Abstract
OBJECTIVE
To study the relationship between HIV-1 subtype C genetic diversity and mother-to-child transmission and to determine if transmission of HIV-1C V1/V2 env variants occurs stochastically.
DESIGN
Case-case-control study of Malawian mother-infant pairs consisting of 32 non-transmitting women, 25 intrauterine (IU) transmitters and 23 intrapartum (IP) transmitters in Blantyre, Malawi.
METHODS
A heteroduplex tracking assay against the highly variable HIV env V1/V2 region was used to characterize the relationship between HIV diversity and HIV-1 MTCT. The relative abundance of the maternal env variants was quantified, and based on the env variants detected in the infant plasma, categorized as transmitted or untransmitted. The V1/V2 region was sequenced from two mother-infant pairs and a phylogenetic tree was built.
RESULTS
No relationship was found between transmission and overall maternal env diversity. Infants had less diverse HIV-1 populations than their mothers, and IU-infected infants had fewer V1/V2 variants and were more likely to harbor a homogeneous V1/V2 population than infants infected IP. V1/V2 sequences cloned from two mother-infant transmission pairs support multiple env variant transmission when multiple variants are detected, rather than single variant transmission followed by diversification. Almost 50% of the HIV-infected infants contained V1/V2 env variants that were not detected in maternal plasma samples, and, transmission of env variants was not related to their abundance in maternal blood.
CONCLUSIONS
These data suggest that the predominant mechanism(s) of HIV-1 subtype C MTCT differs by the timing of transmission and is unlikely to be explained by a simple stochastic model.
INTRODUCTION
HIV-1 subtype C is the most prevalent subtype worldwide, and it is the dominant subtype in Sub-Saharan Africa, where one-half of all infected women and children live [1,2]. Approximately 30% of infants born to untreated HIV-positive women will become infected with the virus, of whom approximately 20% will become infected in utero, 50% intrapartum, and the remaining 30% through breastmilk [3]. Little is known about the mechanism of transmission among these distinct groups, but several maternal characteristics are associated with increased rates of MTCT, including high viral RNA load, advanced disease status, and low CD4+ T-cell count [4].
One consistent feature of vertical HIV-1 transmission is a viral genetic bottleneck from mother to infant [5–8], whereby the genetic diversity of HIV-1 in the maternal viral population is greater than that in their infected infants. The bottleneck has been attributed to various factors involving selection, including some that are virus-specific [7] while others the result of donor/recipient immune responses [9,10]. In contrast to selective mechanisms, vertical transmission could also be a stochastic event, dependent solely on the donor’s viral burden with chance favoring the most abundant maternal variant for transmission, as suggested by at least one study [11].
Most previous studies of HIV-1 MTCT have involved small numbers of mother-infant pairs; in this report, we used a heteroduplex tracking assay (HTA) and phylogenetic analyses to study the viral diversity of the HIV-1 subtype C in 25 IU mother-infant pairs (MIPs), 23 IP MIPs, and 32 nontransmitting HIV-1-positive mothers. In addition, we used a mathematical simulation in an attempt to fit our data to a stochastic model of transmission.
MATERIALS AND METHODS
Study Participants
The HIV-1 seropositive pregnant women and their infants included in this study were participants in the Malaria and HIV-1 in Pregnancy (MHP) prospective cohort [12–14]. This study was approved by both the Malawi College of Medicine Research Committee and the UNC IRB. Informed consent was obtained from the participants.
Plasma was isolated from maternal blood collected at labor-ward admission, from the umbilical cord at delivery, and from infant heel-sticks at three time-points: within 48 hours of birth, 6-weeks, and 12-weeks of age. Women and their newborn infants received single-dose nevirapine according to the HIVNET 012 protocol [15]. The timing of HIV-1 transmission was categorized according to the definitions of Bryson et al. [16], as follows: infants HIV-1 DNA negative by real-time PCR [17] at 0 and 6 weeks had their mothers defined as non-transmitters (NT); infants HIV-1 DNA positive at birth were defined as in utero (IU) infections; and infants HIV-1 DNA negative at birth but DNA positive at 6 weeks were defined as intrapartum infections (IP). Owing to the timing of the infant blood sampling, it is likely that the IP definition cannot resolve late IU and early breast-feeding transmissions from true IP-infections. Four infants who were HIV-1 DNA negative but HIV-1 RNA positive by reverse-transcriptase polymerase chain reaction (RT-PCR) at birth were classified as IU.
