a metabolic network with one or multiple incomplete biochemical pathways (from substrate to biomass building blocks) was found to result in a maximized specific growth rate of zero (no growth in silico). This approach is illustrated by a flow diagram of Figure 2. Our reverse engineering algorithm requires a set of biomass constituting equations (see Supplementary Appendix 1) and a metabolic network (complete or incomplete). The set of membrane transporters required for minimal medium (Monot et al., 1982) (see Supplementary Appendix 1) were used here as well. If the application of FBA to the existing metabolic network does not yield the production of biomass in silico, biomass transfer equations are added to the metabolic network. These equations are listed in Supplementary Appendix 2 and consist of the individual components comprising biomass (e.g., RNA, DNA, protein, lipids, cell wall, and pooled solutes) and which are separately transported into an incomplete metabolic network. The addition of biomass transfer equations results in a positive specific growth rate in silico when FBA is applied. It is noted that biomass transfer equations and component transfer equations (discussed later) are arbitrary membrane transport equations used to identify metabolic network discrepancies only. These equations are not present in the final version of the metabolic network reconstruction. Following their addition, one-by-one the biomass transfer equations are eliminated. Once the elimination of a biomass transfer equation results in a specific growth rate of zero (arrested growth in silico), that broadly defined component of biomass is broken down into its constituents. For example, the biomass component RNA is composed of genome-specific stoichiometric amounts of ATP, CTP, GTP, and UTP. In this case, the RNA biomass transfer equation would be removed and ATP, CTP, GTP, and UTP would be added to the metabolic network by separate equations termed component transfer equations. The full list of component transfer equations used in the model-building process is given in Supplementary Appendix 2. In a similar procedure, the component transfer equations are systematically eliminated until a specific growth rate of zero is realized. The component responsible for arresting growth in silico is recognized as being inadequately synthesized/degraded in the existing metabolic network. Upon identification of this type of discrepancy in the metabolic network, iterative measures, as shown in Figure 1, are implemented to resolve the network connectivity.
Figure 2.
Flow diagram for reverse engineering of a metabolic network reconstruction. Complete lists of biomass transfer equations and component transfer equations are presented in Supplementary Appendix 2.
Thermodynamic Analysis of Proposed Pathways
We also assessed the thermodynamic feasibility of proposed metabolic pathways (e.g., the reverse partial TCA cycle) for C. acetobutylicum that are not common to reaction network databases. This was done by calculating the Gibbs free energy of all reactions of the pathway using previously published methods and estimated values for the standard Gibbs free energy of formation, , and estimated standard Gibbs free energy of reaction, (Henry et al., 2006, 2007).
| (1) |
A negative Gibbs free energy of reaction,
| (2) |
is required for a metabolic reaction to occur and was calculated given m compounds of a chemical reaction with stoichiometric coefficients n, where R is the ideal gas constant, and an assumed temperature, T, of 298 K. Millimolar concentrations, ci, of reaction components (Henry et al., 2006) and dimensionless activity coefficients, γi were used to calculate the concentration-dependent term of the Gibbs free energy of reaction equation (Eq. 2). As shown previously (Henry et al., 2007), the standard error in and terms calculated from group contribution theory (Mavrovouniotis, 1990) outweighed the influence of ionic strength, despite the illustration of its strong influence on ΔrG′ (Maskow and von Stockar, 2005). Given these results, activity coefficients were set to 1 for our calculations. For proposed pathways in C. acetobutylicum not native to reaction network databases (e.g., KEGG), combinations of metabolite concentrations yielding negative ΔrG′ values for every reaction in the pathway were calculated. Pathways incapable of producing negative ΔrG′ values for every reaction are thermodynamically infeasible. Resulting metabolite concentrations were compared to measured physiological metabolite concentrations of C. acetobutylicum (when available) to assess the practicality of the proposed reaction, similar to that done for glycolysis (Maskow and von Stockar, 2005). For cases in which not all metabolite data were available, ranges of metabolite concentrations at which a proposed pathway is feasible were calculated. It is noted that a wide range of short-comings currently exist for the thermodynamic analysis of metabolic pathways (Maskow and von Stockar, 2005). Aside from the obvious pitfalls of accurate and cytoplasm ionic strength calculations, the influence of intracellular pH changes on is unaccounted for in our calculations as pH 7 was assumed for all cases. In addition, charge balances were carried-out using only the major protonated species at pH 7 to remain consistent with calculations (Henry et al., 2006, 2007).
Results and Discussion
Construction of the Genome-Scale Model
The genome-scale metabolic network for C. acetobutylicum was constructed using the iterative methods of pathway construction shown in Figure 1 and the reverse engineering algorithm of Figure 2. The network consists of 422 metabolites involved in 552 reactions, including 80 metabolite transport reactions across the cell membrane. Simulation of the genome-scale model produced a positive specific growth rate for the wild-type genome with the complete set of transporter reactions. The buk gene knock-out mutant (Green and Bennett, 1998; Harris et al., 2000) was simulated by restricting flux through the butyrate kinase enzyme (Buk, EC 2.7.2.7, CAC3075) to zero using constraints. In addition, the pSOL1 megaplasmid degenerate M5 strain (Tomas et al., 2003) was simulated by restricting flux through enzymes encoded by megaplasmid genes. These reactions are specifically labeled in Supplementary Appendix 1. The qualitative results of these simulations are given in Table I. Resulting specific growth rates of these simulation studies did not match experimental observations due to the lack of regulatory mechanisms and large number of reversible reactions in this initial version of the genome-scale model. We further investigated the capabilities of the genome-scale model to simulate growth on the published minimal medium formulation for C. acetobutylicum (Monot et al., 1982) and a glycerol-containing synthetic medium (Vasconcelos et al., 1994). These results are also summarized in Table I. In all cases, growth in silico was successful without adding further additional transport equations to provide metabolites or macromolecules not adequately synthesized or effectively degraded by the metabolic network. In addition, observed phenotypes of knock-out strains were obtained in silico, suggesting that the network is complete and represents C. acetobutylicum metabolism. The number of reactions in the reconstructed metabolic network used to represent specified metabolic functions is shown in Table II. This table also provides statistics that relate the completed metabolic network to the genomic annotation used to reconstruct it.
Table I.
In silico growth results of C. acetobutylicum genome-scale model given genetic and environmental manipulations.
| In silico strain | Medium | Additional transport reactions required |
Comment | Growth in silico? |
|---|---|---|---|---|
| Wild-typea | Complexd | None | Acidogenic and solventogenic | Yes |
| Wild-typea | Minimale | None | Acidogenic and solventogenic | Yes |
| Wild-typea | Syntheticf | None | Acidogenic and solventogenic | Yes |
| buk knock-outb | Minimale | None | No production of butyrate | Yes |
| M5c | Minimale | None | No production of acetone or butanol; decreased ethanol production |
Yes |
The wild-type in silico strain contains all reactions and constraints as listed in the metabolic network in Supplementary Appendix 1.
The buk knock-out in silico strain was created by constraining the reaction catalyzed by the butyrate kinase (Buk, EC 2.7.2.7, CAC3075) to zero.
The M5 in silico strain was created by constraining reactions only catalyzed by megaplasmid genes to zero. Megaplasmid genes contain locus number beginning with the CAP prefix.
We simulated a complex medium by leaving all membrane transport reactions unconstrained.
A minimal medium (Monot et al., 1982) was simulated by constraining all membrane transport reactions of Supplementary Appendix 1 to zero except those given the label Minimal.
The synthetic medium (Vasconcelos et al., 1994) is similar to the minimal medium except that it contains glycerol and ammonium chloride replaced ammonium acetate. The membrane transport reactions needed in addition to the Minimal transport reactions to support this medium are labeled Synthetic in Supplementary Appendix 1.
Table II.
Summary of the metabolic network reconstructed for C. acetobutylicum.
| Total number of protein-encoding genes in C. acetobutylicum genome (including pSOL1 megaplasmid) | 3,748 (including 176 from pSOL1) |
| Number of protein-encoding genes with role in reconstructed metabolic network | 474 (12.6% of protein-encoding genes) |
| Number of enzyme-encoding genes excluded from the metabolic network | 366 (9.77% of protein-encoding genes) |
| DNA polymerases | 31 |
| tRNA ligases | 46 |
| Number of genes associated with uncharacterized membrane transporters | 242 (6.46% of protein-encoding genes) |
| Number of intracellular metabolites | 422 |
| Number of reactions (non-redundant)a | 552 |
| Carbohydrate metabolism | 125 (28) |
| Energy metabolism | 11 (3) |
| Lipid metabolism | 46 (22) |
| Nucleotide metabolism | 82 (14) |
| Amino acids metabolism | 125 (13) |
| Metabolism of cofactors and vitamins | 76 (24) |
| Membrane transport | 80 (20) |
| Biomass and maintenance | 20 (20) |
| Number of irreversible metabolic reactions (excluding biomass equations and membrane transporters) | 26 |
Some reactions are included in more than one classification and some classifications are not included in this table. Numbers in parentheses represent the number of reactions in the pathway not currently assigned to a specific gene. These reactions were constructed from pathway completion methods given physiological data of C. acetobutylicum.
