Abstract
Double strand breaks (DSBs) are deleterious DNA lesions and if left unrepaired result in severe genomic instability. Cells use two main pathways to repair DSBs: homologous recombination (HR) or non-homologous end joining (NHEJ) depending on the phase of the cell cycle and the nature of the DSB ends. A key step where pathway choice is exerted is in the ‘licensing’ of 5′-3′ resection of the ends to produce recombinogenic 3′ single-stranded tails. These tails are substrate for binding by Rad51 to initiate pairing and strand invasion with homologous duplex DNA. Moreover, the single-stranded DNA generated after end processing is important to activate the DNA damage response. The mechanism of end processing is the focus of this review and we will describe recent findings that shed light on this important initiating step for HR. The conserved MRX/MRN complex appears to be a major regulator of DNA end processing. Sae2/CtIP functions with the MRX complex, either to activate the Mre11 nuclease or via the intrinsic endonuclease, in an initial step to trim the DSB ends. In a second step, redundant systems remove long tracts of DNA to reveal extensive 3′ single-stranded tails. One system is dependent on the helicase Sgs1 and the nuclease Dna2, and the other on the 5′-3′ exonuclease Exo1.
Keywords: Double-strand break, Repair, Recombination, Mre11, Sae2/CtIP, Exo1, Sgs1/BLM
1. Introduction
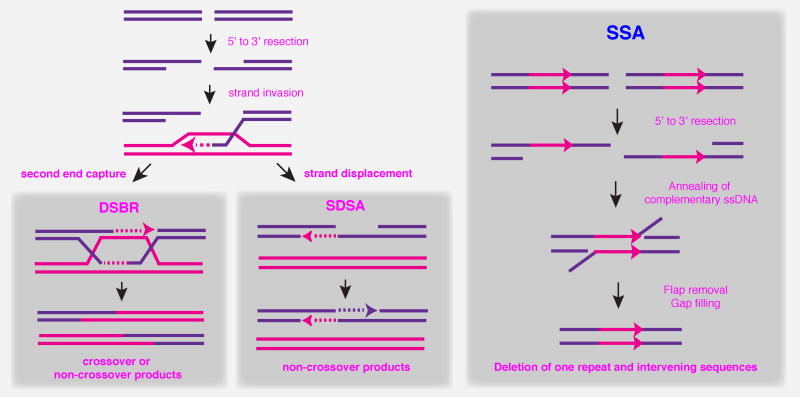
DSBs are potentially lethal lesions that can occur spontaneously during normal cell metabolism or by treatment of cells with DNA-damaging agents. DSBs are however normal intermediates in meiotic recombination and in programmed genome rearrangements, such as mating-type switching in budding yeast and V(D)J recombination in lymphocytes. If unrepaired or repaired inappropriately, DSBs can lead to mutagenic events such as chromosome loss, deletions, duplications or translocations. Several highly conserved proteins are recruited to DSBs for checkpoint activation and subsequent repair by non-homologous end joining (NHEJ) or homologous recombination (HR). NHEJ involves the religation of the two ends of the broken chromosome and can occur with high fidelity, or be accompanied by gain or loss of nucleotides at the junction [1]. HR relies on the presence of a homologous duplex to template repair of the broken chromosome. Several sub-pathways of HR have been defined; these include DSBR, SDSA and SSA (Figure 1). At the molecular level, all three mechanisms are initiated by 5′ to 3′ degradation of the broken DNA ends to create 3′ single-stranded DNA (ssDNA) tails. For repair by DSBR or SDSA, Rad51 binds to the resulting ssDNA tails to initiate pairing and strand invasion with homologous duplex DNA. The 3′ end from the broken chromosome is used to prime leading strand DNA synthesis templated by the donor duplex. According to the canonical DSBR model, the other end of the break interacts with the displaced strand from the donor duplex (D-loop) to prime DNA synthesis from the other end of the break [2]. During SDSA, the invading strand that has been extended by DNA synthesis is displaced and anneals to complementary sequences exposed by 5′-3′ resection of the other side of the break [3,4]. The remaining gaps can be filled by DNA synthesis and the nicks ligated. Under some circumstances a broken chromosome may present only one end for repair, for example, when a telomere becomes uncapped and is degraded. The chromosome end can then invade homologous sequences and initiate DNA synthesis from the site of strand invasion to the telomere, a process known as break-induced replication [5]. Single-strand annealing (SSA) is restricted to DSBs that occur within a repeat or between direct repeats. Rad52 anneals complementary single-strand regions revealed by extensive resection of the DNA ends. The 3′ single-strand tails are removed by the Rad1/10 nuclease, together with Slx4 and Saw1, and the resulting gaps/nicks filled by DNA repair synthesis and ligation [6]. This process is accompanied by deletion of one of the repeats and the intervening DNA is therefore considered to be mutagenic.