As reported elsewhere, there were 65 infants infected IU, 65 infants infected IP (not including those infected post-partum), and 418 infants HIV-free at 12 weeks[13]. Samples were chosen from these participants based on availability, and after RT-PCR, a total of 32/418 NT, 25/65 IU, and 23/65 IP samples were included in the dataset. Samples were excluded from this study if there was insufficient maternal/infant plasma, the RT-PCR was negative where the DNA was positive, or the HTA patterns were not reproducible due to poor sampling of low abundance variants. Compared to the included samples, the median log10 RNA copies/ml was significantly lower in the excluded samples (data not shown).
Laboratory Tests
Genomic DNA was analyzed for the presence of HIV-1 DNA by real-time PCR [17]. Plasma HIV-1 RNA was quantified using Amplicor HIV-1 Monitor v1.5 (Roche Diagnostics). CD4+ T-cells were quantified by FACScan (Becton Dickinson).
Viral RNA Isolation
Viral RNA was isolated from peripheral blood plasma using the QIAmp viral RNA kit (Qiagen). Plasma from 6 women whose RT-PCR reaction was negative was concentrated by centrifugation and amplicons were obtained from 5.
RT-PCR
The Titan One-Tube RT-PCR system (Roche) or the Stratagene Accuscript RT-PCR system was used to amplify the HIV-1 subtype C V1/V2 region of the env gene as previously described [18]. All samples were RT-PCR amplified in two independent reactions to allow assessment of the quality of sampling.
Heteroduplex Tracking Assay (HTA)
The HTA [18–20] was used to document viral diversity as previously described [21] using a subtype C V1/V2 env probe derived from the DU151 clone [18,22].
Data Analysis
ImageQuant TL software (Molecular Dynamics/GE Healthcare) was used to quantify the intensity of each heteroduplex band and calculate the percent abundance. An HTA band was included as an env variant if: 1) it was not present in the probe alone lane; 2) on average it comprised greater than 2 % of the total viral population; and 3) it was present in both PCR replicates. Reproducibility in sampling of the population of HIV-1 variants was determined using the percent change between duplicates, as previously described [23]. The maternal replicates had a median 7 % (IQR:3,10) difference; the reproducibility of the replicates among the first positive infant samples was ~1%, perhaps due to presumed high RNA viral loads and observed low viral complexity among the infants. Validation of proper sampling is done to limit the appearance of population differences where none exists [21]. Infant V1/V2 env variant bands with a corresponding band in the maternal sample (determined by migration in the gel) were defined as “detected.” If an infant V1/V2 env variant had no corresponding band in the maternal sample (or the band was below the level of detection, as defined above), the band was classified as “undetected.”
Sequencing of V1/V2
RT-PCR products were amplified and cloned into a plasmid vector [24]. V1/V2 sequences from individual clones were manually edited and aligned with MAFFT version 5.8, using the L-INS-i method [25]. A maximum likelihood phylogenetic tree was constructed using Tree-puzzle (version 5.2) with a gamma time-reversible (GTR) evolutionary model [26]. Phylogenetic trees were subjected to 1000 puzzling steps, with reliability values greater than 0.70 considered significant. According to the Los Alamos HIV geography database, approximately 96% of the HIV-1 sequences deposited from Malawi are subtype C (accessed 11 November 2007). V1/V2 sequences from this study were aligned with the Los Alamos HIV subtype reference sequences (HXB2 coordinates 6555–6980) and a simple neighbor joining tree was constructed. All of the sequences clustered exclusively with the HIV-1 subtype C reference sequences (data not shown).
Mathematical Modeling
Transmission was modeled as a multinomial experiment, where variants were selected from the mother, with replacement, at probabilities equal to their observed frequency in the maternal viral population. For each mother-infant pair, the number of viruses sampled was equal to the number of variants observed in the infant. We simulated 10,000 multinomial transmission events for each pair and recorded the probability of each event depending on whether the infant received the mother’s most frequent variant. The probability of the infant not receiving the most abundant maternal variant is the binomial probability of 0 successes:
| (1) |
and the probability of receiving it is the probability of 1 or more success:
| (2) |
where n = the number of variants in the infant and p = the proportion of the mother’s most abundant variant. We calculated the joint probability for all IP (or IU) transmissions as the sum of the log probabilities, since transmission events for each pair are independent. The significance of the observed probability value is equal to the fraction of the random simulations that generated a probability equal to or less than the probability of the observed data. A low value rejects the hypothesis of stochastic transmission for the observed data. All modeling was conducted in R [27], and scripts are available on request.
Statistical Methods
Normally distributed, continuous variables were compared using a one way ANOVA. Non-normally distributed, continuous variables were compared using the Mann-Whitney or Kruskal-Wallis statistic. Paired non-normally distributed continuous variables were compared with the Wilcoxon matched-pairs signed-ranks test. A chi-squared or two-sided Fisher’s exact statistic was used to compare proportions. All calculations were done using STATA v.8.2.