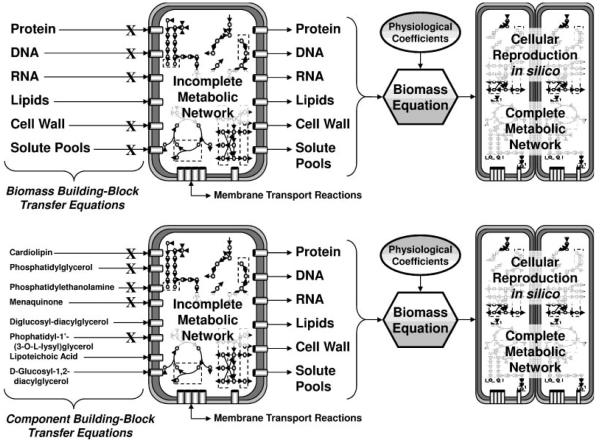
An example of one iteration of the semi-automated reverse engineering process for completing the genome-scale metabolic network is shown in Figure 3, whereby deficient lipid biosynthesis of lipoteichoic acid, diglucosyl diacylglycerol and d-glucosyl-1,2-diacylglycerol were found responsible for arresting cell growth when the metabolic flux profile was optimized. The metabolic pathways for these precursors were investigated and manually rectified. Employing the reverse engineering procedure iteratively was necessary for identifying and correcting these growth-preventing errors in the metabolic network. Application of the reverse engineering algorithm of Figure 2 to an initial draft of the C. acetobutylicum metabolic network largely created from the KEGG database, revealed reaction network discrepancies beyond simply missing enzymes. These discrepancies are shown as Supplementary Appendix 3 and include a list of aerobic reactions annotated in KEGG to belong to C. acetobutylicum, a strict anaerobe.
Figure 3.
Reverse engineering of metabolic network completion through the inclusion of additional biomass building-block transfer reactions. The procedure initiated with an incomplete metabolic network (incapable of producing biomass). All components of the biomass constituting equation were imported (called building-block transfer equations). One-by-one, building-block transfer equations were eliminated until those required for in silico biomass growth were located. Then, transfer equations of generalized biomass building-blocks were evaluated in order to identify specific regions of the metabolic network impeding biomass growth. The following example is shown for a case in which deficiencies in lipoteichoic acid, diglucosyl-diacylglycerol and d-glucosyl-1,2-diacylglycerol biosynthesis pathways in the metabolic network impeded biosynthesis of the lipids block of the biomass constituting equation, resulting in impeded growth. This procedure was developed to quickly locate incomplete metabolic pathways (e.g., from missing or unidentified enzymes) in the metabolic network.
Representation of Lipid Biosynthesis
Total lipids in C. acetobutylicum have been found to account for 5–6% of the dry cell weight (Lepage et al., 1987). It has been also reported that solvent exposure leads to an increase in the ratio of saturated and cyclopropane fatty acids to unsaturated membrane fatty acids (Baer et al., 1987; Vollherbst-Schneck et al., 1984; Zhao et al., 2003), changes in the mean fatty acid acyl chain length (Lepage et al., 1987; Vollherbst-Schneck et al., 1984; Zhao et al., 2003) and changes in the membrane phospholipid composition (Johnston and Goldfine, 1992; Lepage et al., 1987; MacDonald and Goldfine, 1991). Nevertheless, due to the absence of specific compositional information about these changes, a single lipid biosynthesis equation (see Supplementary Appendix 1) was used in the calculation of biomass composition over the entire course of exponential growth. The relative amounts of lipids and phospholipids of the lipids biosynthesis equation was derived based on a consensus of the cited literature data corresponding to exponential growth. The fatty acid composition in all cases was also held constant at 16:0 (carbon chain length: number of double-bonds), which is a dominant experimental observation (Lepage et al., 1987; Vollherbst-Schneck et al., 1984). For the lipid-equation component of lipoteichoic acid (LTA), literature data specific to B. subtilis (Neuhaus and Baddiley, 2003; Perego et al., 1995) were used, due to insufficient data available for C. acetobutylicum. The average LTA composition of 29 glycerophosphate units per chain was used. Also, an average of 13 glycerophosphate units per chain were substituted with d-alanine esters (d-alanylation) in B. subtilis (Neuhaus and Baddiley, 2003; Perego et al., 1995). The process of d-alanylation was ignored in the C. acetobutylicum model due to the absence of a dlt operon (Kiriukhin and Neuhaus, 2001; Perego et al., 1995).
Cell-Wall Composition
Cell wall is made up of crosslinked peptidoglycan and wall teichoic acid (WTA). Due to the lack of information specific to C. acetobutylicum, in the cell-wall equation (see Supplementary Appendix 1), the stoichiometric coefficients of these components were kept identical to those found for S. aureus N315 (Heinemann et al., 2005). At the time of model construction, the genome-scale model of B. subtilis (Oh et al., 2007) had not yet been published, and thus information from B. subtilis was not employed in our model. Modifications of peptidoglycan structures and amino acids of the interpeptide bridge have been observed as a result of environmental changes (Schleifer and Kandler, 1972), and large differences exist between the peptidoglycan structures of vegetative cells and spores (Atrih and Foster, 2001; Makino and Moriyama, 2002). However, a single description of crosslinked peptidoglycan (Cummins and Johnson, 1971; Schleifer and Kandler, 1972) (see Supplementary Appendix 1) was used for model development of C. acetobutylicum vegetative growth. In addition, a model of WTA from B. subtilis (Neuhaus and Baddiley, 2003; Perego et al., 1995) was used, in absence of specific literature data for C. acetobutylicum. As with LTA, the cellular process of d-alanylation of WTA was ignored for the C. acetobutylicum model.
The Urea Cycle to Complete the TCA Cycle in the Metabolic Network
The TCA cycle of C. acetobutylicum is incomplete, lacking the necessary enzymes to support the biochemical conversions of: (i) oxaloacetate to citrate; (ii) succinyl-CoA to succinate; and (iii) succinate to fumarate. It has been proposed that fumarate is synthesized from oxaloacetate (with malate as an intermediate) through a reverse TCA pathway involving malate dehydrogenase (EC 1.1.1.37, CAC0566) and fumarate dehydratase (FumA, EC 4.2.1.2, CAC3090, CAC3091) (Nolling et al., 2001). Calculation of the estimated standard Gibbs free energy of reaction () using published values (Henry et al., 2007) yielded a thermodynamically favorable production of malate from oxaloacetate (). It is noted that significantly different standard Gibbs free energy of formation values have been published recently (Alberty, 2004). With an intracellular NAD+/NADH ratio of 10, this reaction is favorable (for the production of malate) given a malate to oxaloacetate ratio of less than ∼300. If the NAD+/NADH ratio drops to 0.1, the reaction is favorable for malate to oxaloacetate ratios less than ∼3 × 104. Fumarate production from malate through FumA is less thermodynamically favorable (); however, this reaction has a negative ΔrG′ when the intracellular ratio of malate to fumarate is greater than ∼2.
With an incomplete TCA cycle, biosynthesis of the amino acid precursor 2-oxoglutarate remained unresolved in initial versions of the metabolic network but it was suggested to include a previously unresolved pathway involving the urea cycle (Nolling et al., 2001). Since growth was observable in a minimal medium containing no free amino acids or peptides (Monot et al., 1982), C. acetobutylicum must be capable of synthesizing all necessary l- and d-amino acids. To resolve this previously undefined pathway, the biosynthesis of 2-oxoglutarate without an l-glutamate precursor was explored through several possible mechanisms. One investigated pathway requires the conversion of l-ornithine to l-proline by ornithine cyclodeaminase (EC 4.3.1.12), an enzyme and metabolic reaction known to exist in S. aureus N315 (SAU0113). However, BLASTP results of this encoded protein sequence against that of the C. acetobutylicum genome returned a bit-score insufficient for orthology (Paredes et al., 2005; Pearson, 1996). Other investigated pathways require a 2-oxoglutarate to l-glutamate conversion as part of an intermediate reaction step. This requires a pre-existing l-glutamate solute pool for l-glutamate biosynthesis through these pathways. However, the investigation of known enzymatic mechanisms in available public databases and known pathways of similar organisms revealed an alternative pathway through enzyme homology. Although the genome annotation did not identify an ORF of C. acetobutylicum encoding a bacterial ornithine-oxo-acid transferase enzyme (RocD, EC 2.6.1.13), this enzyme was investigated using the BRENDA enzymatic database. Several researchers have identified the inclusion of 2-substituted oxo-acids (e.g., pyruvate, 2-oxoglutarate, oxaloacetate, 2-oxobutanoate, methylglycoxyl) as viable substrates in the conversion of l-ornithine to l-glutamate 5-semi-aldehyde as catalyzed by a Bacillus sp. YM-2-isolated ornithine-oxo-acid transferase (RocD, EC 2.6.1.13) (Jhee et al., 1995). Among microorganisms similar to C. acetobutylicum, both B. subtilis and S. aureus N315 are known to contain both RocD and ArgD (EC 2.6.1.11) enzymes. Both RocD and ArgD of B. subtilis (and S. aureus) were analyzed against the C. acetobutylicum genome using BLASTP. Bit-scores suggested orthology (Pearson, 1996) of both enzymes (of both organisms) with CAC2388. When further analyzed in the BRENDA enzyme database, multiple researchers have noted that acetylornithine transaminase (EC 2.6.1.11) may exhibit very similar enzymatic activity to ornithine-oxo-acid transferase (EC 2.6.1.13) (Billheimer et al., 1976; Friedrich et al., 1978; Voellmy and Leisinger, 1975). Because of the homology observed between the two enzymes and the availability of pyruvate through glycolysis (Desai et al., 1999; Papoutsakis and Meyer, 1985), we propose the following biochemical reaction catalyzed by the enzyme of CAC2388, which is currently annotated as an acetylornithine transaminase (ArgD, EC 2.6.1.11) in C. acetobutylicum:
| (3) |
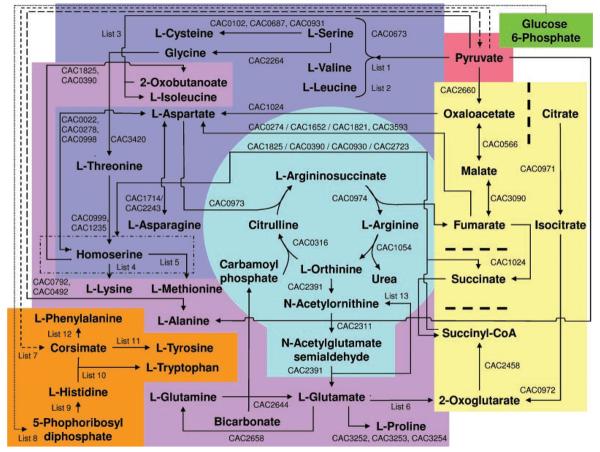
The estimated standard Gibbs free energy () of the reaction shown as Equation (3) was calculated as 2.19 kcal mol−1 using published values (Henry et al., 2007) of charged species at pH 7. Intracellular pyruvate levels were estimated as 5 mM based on E. coli data (Yang et al., 2001), and an intracellular concentration of l-alanine of 1.3 mM was assumed (from S. aureus data) (Heinemann et al., 2005). Intracellular l-ornithine levels were estimated at 10 mM, based on measurements in other bacteria (Poolman et al., 1987). Given these values, a ratio of intracellular concentration of l-ornithine to l-glutamate 5-semi-aldehyde greater than or equal to 10.6 is required for the reaction of Equation (3) to proceed in the forward direction as written. Finally, l-glutamate is synthesized by the metabolic network from l-glutamate 5-semi-aldehyde by γ-glutamyl-phosphate reductase (ProA, EC 1.2.1.41, CAC3254) and by glutamate 5-kinase (ProB, EC 2.7.2.11, CAC3253). Using this mechanism of l-glutamate biosynthesis, a proposed amino acid biosynthesis and the linking of the degenerate TCA cycle through the urea cycle are shown in Figure 4. The proposed pathway yields a net production of l-glutamate because l-ornithine does not require an l-glutamate precursor in C. acetobutylicum. Given the incomplete TCA cycle of C. acetobutylicum, the pathway of l-ornithine biosynthesis is as follows (also shown in Fig. 4): (i) pyruvate to oxaloacetate (PykA, EC6.4.1.1, CAC2660); (ii) oxaloacetate to l-aspartate (NadB, EC 1.4.3.16, CAC1024); (iii) l-asparate to l-argininosuccinate (ArgG, EC 6.3.4.5, CAC0973); (iv) l-argininocussinate to l-arginine (ArgH, EC 4.3.2.1, CAC0974); and (v) l-arginine to L-ornithine (EC 3.5.3.1, CAC1054). In absence of an l-glutamate intracellular solute pool, a citrulline pool is required of this pathway. Citrulline solute pools, up to 1 mM, have been measured in clostridia (Kleiner, 1979). Based on current annotation and available enzymatic studies, the pathway presented is the most probable mode of l-glutamate biosynthesis in absence of a complete TCA cycle and intracellular l-glutamate (or l-glutamine) solute pool.
Figure 4.
Reconstructed pathways of l-amino acids biosynthesis in C. acetobutylicum in view of the incomplete TCA cycle. The diagram assumes l-glutamate biosynthesis from pyruvate, rather than through ammonia assimilation, which requires an intracellular l-glutamate pool. The following pathways and sub-categories are highlighted within the broadly defined pathway of amino acids biosynthesis. The links to carbohydrate metabolism, pyruvate and glucose-6-phosphate (which are linked through glycolysis) are shown in red and green, respectively. The incomplete TCA cycle (light yellow) contains separators (broad dashed lines) to show locations of missing conventional TCA cycle enzymes. l-amino acids synthesized directly from pyruvate, oxaloacetate or malate precursors are shown in dark blue. Other TCA cycle l-amino acid precursors (2-oxoglutarate, succinyl-CoA and succinate) are connected to oxaloacetate, malate and fumarate through the urea cycle (shown in light blue). Shown in purple are l-amino acids, which are (i) synthesized from 2-oxoglutarate or l-glutamate, (ii) require the 2-oxoglutarate/l-glutamate interconversion during a processing step or (iii) require a succinyl-transferase during synthesis. Pathways leading to the biosynthesis of aromatic l-amino acids and l-histidine are shown in orange. ORFs of C. acetobutylicum encoding required enzymes of a particular pathway are given. Gene names separated by commas designate sequential processing steps. A slash (/) separator identifies genes encoding enzymes capable of parallel operation. Pathways involving a large number of genes are labeled in the figure as Lists. The corresponding genes for these pathways are given below. List 1: CAC3169/CAC3176/CAC3652, CAC0091, CAC3170/CAC3604, CAC1479, CAC2399. List 2: CAC3169/CAC3176/CAC3652, CAC0091, CAC3170/CAC2604, CAC0273, CAC3172/CAC3173, CAC3171, CAC1479. List 3: CAC3169/CAC3176/ CAC3652, CAC0091, CAC3170/CAC3604, CAC1479, CAC3038. List 4: CAC0998, CAC2378/CAC3600, CAC2379, CAC2381, CAC2380, CAC2723, CAC2624, CAC0608. List 5: CAC1825, CAC0390, CAC0391, CAC3348/CAC0578. List 6: CAC0737/CAC1001/CAC1819/CAC2832/CAC0764/CAC1673/CAC1674. List 7: (from DAHP) CAC0894, CAC0713/CAC0899, CAC0897, CAC0898, CAC0895, CAC0896. List 8: CAC2680, CAC0944, CAC1348, CAC1730, CAC0726, CAC0819/CAC3221. List 9: CAC0936, CAC0943, CAC0942, CAC0940, CAC0939, CAC0938, CAC1369, CAC2727, CAC0937. List 10: CAC3162/CAC3163, CAC3161, CAC3159, CAC3160, CAC3157/CAC3158. List 11: CAC1234, CAC0893, CAC1369/CAC3031/CAC1001/CAC1819/CAC2832. List 12: CAC1234, CAC0217, CAC1369/CAC3031/CAC1001/CAC1819/CAC2832. List 13 (also shown in Fig. 5): CAC2391/CAC3020, CAC2388, CAC2389, CAC2390.