RESULTS
Participant Characteristics
In this study we analyzed plasma from 32 non-transmitting mothers (NT), 25 transmitting mother-infant pairs (MIPs) whose infants were infected with HIV-1 in utero (IU), and 23 transmitting MIPs whose infants were infected intrapartum (IP) [12]. Baseline characteristics of the subset of mothers selected for the three groups (NT, IU, and IP) are outlined in Table I.
TABLE I.
Participant Characteristics
| Non-transmitters (N=32) | IU MTCT (N = 25) | IP MTCT (N = 23) | P | |
|---|---|---|---|---|
| Maternal Age median, (range) | 23.5 (16,31) | 24 (18,32) | 25 (16,37) | 0.2 * |
| Maternal CD4 T-cells ≤ 200/μL | 9 (29%) | 4 (16%) | 13 (57%) | 0.01 § |
| Duration of membrane rupture (hrs.) median(IQR) | 1(0.17, 4) | 1.0 (0.33, 13) | 1.8 (0.1, 2.5) | >0.5 * |
| Gestation (weeks) median(IQR) | 40 (38, 40) | 38 (37, 39) | 38 (36, 40) | 0.01 |
| Maternal HIV-1 RNA load (copies/mL) median, (IQR) | 4.9 (4.5, 5.3) | 4.8 (4.4, 5.3) | 5.3 (4.9, 5.5) | 0.1 † |
| Maternal NVP dose taken | 30 (94%) | 24 (96%) | 22 (96%) | >0.5 § |
| Infant NVP dose taken | 32 (100%) | 25 (100%) | 23 (100%) | >0.5 § |
| Primigravidae | 9 (28%) | 7 (28%) | 2 (9%) | 0.2 § |
| Spontaneous vertex delivery | 20(63%) | 17 (68%) | 18 (78%) | 0.4 § |
| Maternal V1/V2 variants | 3 (2,4) | 3 (2,4) | 4 (2,6) | >0.5* |
| Infant V1/V2 variants | n/a | 1 (1,2) | 2 (1,4) | 0.04* |
| Infants initially infected with a single V1/V2 variant | n/a | 14 (56%) | 6 (26%) | 0.05* |
|
Transmitted virus†† Only detected variants Any undetected variant |
n/a |
16 (64%) 9 (36%) |
9 (39%) 14 (61%) |
0.15§ |
Data are n (%) unless listed otherwise.
NT sample was missing CD4 T-cell data
Kruskal-Wallis test for equality of populations
Fisher’s exact statistic
One-way ANOVA
4 IU and 6 IP participants did not have maternal viral load data
Any detected includes pure populations of undetected maternal variants as well as mixtures of detected and undetected maternal variants
V1/V2 env Diversity in Mothers
The number of unique HIV-1 variants in each subject was determined using a heteroduplex tracking assay (HTA) querying the HIV-1 env variable regions 1 and 2 (V1/V2). Representative HTA autoradiographs are shown in Figure 1. Among the 80 pregnant women characterized, we detected a median of 3 V1/V2 env variants per subject (interquartile range [IQR]: 2, 4.5). There was a weak positive correlation between the number of maternal V1/V2 env variants and log10 HIV-1 RNA copies (correlation coefficient = 0.23, p=0.05). CD4 T-cell counts below 200 cells/ml were associated with a greater number of maternal V1/V2 env variants (p=0.01). Table I shows that the transmission groups had a similar number of maternal V1/V2 env variants. This suggests that differences in maternal V1/V2 env diversity are not significantly associated with vertical HIV-1 transmission.
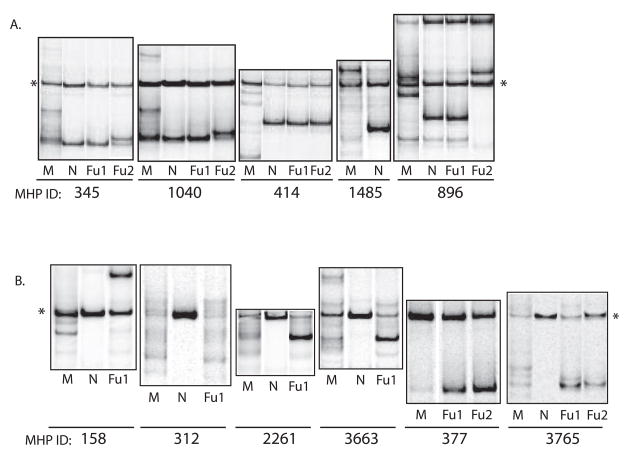
Figure 1. Heteroduplex Tracking Assays.
Autoradiographs of V1/V2 heteroduplex tracking assays against plasma associated HIV-1 isolated from mother-infant pairs (MIPS). Panel A shows examples of IU MIPs who transmitted a single variant (MHP: 345, 1040, 414), an undetected maternal variant (MHP:1485), a detected maternal variant (MHP:345, 414), and a mixture of detected/undetected maternal variants (MHP: 896). Panel B shows examples of IP MIPS who transmitted multiple variants (MHP:158,312,2261,3663,3765), detected variants (MHP:312,377,3765) and mixtures of detected/undetected (MHP:158, 3663). M=maternal plasma at delivery, N= infant plasma at birth, Fu1= infant plasma 6 weeks post-partum, Fu2= infant plasma 12 weeks post-partum,*= single stranded probe.
V1/V2 env Diversity in Infected Infants
In order to characterize the transmission of HIV-1 variants, we compared the V1/V2 env variants present in the maternal plasma at enrollment with the variants detected in the infant’s first HIV-1-positive plasma sample: at birth for the children infected IU and at 6 weeks for the children infected IP (Table I). Fewer V1/V2 env variants were detected in the IU- and IP-infected infants than in their mothers (IU p=0.0006, IP p=0.005). Thus, during vertical HIV-1 transmission a restricted number of variants are transmitted from mother to child, representing a genetic bottleneck.
We observed a contrast between the infant V1/V2 env diversity patterns during IU and IP transmission, suggesting a qualitative difference in HIV-1 transmission: IU-infected infants tend to be infected with single variants that are more often detected in the maternal plasma, while IP-infected infants tend to be infected with multiple V1/V2 env variants typically composed of a mixture of detected and undetected maternal variants (Table I). Overall, there was no association between the number of variants transmitted and maternal CD4+T cell count less than 200 cells/ml (p=0.2).
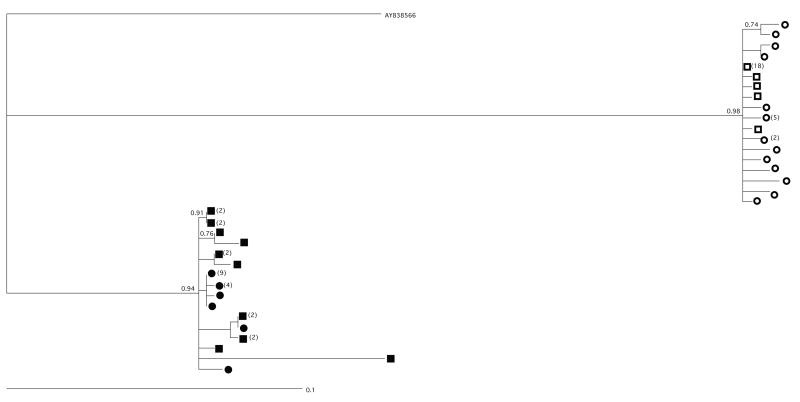
To confirm whether the multiple HTA bands in the infant correspond to the transmission of multiple maternal variants, as opposed to the rapid diversification of a single transmitted variant, we created a phylogenetic tree of V1/V2 env region sequences from two mother-infant pairs whose infant samples harbored multiple variants (Fig. 2). If multiple maternal variants were transmitted we would expect multiple branches of intermingled maternal and infant sequences, while if transmission of a single maternal variant were followed by outgrowth and diversification in the infant then the infant samples would cluster together on the same branch. In the MHP-2017 transmission pair, the HTA indicated that the infant was infected with one detected and one undetected maternal variant that composed 86% and 14% of the infant viral population, respectively. In the tree, a majority of the maternal and infant sequences cluster together, likely representing the variant with high abundance, while a separate branch at the top of the tree likely represents the low abundance variant. In MHP-3765, the HTA indicated that the infant was infected with two env variants, composing 84% and 16% of the infant viral population, both detected in the maternal plasma. The phylogenetic tree for this pair shows that maternal and infant sequences are commingled on multiple branches, suggesting transmission of multiple maternal variants. Therefore, in the two mother-infant pairs that were sequenced, the phylogenetic trees are consistent with the HTA data and support the transmission of multiple variants.
Figure 2. Phylogenetic Tree.
V1/V2 sequences from two mother-infant pairs whose infants were infected intrapartum. A total of 24–30 V1/V2 env clones from each mother and infant were sequenced, aligned using MAFFT, and used to build a maximum likelihood phylogenetic tree. Maternal samples are represented by squares and infant samples are represented by circles (MHP 2017 = open shapes; MHP 3765=filled shapes). Parenthesis represent the number of clones with identical sequences.