l-glutamate Biosynthesis Originating From the Intracellular Solute Pool
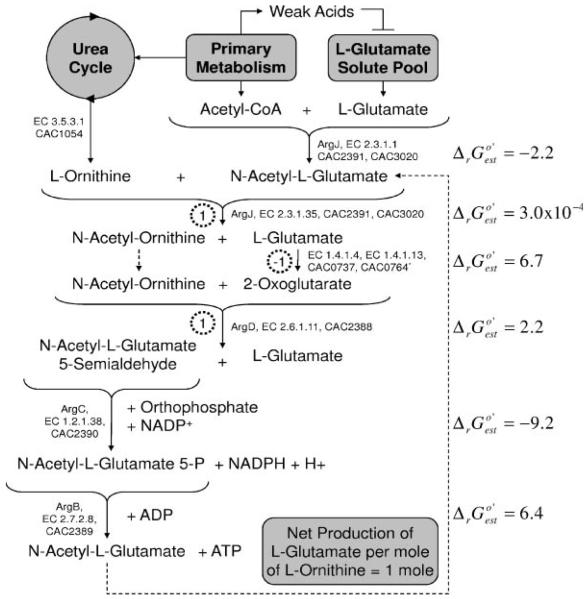
We have proposed a pathway (Eq. 3) for the biosynthesis of l-glutamate. However, in the presence of an l-glutamate solute pool, the arginine biosynthesis pathway (operating in reverse) may contribute to l-glutamate biosynthesis. Here, we further discuss this pathway, the thermodynamic feasibility of its operation in reverse, and the physiological characteristics (metabolic programs and environmental conditions) that have been observed to alter the size of the intracellular l-glutamate solute pool in other clostridia. The production of the l-glutamate biosynthesis precursor through ArgD (CAC2388) (Eq. 3) suggests that l-glutamate biosynthesis through this (single) enzyme may be a bottleneck of protein biosynthesis. The proposed biosynthesis of l-glutamate from l-ornithine with calculated values is illustrated in Figure 5. l-glutamate, from the intracellular solute pool, and acetyl-CoA, from the primary metabolism of glucose, are used by the glutamate N-acetyltransferase (ArgJ, EC 2.3.1.1, CAC2391, CAC3020) to produce N-acetyl-l-glutamate, which is then combined with l-ornithine (from the urea cycle) to form N-acetyl-ornithine and l-glutamate by ArgJ. l-glutamate is converted to 2-oxoglutarate by a host of enzymes in C. acetobutylicum; the glutamate dehydrogenase (EC 1.4.1.4, CAC0737) and the NADPH-dependent glutamate synthase (EC 1.4.1.13, CAC0764) are listed in Figure 5. The 2-oxoglutarate and N-acetyl-ornithine are converted to N-acetyl-l-glutamate 5-semi-aldehyde and l-glutamate by the N-acetylornithine aminotransferase (ArgD, EC 2.6.1.11, CAC2388). Further processing back to N-acetyl-l-glutamate occurs through the N-acetyl-γ-phosphate-reductase (ArgC, EC 1.2.1.38, CAC2390) and the acetylglutamate kinase (ArgB, EC 2.7.2.8, CAC2389). This cycle results in the net production of 1 mole of l-glutamate per mole of l-ornithine processed by this pathway. The net biochemical reaction (not charge-balanced) of the cycle, starting and ending with N-acetyl-l-glutamate, as shown in Figure 5, is given as Equation (4).
Figure 5.
Proposed pathway of l-glutamate biosynthesis in C. acetobutylicum given: (i) an intracellular l-glutamate solute pool and (ii) reversibility of the l-arginine biosynthesis pathway in C. acetobutylicum. The pathway produces 1 mol of l-glutamate per mole of l-ornithine processed. Moles of l-glutamate produced (positive number) or consumed (negative number) at each step are shown in dashed circles. The pathway loop (closed by dashed line) begins and terminates with N-acetyl-l-glutamate. The primary metabolism of C. acetobutylicum produces weak acids (notably acetate and butyrate) during exponential growth. Weak acids have been shown to inhibit intracellular l-glutamate solute pool accumulation in other clostridia (Flythe and Russell, 2006). Not all reaction details, including charge-balances, are shown here.
| (4) |
Also shown in Figure 5 is the observed inhibitory relationship between weak acids and the accumulation of an intracellular l-glutamate solute pool. Recent studies have suggested that the intracellular accumulation of weak acids at low extracellular pH disrupts the cellular anion balance and results in a largely diminished l-glutamate solute pool (Flythe and Russell, 2006; Roe et al., 1998). Although the governing mechanisms behind this observation remain unclear, intracellular l-glutamate pools approached ∼0 mM in the presence of 100 mM sodium acetate and 100 mM sodium lactate (pH 5.0) in C. sporogenes MD1 (Flythe and Russell, 2006). The effects of fermentation acids on the intracellular pools of TCA cycle component 2-oxoglutarate remain unknown. However, the glutamate dehydrogenase (EC 1.4.1.4, CAC0764) strongly favors the formation of l-glutamate from 2-oxoglutarate (), and intracellular measurement of 2-oxoglutarate, investigating its involvement in nitrogen sensing in various bacteria, yielded pool levels orders of magnitude lower than that of l-glutamate (Muller et al., 2006; Muro-Pastor et al., 2001). Thus, a metabolic reaction (Eq. 3) ultimately leading to the production of l-glutamate or 2-oxoglutarate without the involvement of the intracellular l-glutamate solute pools or direct transport into the cell was required of the genome-scale metabolic network for C. acetobutylicum. Other specific cases of specialized clostridial metabolic network-building, related to anaerobic pathways, are discussed in the following sections.
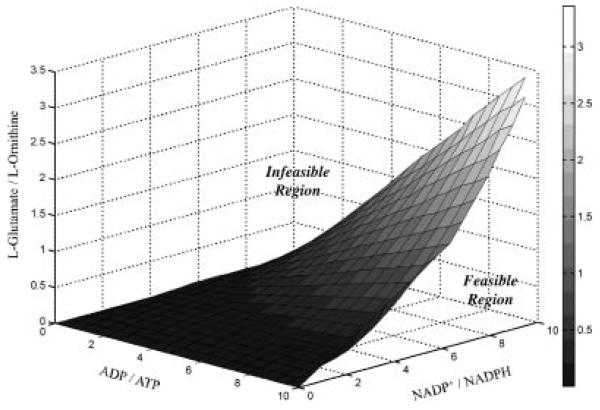
A thermodynamic analysis was performed to assess the feasibility of the proposed reverse function of the arginine biosynthesis pathway, shown in Figure 5. This was done by calculating the ΔrG′ for all reactions of the pathway. It is noted that for a pathway to be thermodynamically possible, all reactions of the pathway (not the net reaction shown by Eq. 4) must have negative ΔrG′ values. Given the large positive values of (shown in Fig. 5) for reactions catalyzed by glutamate dehydrogenase (EC 1.4.1.4, CAC0737) and acetylglutamate kinase (ArgB, EC 2.7.2.8, CAC2389), the proposed pathway requires significant concentration gradients to be thermodynamically feasible. The effect of the size of the l-glutamate solute pool is two-fold in this case. On one hand, the l-glutamate concentration must be large enough to provide the concentration gradient needed to overcome the large of the glutamate dehydrogenase, yet a smaller l-glutamate solute pool favors the reverse function of the N-acetylornithine aminotransferase (ArgD, EC 2.6.1.11, CAC2388), as shown in Figure 5. In addition, a thermodynamic analysis of this reaction pathway requires inputs of the intracellular nucleotide levels of ATP, ADP, NADPH, and NADP+; however, it has been shown extensively in C. acetobutylicum that these levels vary widely in response to substrate limitation and dominant metabolic programs (e.g., acid-ogenesis or solventogenesis) (Girbal and Soucaille, 1994; Grupe and Gottschalk, 1992; Meyer and Papoutsakis, 1989). Thus, the thermodynamic feasibility of the proposed reverse arginine biosynthesis pathway was studied as a function of the intracellular concentration ratios of ADP/ATP, NADP+/NADPH, and l-glutamate/l-ornithine. The following intracellular metabolite concentrations were used as constants in the analysis: 0.4 mM acetyl-CoA (Boynton et al., 1994); 0.1 mM CoA (Boynton et al., 1994); 1 mM ammonia (Schreier et al., 1982); and 10 mM orthophosphate (Heinemann et al., 2005). The concentration ratios mentioned above were adjusted to yield ΔrG′ values of zero for all reactions of the pathway (Fig. 5). These calculations produced a 3-dimensional plane (ΔrG′ = 0), shown as Figure 6. Above the plane of Figure 6, reverse operation of the arginine biosynthesis pathway is thermodynamically infeasible, and below the plane all ΔrG′ values of the pathway are negative. For example, given ADP/ATP and NADP+/NADPH ratios of 10 (respectively), the ratio of l-glutamate to l-ornithine intracellular solute pool levels must be less than 3 for l-glutamate production from the reverse arginine biosynthesis pathway to be possible. Even though the intracellular nucleotide levels have been widely studied, metabolomic data of the l-glutamate/l-ornithine ratio in C. acetobutylicum remain unknown; however, this analysis provides the necessary physiological conditions to allow the arginine biosynthesis pathway to contribute to the intracellular l-glutamate solute pool.