Modeling the Genetic Bottleneck at Vertical Transmission
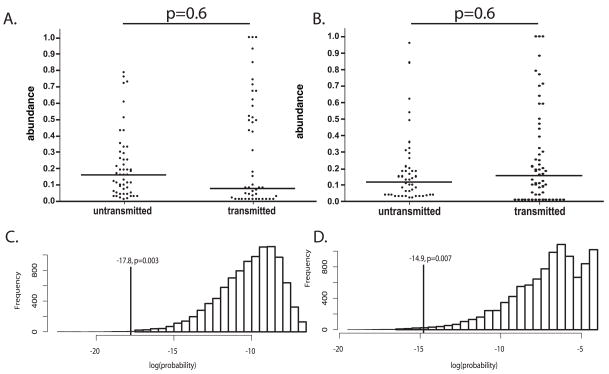
We determined the relative abundance of each maternal V1/V2 env variants within the sample population, and used that information to determine if our data were consistent with a stochastic mechanism of transmission. Transmitted variants that were undetected in the maternal peripheral plasma viral population were assigned an abundance of 1%. As seen in Fig. 3A & B, both high and low abundance maternal variants were detected in the first positive infant sample; this suggests variant abundance was not strongly associated with either IU or IP transmission (IU, p=0.6; IP, p=0.6). The probability of IU or IP transmission of the observed variants, according to their abundance in maternal plasma, was compared to a set of 10,000 simulated transmissions where the maternal variants were sampled based on abundance (Fig. 3C & D). When the observed data are compared to the simulated data sets, they do not support the bottleneck being generated by random sampling of plasma-associated maternal variants based on abundance; in other words, the observed data correspond to an uncommon outcome (IU p=0.003, IP p=0.007). In order to exclude the possibility that our observed transmission pattern was skewed by the inclusion of the undetected maternal variants, we repeated the simulation using only the detected maternal variants. Similar to the previous simulation, the observed transmission pattern remained an uncommon outcome (IU p=0.02, IP p=0.006), providing further support for a non-stochastic bottleneck mechanism.
Figure 3. HIV-1 envelope V1/V2 Variant Transmission.
The proportional abundance of each maternal V1/V2 variant was determined by the heteroduplex tracking assay and quantified by phoshorimaging. Graphed are the abundances of the transmitted versus untransmitted variants in the A) IU mother-infant pairs, and B) IP mother-infant pairs. Transmitted variants that were undetected in the maternal viral population were assigned a relative abundance of 0.01. The solid horizontal line represents the median abundance per group. Histogram of the probability of transmission obtained from random sampling, for both C) IP and D) IU transmissions, we sampled from the variant distributions described by the mothers’ HTAs according to their abundance in order to mimic a stochastic transmission process. Shown are the summed log probabilities (see Methods) for 10,000 such samples. The probability of the observed data is indicated by a vertical line. The fraction of the random samples attaining a probability equal to or less than the observed data is indicated.
Umbilical Cord Plasma
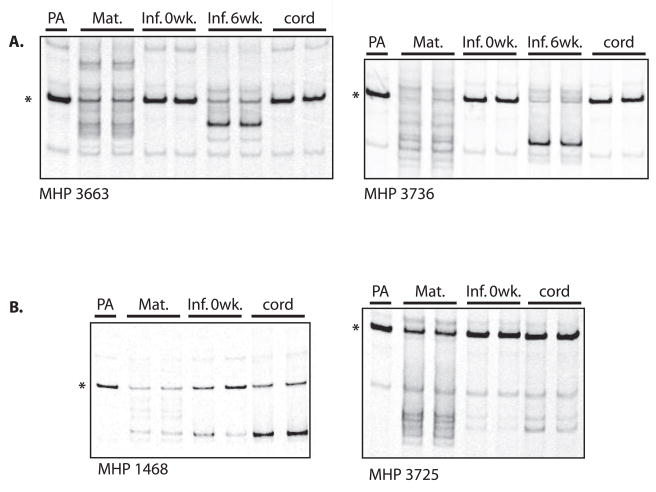
Finally, we used the V1/V2 env HTA to determine if HIV-1 isolated from umbilical cord plasma more closely resembles the infant or the maternal viral population. Umbilical cord plasma samples from the six NT women examined were V1/V2 env RT-PCR negative (data not shown). Similarly, for three infants infected IP, the cord blood V1/V2 env RT-PCR reaction was negative (Fig. 4). In contrast, in four infants infected IU the cord blood sample had a viral population that was indistinguishable from the infant birth sample but distinct from the mother’s sample. These results show that cord blood plasma represents the HIV-1 population present in the infant and suggests that cord blood plasma could be a readily accessible source of the HIV-1 population present at birth in IU-infected infants.
Figure 4. Umbilical Cord Plasma.
Autoradiographs of V1/V2 heteroduplex tracking assays against plasma-associated HIV-1 isolated from mother-infant pairs whose infants became HIV-infected: A) in utero, B) intrapartum. PA= probe alone, Mat=maternal plasma at delivery, N= infant plasma at birth, Fu1= infant plasma 6 weeks post-partum, cord= venous umbilical cord plasma, * = single stranded probe.
DISCUSSION
In this report, we describe the relationship between genetic diversity in HIV-1 env V1/V2 region and subtype C HIV-1 MTCT in NT mothers, and IU- and IP-transmitting mother-infant pairs. We found no relationship between the amount of maternal env diversity and the rate of MTCT, but we did observe a significant genetic bottleneck between the matched maternal and infant infections. The pattern of transmitted V1/V2 variants differed by the timing of HIV-1 transmission: infants infected IU frequently harbored single variants which were more often detected in the maternal plasma, and infants infected IP frequently harbored multiple variants that were more often a mixture of detected and undetected maternal variants. Finally, modeling of our data showed that on average MTCT did not favor transmission of the most abundant env variants present in maternal plasma, arguing against a stochastic model of vertical transmission.
These conclusions are based on data generated with a HTA against the env V1/V2 region, which could have several limitations. First, although the HTA cannot reliably sample genomic variants composing less than 1% of the viral population, sampling of these low abundance variants with DNA sequencing would require a minimum of 300 cloned env genes per sample. Second, it could be argued that a measure of HIV-1 diversity should sample larger regions than the approximately 400 base-pairs sampled with our assay. However, the HTA is most sensitive to sequence and size changes on genomic regions of this size, and this region is one of the most heterogeneous region in the HIV-1 genome [28]. These limitations must be balanced against the resources required to generate similar data via DNA sequencing, and owing to this constraint, we have chosen to sample a larger number of mother-infant pairs, in a hypervariable region of the env gene, rather than report diversity of longer regions of env in fewer mother-infant pairs. Finally, any misclassification of population diversity derived from using the V1/V2 region as a surrogate for actual diversity is likely to be non-differential, and is unlikely to bias our comparisons.
The observation of similarity in HIV-1 env diversity in women in this study is different from the findings of Dickover and colleagues [29], who examined HIV-1 subtype B env diversity (using a heteroduplex mobility assay approach). Dickover et al. observed that women who transmitted IU had lower V3/V4 diversity (and lower CD4+ T cell counts), suggesting that women who transmit IU have poor immunologic control of their HIV-1. In our study women in all groups had similar diversity. There are, however, many differences between that US-based cohort and our Malawi-based cohort, such as coinfections, the region of the env gene examined, the sensitivity of the HTA as compared to the HMA, and subtype B versus C HIV-1 [30].
The bottleneck of population diversity during vertical HIV-1 transmission seen here has been previously reported [5–8]. In addition to the reduction in viral diversity in infants, we found that the pattern of transmitted V1/V2 variants differed by the timing of HIV-1 transmission, with IU transmission more often representing a single variant, and IP transmission more often involving multiple variants. The results agree with a previous study of 10 IU-infected infants and their mothers by Fischetti and colleagues, where the majority of infants harbored a homogeneous virus population that consisted of both major and minor maternal variants [32]. A confounder of this result could be that the first positive sample from infants infected IU was collected within 48 hours of single-dose nevirapine treatment, which may have lowered viral RNA load and created an artificial bottleneck. However, the turnover rate of HIV-infected cells, the slow decline of viral RNA in the presence of a single dose of nevirapine relative to the timing of sampling [31], and the relative ease of HIV amplification in these samples from small volumes of infant plasma suggests high viral RNA loads. Nevirapine could also have confounded IP transmission through lowered maternal diversity prior to delivery. Given the greater diversity in IP-infected infants compared to IU, and the frequent transmission of low abundance maternal variants, we suggest if there was a NVP effect it would have dampened the difference between groups which only emphasizes the suggested difference in the mechanism of transmission IU versus IP.
Among the 48 transmission events examined in this study, nearly 50% included the transmission of variants we were unable to detect in the mother’s blood plasma. While the origin of these undetected variants is unknown, there are several possibilities, including the following: a compartmentalized HIV-1 population that was not in equilibrium with the sampled peripheral blood; low-abundance maternal variants; or variants that arose in the infant de novo, as the virus evolved in response to the single dose nevirapine exposure or its new environment. If infants are being infected with compartmentalized viruses, it remains possible that the transmitted viruses were the most abundant variants in those compartments. Regardless, on average, in our data set, the most abundant maternal variant observed in the blood plasma was not the most frequently transmitted variant in the infant by either time window of transmission (IU or IP). Therefore, the most plausible mechanisms for the bottleneck are either transmission of many variants followed by selective amplification of the detected variants, or selective transmission [33]. Distinguishing between these mechanisms is difficult as variants in the infant need to undergo amplification before they can be detected.