Figure 6.
Intracellular metabolite ratios resulting in reversibility (ΔrG′0) for all reactions of the reverse arginine biosynthesis pathway. The pathway is thermodynamically feasible given metabolite ratios falling below the plane shown above.
Other Resolved Pathways of Anaerobic Metabolism
Development of a genome-scale model for a strict anaerobe, such as C. acetobutylicum, from reaction network databases and enzyme homology yielded multiple aerobic reactions that were further resolved using the BRENDA database to locate anaerobic reactions catalyzed by available enzymes. The list of aerobic reactions assigned to the C. acetobutylicum genome in the KEGG database (as of August 2007) is presented in Supplementary Appendix 3. It is possible that many of the enzymes identified through homology searches that catalyze aerobic reactions also catalyze anaerobic reactions that remain uncharacterized. Two examples are: (i) the NAD biosynthesis pathway; and (ii) anaerobic biosynthesis of l-isoleucine.
Anaerobic NAD Biosynthesis
The quinolinate precursor of NAD is commonly synthesized in vivo from l-aspartate through an iminoaspartate intermediate by l-aspartate oxidase (NadB, EC 1.4.3.16, CAC1024) and quinolinate synthase (NadA, EC 3.2.2.5, CAC1025). Alternatively, quinolinate is synthesized from the metabolism of l-tryptophan. However, with current genome annotation of C. acetobutylicum, the pathway of possible l-tryptophan utilization, yielding quinolinate, is largely uncharacterized. This biochemical process requires, at minimum, five enzymes, and none have been identified in C. acetobutylicum through gene homology. Since a minimal medium (Monot et al., 1982), that contained no amino acids or peptides was used, the assumption was made that amino acids were synthesized in vivo for incorporation into protein and as precursors of other biological macromolecules. Thus, quinolinate biosynthesis from l-tryptophan was not considered a feasible pathway of biosynthesis in a minimal medium. Thus, a feasible pathway of NAD biosynthesis requires the conversion of l-aspartate to iminoaspartate by l-aspartate oxidase (NadB, EC 1.4.3.16, CAC1024) under anaerobic conditions. Incidentally, l-aspartate oxidase is also one of multiple catalysts for the conversion between l-aspartate and oxaloacetate. However, reaction mechanisms catalyzed by l-aspartate oxidase currently available in the KEGG database are aerobic. Through the BRENDA database and a further literature investigation, fumarate was identified as a possible electron acceptor for the conversion of l-aspartate to oxaloacetate catalyzed by l-aspartate oxidase under anaerobic conditions (Messner and Imlay, 2002; Tedeschi et al., 1996). Further, an l-asparate oxidase has been identified in an anaerobic hyperthermophilic bacterium and has been found to catalyze anaerobic l-aspartate dehydrogenation (Sakuraba et al., 2002). Thus, we propose the conversion of l-aspartate to iminoaspartate by l-asparate oxidase (NadB, EC 1.4.3.16, CAC1024) in the C. acetobutylicum metabolic network through the use of fumarate as a terminal electron acceptor, resulting in the production of succinate as well as iminoaspartate, as shown by Equation (5).
| (5) |
Anaerobic l-Isoleucine Biosynthesis
The biosynthesis pathway of l-isoleucine in C. acetobutylicum was found not to include l-threonine (Nolling et al., 2001). Homology analysis of the threonine dehydratase from B. subtilis (IlvA, EC 4.3.1.19, BG10673), which catalyzes the reaction of l-threonine to 2-oxobutanoate, yielded a low bit-score (Pearson, 1996) when compared to ORFs of C. acetobutylicum. The biosynthesis of 2-oxobutanoate through a 2-methylmaleate intermediate was investigated since this pathway was suggested for M. thermaautotrophicum (Eikmanns et al., 1983). However, a homology search of the B. subtilis l-serine dehydratase (SdaAA, SdaAB; EC 4.3.1.17; BG13397, BG13398) against proteins of the C. acetobutylicum genome using BLASTP returned low bit-scores as well. Finally, biosynthesis was traced from l-aspartate to homoserine to 2-oxobutanoate through homoserine-O-succinyl-transferase (MetB, EC 2.3.1.46, CAC1825) and cystathione-γ-synthase (EC 2.5.1.48, CAC0390). This metabolic route of l-isoleucine biosynthesis is inefficient as MetB requires succinyl-CoA as a substrate. This TCA cycle component is derived from 2-oxoglutarate at the bottom of the incomplete TCA cycle of C. acetobutylicum (Fig. 4).
Utilization of Succinate
In the current model, succinate is produced from succinyl-CoA in the biosynthesis of homoserine and from the anaerobic biosynthesis of NAD. However, a clear path for its degradation remains elusive due to the incomplete TCA cycle (Fig. 4). Utilization of succinate through the reverse reaction of Equation (5) is infeasible since iminoaspartate is consumed by NAD biosynthesis. Other possibilities for succinate assimilation exist: (i) it is transported out of the cell, (ii) it is converted back to succinyl-CoA by a CoA transferase, or (iii) it is processed to butyric acid through a crotonyl-CoA intermediate by a pathway similar to that observed for C. kluyveri (Sohling and Gottschalk, 1996). The conversion of succinate to succinyl-CoA was chosen for the genome-scale model for the following reasons: (i) the primary metabolism of C. acetobutylicum is well-established and does not support butyric acid production from succinate, (ii) succinate is not a byproduct commonly found in C. acetobutylicum fermentation broths, (iii) several CoA transferase enzymes exist in the genome (see Supplementary Appendix 1), and (iv) a purified CoA transferase from C. acetobutylicum demonstrated reversibility and specificity for multiple substrates in previous kinetic studies (Wiesenborn et al., 1989). Therefore, we realize that the proposed pathway of succinate assimilation to succinyl-CoA is an approximation based on the best available data at this time.
Identification of Growth-Arresting Knock-Outs In Silico
The reconstructed metabolic network for C. acetobutylicum was used with FBA and systematic gene knock-outs to identify those enzymes (and their encoding genes) that will prevent growth when knocked-out in silico. One goal of this computational study is to identify gene knock-outs that arrest growth but do not disrupt the primary metabolism of C. acetobutylicum. Cells were grown in silico on three different media in this study, given the developed genome-scale model for C. acetobutylicum: (i) the minimal medium extracellular environment (Monot et al., 1982), (ii) minimal medium supplemented with l-glutamine, l-asparagine, l-histidine and l-cysteine (called partially supplemented medium), and (iii) minimal medium supplemented with all l-amino acids as well as d-ribose and glycerol 3-phosphate (called supplemented medium). It is noted that the energetics and metabolic capacities of these in silico knock-out strains were not probed in depth. Only the ability of the altered metabolic network to produce biomass in silico was investigated, so the underlying membrane transport mechanisms of supplemented media nutrients and details of resulting metabolic capacity were ignored for these simulations. Reactions resulting in arrested growth in silico of C. acetobutylicum for each medium are included in Supplementary Appendix 1. Table III contains a summary of the number of reactions arresting growth in silico, broken-down into broadly defined metabolic pathways. In particular, in the absence of an extracellular source of amino acids (minimal medium), the pathways of amino acids biosynthesis (e.g., aromatic amino acids biosynthesis) contained a large number of reactions that arrested growth in silico when knocked-out. In the presence of supplemented media, predictably, these pathways did not arrest growth in silico when knocked-out. However, four reactions in amino acids metabolism did arrest growth in this medium following in silico knock-outs. These particular enzymes are responsible for processing amino acids into precursors of other pathways. One member of this group is the d-alanine-d-alanine ligase (ddlA, EC 6.3.2.4, CAC2895) that produces d-alanyl-d-alanine, which is vital to peptidoglycan biosynthesis. Conversely, in the presence of supplemented media, the large numbers of related reactions leading to arrested growth in silico were in the biosynthesis of steroids, riboflavin, purine and glycerolipids.