Despite the strong bottleneck, more than one variant was often seen in infant viral populations. Multiple mechanisms could account for the presence of multiple variants in infant infections, including: 1) multiple transmissions of a single variant, 2) a single transmission of multiple variants, 3) a single transmission event with a multiply-infected cell, or 4) a single transmission event with rapid diversification, or evolution, between the transmission event and the time of population sampling. We examined the potential for early evolution after transmission by comparing the viral population in the first positive infant sample with the subsequent samples collected at 6-week intervals (data not shown). Using changes in env diversity as a measure of viral evolution, we observed diversification in many of the IU- and IP-infected children following transmission. To begin to address the question of early evolution generating apparent diversity, we selected two mother-infant pairs, whose infant’s viral populations were comprised of two variants, and subjected the viral populations to sequence analysis. Although we cannot exclude rapid diversification in the infant after transmission, the phylogenetic trees for these two transmission pairs are most consistent with transmission of multiple variants from the mother.
In summary, these findings argue for a mother-to-child transmission model involving selection or selective outgrowth. These results are similar to the subtype B data published by Dickover and colleagues [29], thus extending the MTCT paradigm to subtype C.
Acknowledgments
We thank the Malawian mothers and infants for their participation; the MHP Nursing Staff and technicians for excellent logistical and technical support; Milloni Patel and Janera Harris for help with sample extraction. This work was presented, in part, at the 13th Conference on Retroviruses and Opportunistic Infections. This research was supported by NIH awards to JJK (K99-HD056586), SRM (R01-AI49084; R21-AI065369), RS (R37-AI44667), and by the UNC CFAR (P30-AI50410).
References
- 1.Geretti AM. HIV-1 subtypes: epidemiology and significance for HIV management. Curr Opin Infect Dis. 2006;19:1–7. doi: 10.1097/01.qco.0000200293.45532.68. [DOI] [PubMed] [Google Scholar]
- 2.UNAIDS/WHO. 2006 Report on the Global AIDS Epidemic:May 2006. 2006 Available: www.unaids.org via the Internet.
- 3.De Cock KM, Fowler MG, Mercier E, de Vincenzi I, Saba J, et al. Prevention of mother-to-child HIV transmission in resource-poor countries: translating research into policy and practice. JAMA. 2000;283:1175–1182. doi: 10.1001/jama.283.9.1175. [DOI] [PubMed] [Google Scholar]
- 4.Scarlatti G. Mother-to-child transmission of HIV-1: advances and controversies of the twentieth centuries. AIDS Rev. 2004;6:67–78. [PubMed] [Google Scholar]
- 5.Briant L, Wade CM, Puel J, Brown AJ, Guyader M. Analysis of envelope sequence variants suggests multiple mechanisms of mother-to-child transmission of human immunodeficiency virus type 1. J Virol. 1995;69:3778–3788. doi: 10.1128/jvi.69.6.3778-3788.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfs TF, Zwart G, Bakker M, Goudsmit J. HIV-1 genomic RNA diversification following sexual and parenteral virus transmission. Virology. 1992;189:103–110. doi: 10.1016/0042-6822(92)90685-i. [DOI] [PubMed] [Google Scholar]
- 7.Wolinsky SM, Wike CM, Korber BT, Hutto C, Parks WP, et al. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science. 1992;255:1134–1137. doi: 10.1126/science.1546316. [DOI] [PubMed] [Google Scholar]
- 8.Zhang LQ, MacKenzie P, Cleland A, Holmes EC, Brown AJ, et al. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol. 1993;67:3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickover R, Garratty E, Yusim K, Miller C, Korber B, et al. Role of maternal autologous neutralizing antibody in selective perinatal transmission of human immunodeficiency virus type 1 escape variants. J Virol. 2006;80:6525–6533. doi: 10.1128/JVI.02658-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu X, Parast AB, Richardson BA, Nduati R, John-Stewart G, et al. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J Virol. 2006;80:835–844. doi: 10.1128/JVI.80.2.835-844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verhofstede C, Demecheleer E, De Cabooter N, Gaillard P, Mwanyumba F, et al. Diversity of the human immunodeficiency virus type 1 (HIV-1) env sequence after vertical transmission in mother-child pairs infected with HIV-1 subtype A. J Virol. 2003;77:3050–3057. doi: 10.1128/JVI.77.5.3050-3057.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwiek JJ, Mwapasa V, Milner DAJ, Alker AP, Miller WC, et al. Maternal-fetal microtransfusions and HIV-1 mother-to-child transmission in Malawi. PLoS Med. 2006;3:e10. doi: 10.1371/journal.pmed.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mwapasa V, Rogerson SJ, Kwiek JJ, Wilson PE, Milner D, et al. Maternal syphilis infection is associated with increased risk of mother-to-child transmission of HIV in Malawi. AIDS. 2006;20:1869–1877. doi: 10.1097/01.aids.0000244206.41500.27. [DOI] [PubMed] [Google Scholar]