Conclusions
Semi-automated reverse engineering of a genome-scale reaction network using building-block transfer equations was developed and coupled with iterative measures of network-building through database and literature mining resulting in the first genome-scale reaction network for C. acetobutylicum. This is the first genome-scale model for any of the clostridia. Thus, several examples of the use of reaction and enzyme databases to characterize anaerobic reactions catalyzed by pathways for several well-known enzymes were presented. In addition, the function of the incomplete TCA cycle, through incorporation of the urea cycle, was resolved in detail based on homology searches and metabolic demands of the genome-scale reaction network. This led to two possible mechanisms of l-glutamate biosynthesis that are influenced by the presence of weak acids and an intracellular l-glutamate solute pool. Thermodynamic analyses determined the physiological conditions under which the proposed pathways are capable of producing l-glutamate. Given the previously observed concentration ratios of ADP/ATP and NAD+/NADH, the levels of the intracellular l-glutamate solute pool (which is also subject to fluctuation) has a large impact on the thermodynamic feasibility of many metabolic pathways in C. acetobutylicum. Our model successfully predicted acidogenesis and solventogenesis of the wild-type strain, the loss of butyric acid production in the buk knock-out, and the loss of butanol and acetone production by the M5 strain. However, due to the lack of regulation, the genome-scale model cannot yet describe the metabolic events of acido-genesis, acid re-uptake, and solventogenesis as a cascading sequence of events governed by the activation of sigma factors.
Supplementary Material
Table III.
Number of reactions preventing growth when knocked-out of reconstructed metabolic network.
| Pathway | Minimal mediuma | Partially supplemented mediumb | Supplemented mediumc |
|---|---|---|---|
| Carbohydrate metabolism | 25 | 10 | 3 |
| Energy metabolism | 7 | 0 | 0 |
| Lipid metabolism | 29 | 27 | 27 |
| Nucleotide metabolism | 22 | 15 | 9 |
| Amino acids metabolism | 71 | 42 | 4 |
| Metabolism of cofactors and vitamins | 41 | 36 | 32 |
| Biomass and maintenance | 12 | 10 | 10 |
| Total number of reactions | 207 | 140 | 85 |
Results are reported in groups of biosynthetic pathways or programs.
Minimal medium for C. acetobutylicum has been published (Monot et al., 1982).
Partially supplemented medium consists of minimal medium plus l-glutamine, l-asparagine, l-histidine, and l-cysteine.
Supplemented medium consists of minimal medium plus all l-amino acids, d-ribose and glycerol 3-phosphate.
Acknowledgments
This work was supported by NSF grant BES-0418157. R.S.S. was supported by NIH NRSA post-doctoral training grant F32GM078947.
Contract grant sponsor: NIH
Contract grant number: GM078947
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Alberty RA. Levels of thermodynamic treatment of biochemical reaction systems. Biophys J. 1993;65(3):1243–1254. doi: 10.1016/S0006-3495(93)81146-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberty RA. Biochemical thermodynamics. Biochim Biophys Acta. 1994;1207(1):1–11. doi: 10.1016/0167-4838(94)90045-0. [DOI] [PubMed] [Google Scholar]
- Alberty RA. Thermodynamics of systems of biochemical reactions. J Theor Biol. 2002;215(4):491–501. doi: 10.1006/jtbi.2001.2516. [DOI] [PubMed] [Google Scholar]
- Alberty RA. Equilibrium concentrations for pyruvate dehydrogenase and the citric acid cycle at specified concentrations of certain coenzymes. Biophys Chem. 2004;109(1):73–84. doi: 10.1016/j.bpc.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Atrih A, Foster SJ. Analysis of the role of bacterial endospore cortex structure in resistance properties and demonstration of its conservation amongst species. J Appl Microbiol. 2001;91(2):364–372. doi: 10.1046/j.1365-2672.2001.01394.x. [DOI] [PubMed] [Google Scholar]
- Baer SH, Blaschek HP, Smith TL. Effect of butanol challenge and temperature on lipid composition and membrane fluidity of butanol-tolerant Clostridium acetobutylicum. Appl Environ Microbiol. 1987;53(12):2854–2861. doi: 10.1128/aem.53.12.2854-2861.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28(1):304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker SA, Palsson BO. Genome-scale reconstruction of the metabolic network in Staphylococcus aureus N315: An initial draft to the two-dimensional annotation. BMC Microbiol. 2005;5(1):8. doi: 10.1186/1471-2180-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billheimer JT, Carnevale HN, Leisinger T, Eckhardt T, Jones EE. Ornithine delta-transaminase activity in Escherichia coli—Identity with acetylornithine delta-transaminase. J Bacteriol. 1976;127(3):1315–1323. doi: 10.1128/jb.127.3.1315-1323.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277(5331):1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Boynton ZL, Bennett GN, Rudolph FB. Intracellular concentrations of coenzyme A and its derivatives from Clostridium acetobutylicum ATCC 824 and their roles in enzyme regulation. Appl Environ Microbiol. 1994;60(1):39–44. doi: 10.1128/aem.60.1.39-44.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W, Saier MH., Jr The transporter classification (TC) system, 2002. Crit Rev Biochem Mol Biol. 2002;37(5):287–337. doi: 10.1080/10409230290771528. [DOI] [PubMed] [Google Scholar]
- Cakir T, Patil KR, Onsan Z, Ulgen KO, Kirdar B, Nielsen J. Integration of metabolome data with metabolic networks reveals reporter reactions. Mol Syst Biol. 2006;2:50. doi: 10.1038/msb4100085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Foerster H, Fulcher CA, Hopkinson R, Ingraham J, Kaipa P, Krummenacker M, Paley S, Pick J, Rhee SY, et al. MetaCyc: A multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2006;34(Database issue):D511–D516. doi: 10.1093/nar/gkj128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert MW, Schilling CH, Palsson B. Regulation of gene expression in flux balance models of metabolism. J Theor Biol. 2001;213(1):73–88. doi: 10.1006/jtbi.2001.2405. [DOI] [PubMed] [Google Scholar]
- Cummins CS, Johnson JL. Taxonomy of clostridia—Wall composition and DNA homologies in Clostridium butyricum and other butyric acid-producing clostridia. J Gen Microbiol. 1971;67:33–46. [Google Scholar]
- Demain AL, Newcomb M, Wu JH. Cellulase, clostridia, and ethanol. Microbiol Mol Biol Rev. 2005;69(1):124–154. doi: 10.1128/MMBR.69.1.124-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai RP, Nielsen LK, Papoutsakis ET. Stoichiometric modeling of Clostridium acetobutylicum fermentations with non-linear constraints. J Biotechnol. 1999;71(1–3):191–205. doi: 10.1016/s0168-1656(99)00022-x. [DOI] [PubMed] [Google Scholar]
- Edwards ES, Ramakrishna R, Schilling CH, Palsson BO. Metabolic flux analysis. In: Lee SY, Papoutsakis ET, editors. Metabolic engineering. Marcel Dekker; New York: 1999. pp. 13–57. [Google Scholar]
- Edwards JS, Ibarra RU, Palsson BO. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat Biotechnol. 2001;19(2):125–130. doi: 10.1038/84379. [DOI] [PubMed] [Google Scholar]
- Eikmanns B, Linder D, Thauer RK. Unusual pathway of isoleucine biosynthesis in Methanobacterium thermoautotrophicum. Arch Micro-biol. 1983;136(2):111–113. [Google Scholar]
- Feist AM, Scholten JC, Palsson BO, Brockman FJ, Ideker T. Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol Syst Biol. 2006;2:0004. doi: 10.1038/msb4100046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flythe MD, Russell JB. Fermentation acids inhibit amino acid deamination by Clostridium sporogenes MD1 via a mechanism involving a decline in intracellular glutamate rather than protonmotive force. Microbiology. 2006;152(Pt 9):2619–2624. doi: 10.1099/mic.0.29006-0. [DOI] [PubMed] [Google Scholar]