- 14.Mwapasa V, Rogerson SJ, Molyneux ME, Abrams ET, Kamwendo DD, et al. The effect of Plasmodium falciparum malaria on peripheral and placental HIV-1 RNA concentrations in pregnant Malawian women. AIDS. 2004;18:1051–1059. doi: 10.1097/00002030-200404300-00014. [DOI] [PubMed] [Google Scholar]
- 15.Guay LA, Musoke P, Fleming T, Bagenda D, Allen M, et al. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet. 1999;354:795–802. doi: 10.1016/S0140-6736(99)80008-7. [DOI] [PubMed] [Google Scholar]
- 16.Bryson YJ, Luzuriaga K, Sullivan JL, Wara DW. Proposed definitions for in utero versus intrapartum transmission of HIV-1. N Engl J Med. 1992;327:1246–1247. doi: 10.1056/NEJM199210223271718. [DOI] [PubMed] [Google Scholar]
- 17.Luo W, Yang H, Rathbun K, Pau C-P, Ou C-Y. Detection of HIV-1 DNA in Dried Blood Spots Using a Duplex Real-Time PCR Assay. J Clin Microbiol. 2005;43:1851–1857. doi: 10.1128/JCM.43.4.1851-1857.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitrinos KM, Hoffman NG, Nelson JA, Swanstrom R. Turnover of env variable region 1 and 2 genotypes in subjects with late-stage human immunodeficiency virus type 1 infection. J Virol. 2003;77:6811–6822. doi: 10.1128/JVI.77.12.6811-6822.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delwart EL, Pan H, Sheppard HW, Wolpert D, Neumann AU, et al. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J Virol. 1997;71:7498–7508. doi: 10.1128/jvi.71.10.7498-7508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delwart EL, Sheppard HW, Walker BD, Goudsmit J, Mullins JI. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson GJ, Hoffman NG, Ping LH, Fiscus SA, Hoffman IF, et al. HIV-1 populations in blood and breast milk are similar. Virology. 2004;330:295–303. doi: 10.1016/j.virol.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Bures R, Morris L, Williamson C, Ramjee G, Deers M, et al. Regional clustering of shared neutralization determinants on primary isolates of clade C human immunodeficiency virus type 1 from South Africa. J Virol S. 2002;76:2233–2244. doi: 10.1128/jvi.76.5.2233-2244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson JA, Baribaud F, Edwards T, Swanstrom R. Patterns of changes in human immunodeficiency virus type 1 V3 sequence populations late in infection. J Virol. 2000;74:8494–8501. doi: 10.1128/jvi.74.18.8494-8501.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritola K, Pilcher CD, Fiscus SA, Hoffman NG, Nelson JA, et al. Multiple V1/V2 env variants are frequently present during primary infection with human immunodeficiency virus type 1. J Virol. 2004;78:11208–11218. doi: 10.1128/JVI.78.20.11208-11218.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 27.Team RDC. R: A Language and Environment for Statistical Computing. 2006 Available: http://www.R-project.org via the Internet.
- 28.Starcich BR, Hahn BH, Shaw GM, McNeely PD, Modrow S, et al. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45:637–648. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 29.Dickover RE, Garratty EM, Plaeger S, Bryson YJ. Perinatal transmission of major, minor, and multiple maternal human immunodeficiency virus type 1 variants in utero and intrapartum. J Virol. 2001;75:2194–2203. doi: 10.1128/JVI.75.5.2194-2203.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang C, Li M, Newman RD, Shi YP, Ayisi J, et al. Genetic diversity of HIV-1 in western Kenya: subtype-specific differences in mother-to-child transmission. AIDS S. 2003;17:1667–1674. doi: 10.1097/01.aids.0000060412.18106.d4. [DOI] [PubMed] [Google Scholar]
- 31.Musoke P, Guay LA, Bagenda D, Mirochnick M, Nakabiito C, et al. A phase I/II study of the safety and pharmacokinetics of nevirapine in HIV-1-infected pregnant Ugandan women and their neonates (HIVNET 006) AIDS. 1999;13:479–486. doi: 10.1097/00002030-199903110-00006. [DOI] [PubMed] [Google Scholar]
- 32.Fischetti L, Danso K, Dompreh A, Addo V, Haaheim L, et al. Vertical transmission of HIV in Ghanaian women diagnosed in cord blood and post-natal samples. J Med Virol. 2005;77:351–359. doi: 10.1002/jmv.20463. [DOI] [PubMed] [Google Scholar]
- 33.Zhu T, Mo H, Wang N, Nam DS, Cao Y, et al. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]