- Forster J, Famili I, Fu P, Palsson BO, Nielsen J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 2003;13(2):244–253. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francke C, Siezen RJ, Teusink B. Reconstructing the metabolic network of a bacterium from its genome. Trends Microbiol. 2005;13(11):50–558. doi: 10.1016/j.tim.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Friedrich B, Friedrich CG, Magasanik B. Catabolic N2-acetylornithine 5-aminotransferase of Klebsiella aerogenes—Control of synthesis by induction, catabolite repression, and activation by glutamine synthetase. J Bacteriol. 1978;133(2):686–691. doi: 10.1128/jb.133.2.686-691.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianchandani EP, Papin JA, Price ND, Joyce AR, Palsson BO. Matrix formalism to describe functional states of transcriptional regulatory systems. PLoS Comput Biol. 2006;2(8):e101. doi: 10.1371/journal.pcbi.0020101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girbal L, Soucaille P. Regulation of Clostridium acetobutylicum metabolism as revealed by mixed-substrate steady-state continuous cultures: Role of NADH/NAD ratio and ATP pool. J Bacteriol. 1994;176(21):6433–6438. doi: 10.1128/jb.176.21.6433-6438.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EM, Bennett GN. Genetic manipulation of acid and solvent formation in Clostridium acetobutylicum ATCC 824. Biotechnol Bioeng. 1998;58(2–3):215–221. doi: 10.1002/(sici)1097-0290(19980420)58:2/3<215::aid-bit14>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Grupe H, Gottschalk G. Physiological events in Clostridium acetobutylicum during the shift from acidogenesis to solventogenesis in continuous culture and presentation of a model for shift induction. Appl Environ Microbiol. 1992;58(12):3896–3902. doi: 10.1128/aem.58.12.3896-3902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LM, Desai RP, Welker NE, Papoutsakis ET. Characterization of recombinant strains of the Clostridium acetobutylicum butyrate kinase inactivation mutant: Need for new phenomenological models for solventogenesis and butanol inhibition? Biotechnol Bioeng. 2000;67(1):1–11. [PubMed] [Google Scholar]
- Heinemann M, Kummel A, Ruinatscha R, Panke S. In silico genome-scale reconstruction and validation of the Staphylococcus aureus metabolic network. Biotechnol Bioeng. 2005;92(7):850–864. doi: 10.1002/bit.20663. [DOI] [PubMed] [Google Scholar]
- Henry CS, Jankowski MD, Broadbelt LJ, Hatzimanikatis V. Genome-scale thermodynamic analysis of Escherichia coli metabolism. Biophys J. 2006;90(4):1453–1461. doi: 10.1529/biophysj.105.071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry CS, Broadbelt LJ, Hatzimanikatis V. Thermodynamics-based metabolic flux analysis. Biophys J. 2007;92(5):1792–1805. doi: 10.1529/biophysj.106.093138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husemann MHW, Papoutsakis ET. Solventogenesis in Clostridium acetobutylicum fermentations related to carboxylic acid and proton concentrations. Biotechnol Bioeng. 1988;32(7):843–852. doi: 10.1002/bit.260320702. [DOI] [PubMed] [Google Scholar]
- Jhee KH, Yoshimura T, Esaki N, Yonaha K, Soda K. Thermostable ornithine aminotransferase from Bacillus sp YM-2—Purification and characterization. J Biochem. 1995;118(1):101–108. doi: 10.1093/oxfordjournals.jbchem.a124863. [DOI] [PubMed] [Google Scholar]
- Johnston NC, Goldfine H. Replacement of the aliphatic chains of Clostridium acetobutylicum by exogenous fatty acids: Regulation of phospholipid and glycolipid composition. J Bacteriol. 1992;174(6):1848–1853. doi: 10.1128/jb.174.6.1848-1853.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Woods DR. Acetone-butanol fermentation revisited. Microbiol Rev. 1986;50(4):484–524. doi: 10.1128/mr.50.4.484-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30(1):42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko P, Chen L, Freund Y, Vitkup D, Church GM. Identifying metabolic enzymes with multiple types of association evidence. BMC Bioinformatics. 2006;7:177. doi: 10.1186/1471-2105-7-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriukhin MY, Neuhaus FC. D-alanylation of lipoteichoic acid: Role of the D-alanyl carrier protein in acylation. J Bacteriol. 2001;183(6):2051–2058. doi: 10.1128/JB.183.6.2051-2058.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner D. Regulation of ammonium uptake and metabolism by nitrogen-fixing bacteria. 3. Clostridium pasteurianum. Arch Microbiol. 1979;120(3):263–270. [Google Scholar]
- Knorr AL, Jain R, Srivastava R. Bayesian-based selection of metabolic objective functions. Bioinformatics. 2007;23(3):351–357. doi: 10.1093/bioinformatics/btl619. [DOI] [PubMed] [Google Scholar]
- Kumar VS, Dasika MS, Maranas CD. Optimization based automated curation of metabolic reconstructions. BMC Bioinformatics. 2007;8(1):212. doi: 10.1186/1471-2105-8-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummel A, Panke S, Heinemann M. Systematic assignment of thermodynamic constraints in metabolic network models. BMC Bioinformatics. 2006;7:512. doi: 10.1186/1471-2105-7-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390(6657):249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357(9264):1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- Lepage C, Fayolle F, Hermann M, Vandercasteele J-P. Changes in membrane lipid composition of Clostridium acetobutylicum during acetone-butanol fermentation: Effects of solvents, growth temperature and pH. J Gen Microbiol. 1987;133(1):103–110. [Google Scholar]
- MacCarthy T, Pomiankowski A, Seymour R. Using large-scale perturbations in gene network reconstruction. BMC Bioinformatics. 2005;6:11. doi: 10.1186/1471-2105-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald DL, Goldfine H. Effects of solvents and alcohols on the polar lipid composition of Clostridium butyricum under conditions of controlled lipid chain composition. Appl Environ Microbiol. 1991;57(12):3517–3521. doi: 10.1128/aem.57.12.3517-3521.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Moriyama R. Hydrolysis of cortex peptidoglycan during bacterial spore germination. Med Sci Monit. 2002;8(6):RA119–RA127. [PubMed] [Google Scholar]
- Maltsev N, Glass E, Sulakhe D, Rodriguez A, Syed MH, Bompada T, Zhang Y, D'Souza M. PUMA2—Grid-based high-throughput analysis of genomes and metabolic pathways. Nucleic Acids Res. 2006;34(Database issue):D369–D372. doi: 10.1093/nar/gkj095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskow T, von Stockar U. How reliable are thermodynamic feasibility statements of biochemical pathways? Biotechnol Bioeng. 2005;92(2):223–230. doi: 10.1002/bit.20572. [DOI] [PubMed] [Google Scholar]
- Mavrovouniotis ML. Group contributions for estimating standard Gibbs energies of formation of biochemical-compounds in aqueous-solution. Biotechnol Bioeng. 1990;36(10):1070–1082. doi: 10.1002/bit.260361013. [DOI] [PubMed] [Google Scholar]
- Messner KR, Imlay JA. Mechanism of superoxide and hydrogen peroxide formation by fumarate reductase, succinate dehydrogenase, and aspartate oxidase. J Biol Chem. 2002;277(45):42563–42571. doi: 10.1074/jbc.M204958200. [DOI] [PubMed] [Google Scholar]
- Meyer CL, Papoutsakis ET. Increased levels of ATP and NADH are associated with increased solvent production in continuous cultures of Clostridium acetobutylicum. Appl Environ Microbiol. 1989;30(5):450–459. [Google Scholar]
- Monot F, Martin JR, Petitdemange H, Gay R. Acetone and butanol production by Clostridium acetobutylicum in a synthetic medium. Appl Environ Microbiol. 1982;44(6):1318–1324. doi: 10.1128/aem.44.6.1318-1324.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya D, Arevalo C, Gonzales S, Aristizabal F, Schwarz WH. New solvent-producing Clostridium sp. strains, hydrolyzing a wide range of polysaccharides, are closely related to Clostridium butyricum. J Ind Microbiol Biotechnol. 2001;27(5):329–335. doi: 10.1038/sj.jim.7000193. [DOI] [PubMed] [Google Scholar]
- Muller T, Strosser J, Buchinger S, Nolden L, Wirtz A, Kramer R, Burkovski A. Mutation-induced metabolite pool alterations in Corynebacterium glutamicum: Towards the identification of nitrogen control signals. J Biotechnol. 2006;126(4):440–453. doi: 10.1016/j.jbiotec.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Muro-Pastor MI, Reyes JC, Florencio FJ. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J Biol Chem. 2001;276(41):38320–38328. doi: 10.1074/jbc.M105297200. [DOI] [PubMed] [Google Scholar]
- Neuhaus FC, Baddiley J. A continuum of anionic charge: Structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67(4):686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev EV, Burgard AP, Maranas CD. Elucidation and structural analysis of conserved pools for genome-scale metabolic reconstructions. Biophys J. 2005;88(1):37–49. doi: 10.1529/biophysj.104.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolling J, Breton G, Omelchenko MV, Makarova KS, Zeng Q, Gibson R, Lee HM, Dubois J, Qiu D, Hitti J, Wolf YI, Tatusov RL, Sabathe F, Doucette-Stamm L, Soucaille P, Daly MJ, Bennett GN, Koonin EV, Smith DR. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J Bacteriol. 2001;183(16):4823–4838. doi: 10.1128/JB.183.16.4823-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notebaart RA, van Enckevort FH, Francke C, Siezen RJ, Teusink B. Accelerating the reconstruction of genome-scale metabolic networks. BMC Bioinformatics. 2006;7:296. doi: 10.1186/1471-2105-7-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh YK, Palsson BO, Park SM, Schilling CH, Mahadevan R. Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J Biol Chem. 2007;282(39):28791–28797. doi: 10.1074/jbc.M703759200. [DOI] [PubMed] [Google Scholar]
- Osterman A, Overbeek R. Missing genes in metabolic pathways: A comparative genomics approach. Curr Opin Chem Biol. 2003;7(2):238–251. doi: 10.1016/s1367-5931(03)00027-9. [DOI] [PubMed] [Google Scholar]
- Papoutsakis ET. Equations and calculations for fermentations of butyric-acid bacteria. Biotechnol Bioeng. 1984;26(2):174–187. doi: 10.1002/bit.260260210. [DOI] [PubMed] [Google Scholar]
- Papoutsakis ET, Meyer CL. Equations and calculations of product yields and preferred pathways for butanediol and mixed-acid fermentations. Biotechnol Bioeng. 1985;27(1):50–66. doi: 10.1002/bit.260270108. [DOI] [PubMed] [Google Scholar]
- Paredes CJ, Alsaker KV, Papoutsakis ET. A comparative genomic view of clostridial sporulation and physiology. Nat Rev Microbiol. 2005;3(12):969–978. doi: 10.1038/nrmicro1288. [DOI] [PubMed] [Google Scholar]
- Pearson WR. Effective protein sequence comparison. Methods Enzymol. 1996;266:227–258. doi: 10.1016/s0076-6879(96)66017-0. [DOI] [PubMed] [Google Scholar]
- Perego M, Glaser P, Minutello A, Strauch MA, Leopold K, Fischer W. Incorporation of D-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J Biol Chem. 1995;270(26):15598–15606. doi: 10.1074/jbc.270.26.15598. [DOI] [PubMed] [Google Scholar]
- Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O. The comprehensive microbial resource. Nucleic Acids Res. 2001;29(1):123–125. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolman B, Driessen AJ, Konings WN. Regulation of arginine-ornithine exchange and the arginine deiminase pathway in Streptococcus lactis. J Bacteriol. 1987;169(12):5597–5604. doi: 10.1128/jb.169.12.5597-5604.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JL, Vo TD, Schilling CH, Palsson BO. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR) Genome Biol. 2003;4(9):R54. doi: 10.1186/gb-2003-4-9-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JL, Patel TR, Chen KH, Joyce AR, Applebee MK, Herring CD, Bui OT, Knight EM, Fong SS, Palsson BO. Systems approach to refining genome annotation. Proc Natl Acad Sci USA. 2006;103(46):17480–17484. doi: 10.1073/pnas.0603364103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q, Chen K, Paulsen IT. TransportDB: A comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 2007;35(Database issue):D274–D279. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe AJ, McLaggan D, Davidson I, O'Byrne C, Booth IR. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J Bacteriol. 1998;180(4):767–772. doi: 10.1128/jb.180.4.767-772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Jr, Tran CV, Barabote RD. TCDB: The Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34(Database issue):D181–D186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba H, Satomura T, Kawakami R, Yamamoto S, Kawarabayasi Y, Kikuchi H, Ohshima T. L-aspartate oxidase is present in the anaerobic hyperthermophilic archaeon Pyrococcus horikoshii OT-3: Characteristics and role in the de novo biosynthesis of nicotinamide adenine dinucleotide proposed by genome sequencing. Extremophiles. 2002;6(4):275–281. doi: 10.1007/s00792-001-0254-3. [DOI] [PubMed] [Google Scholar]
- Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36(4):407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg I, Chang A, Ebeling C, Gremse M, Heldt C, Huhn G, Schomburg D. BRENDA, the enzyme database: Updates and major new developments. Nucleic Acids Res. 2004;32(Database issue):D431–D433. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier HJ, Smith TM, Bernlohr RW. Regulation of nitrogen catabolic enzymes in Bacillus spp. J Bacteriol. 1982;151(2):971–975. doi: 10.1128/jb.151.2.971-975.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz WH. The cellulosome and cellulose degradation by anaerobic bacteria. Appl Microbiol Biotechnol. 2001;56(5–6):634–649. doi: 10.1007/s002530100710. [DOI] [PubMed] [Google Scholar]
- Sohling B, Gottschalk G. Molecular analysis of the anaerobic succinate degradation pathway in Clostridium kluyveri. J Bacteriol. 1996;178(3):871–880. doi: 10.1128/jb.178.3.871-880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephanopoulos G, Aristidou AA, Nielsen J. Metabolic engineering. Principles and metholologies. Academic Press; San Diego: 1998. [Google Scholar]
- Tedeschi G, Negri A, Mortarino M, Ceciliani F, Simonic T, Faotto L, Ronchi S. L-aspartate oxidase from Escherichia coli. II. Interaction with C4 dicarboxylic acids and identification of a novel L-aspartate: Fumarate oxidoreductase activity. Eur J Biochem. 1996;239(2):427–433. doi: 10.1111/j.1432-1033.1996.0427u.x. [DOI] [PubMed] [Google Scholar]
- Tegner J, Yeung MK, Hasty J, Collins JJ. Reverse engineering gene networks: Integrating genetic perturbations with dynamical modeling. Proc Natl Acad Sci USA. 2003;100(10):5944–5949. doi: 10.1073/pnas.0933416100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Mehrotra S, Papoutsakis ET, Hatzimanikatis V. A model-based optimization framework for the inference on gene regulatory networks from DNA array data. Bioinformatics. 2004;20(17):3221–3235. doi: 10.1093/bioinformatics/bth389. [DOI] [PubMed] [Google Scholar]
- Thomas R, Paredes CJ, Mehrotra S, Hatzimanikatis V, Papoutsakis ET. A model-based optimization framework for the inference of regulatory interactions using time-course DNA microarray expression data. BMC Bioinformatics. 2007;8(1):228. doi: 10.1186/1471-2105-8-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormann K, Feustel L, Lorenz K, Nakotte S, Durre P. Control of butanol formation in Clostridium acetobutylicum by transcriptional activation. J Bacteriol. 2002;184(7):1966–1973. doi: 10.1128/JB.184.7.1966-1973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas CA, Alsaker KV, Bonarius HP, Hendriksen WT, Yang H, Beamish JA, Paredes CJ, Papoutsakis ET. DNA array-based transcriptional analysis of asporogenous, nonsolventogenic Clostridium acetobutylicum strains SKO1 and M5. J Bacteriol. 2003;185(15):4539–4547. doi: 10.1128/JB.185.15.4539-4547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos I, Girbal L, Soucaille P. Regulation of carbon and electron flow in Clostridium acetobutylicum grown in chemostat culture at neutral pH on mixtures of glucose and glycerol. J Bacteriol. 1994;176(5):1443–1450. doi: 10.1128/jb.176.5.1443-1450.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voellmy R, Leisinger T. Dual role for N2-acetylornithine 5-aminotransferase from Pseudomonas aeruginosa in arginine biosynthesis and arginine catabolism. J Bacteriol. 1975;122(3):799–809. doi: 10.1128/jb.122.3.799-809.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollherbst-Schneck K, Sands JA, Montenecourt BS. Effect of butanol on lipid composition and fluidity of Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1984;47(1):193–194. doi: 10.1128/aem.47.1.193-194.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenborn DP, Rudolph FB, Papoutsakis ET. Coenzyme A transferase from Clostridium acetobutylicum ATCC 824 and its role in the uptake of acids. Appl Environ Microbiol. 1989;55(2):323–329. doi: 10.1128/aem.55.2.323-329.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YT, Bennett GN, San KY. The effects of feed and intracellular pyruvate levels on the redistribution of metabolic fluxes in Escherichia coli. Metab Eng. 2001;3(2):115–123. doi: 10.1006/mben.2000.0166. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hindorff LA, Chuang A, Monroe-Augustus M, Lyristis M, Harrison ML, Rudolph FB, Bennett GN. Expression of a cloned cyclopropane fatty acid synthase gene reduces solvent formation in Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 2003;69(5):2831–2841. doi: 10.1128/AEM.69.5.2831-2841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Tomas CA, Rudolph FB, Papoutsakis ET, Bennett GN. Intracellular butyryl phosphate and acetyl phosphate concentrations in Clostridium acetobutylicum and their implications for solvent formation. Appl Environ Microbiol. 2005;71(1):530–537. doi: 10.1128/AEM.71.1.530-537.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.