Figure 1. Models for DSB repair by homologous recombination.
The first and common step in all mechanisms is the 5′ to 3′ degradation of the DSB ends, to generate to invasive 3′ ssDNA tails (arrowhead). In the DSBR pathway, after priming DNA synthesis (dashed lines), second end capture allows formation of a double Holliday junction, whose resolution can lead to either crossover or non-crossover products. In the SDSA pathway, the 3′ end is displaced, it pairs with the other 3′ single stranded tail and DNA synthesis completes repair. The SSA pathway is restricted to DSBs that occur between direct repeats. Extensive resection of the ends reveals the complementary sequences, which can then anneal resulting in the deletion of one of the repeats and the intervening sequences.
A key step in HR is the generation of ssDNA, the substrate for Rad51 binding to initiate homologous pairing and strand exchange, and for Rad52-mediated annealing [7]. Mounting evidence indicates that ssDNA is also an important stimulus for activating the DNA damage response (DDR) and arresting the cell cycle in response to DNA damage [8]. In a reciprocal manner, checkpoint proteins not only recognize ssDNA but affect the rate at which ssDNA arises, suggesting that the DDR regulates accumulation of ssDNA and it does so by regulating the activity of various nucleases [9]. This review will focus on nucleases that participate in processing of DNA ends at DSBs to generate recombinogenic 3′-ssDNA tails in the model organism Saccharomyces cerevisiae, with reference to other organisms where applicable.
2. DSB formation and processing in meiosis: why, when and how?
2.1. Initiation of meiotic recombination
During meiosis in most sexually reproducing organisms recombination establishes a physical connection between homologous chromosomes that allows them to orient properly on the spindle and to segregate accurately at the first division. To initiate meiotic recombination, SPO11-dependent programmed DNA DSBs are formed at 150-200 sites within the genome [10,11]. SPO11 encodes a meiosis-specific endonuclease with homology to Top6A, the catalytic subunit of an archaebacterial type 2 topoisomerase [12]. Keeney et al. [13] identified Spo11 as the protein covalently bound to DNA ends at meiotic DSBs in rad50S mutants, providing compelling evidence that Spo11 is the catalytic subunit of the meiotic DNA cleavage activity. It is thought that a tyrosine side chain on Spo11 (Tyr 135) attacks the phosphodiester backbone, generating a phosphodiester linkage between the protein and the 5′ terminal strand, releasing a free 3′-OH terminus. The current model is that a Spo11 dimer coordinately breaks both strands of DNA, creating a DSB with covalent linkages between the newly created 5′ DNA strand ends and the catalytic tyrosine residue on each Spo11 monomer [11]. Spo11 homologs have been identified in many eukaryotes including fission yeast, multicellular fungi, nematodes, fruit flies, plants and mammals, implying that the role of Spo11 in meiotic recombination is highly conserved [10]. Mutations in nine other genes confer a similar phenotype to spo11, suggesting that a multi-protein complex is required to promote the formation of DSBs in meiosis with Spo11 providing the catalytic activity [10,11,14].
2.2. Processing of meiotic breaks
Spo11 must be removed from the DSB ends to allow further processing by 5′ to 3′ resection in order to initiate HR. The isolation of RAD50 and MRE11 separation-of-function alleles that result in the accumulation of unresected, Spo11-bound meiotic DSBs suggested a role for the Mre11-Rad50-Xrs2/NBS1 (MRX/MRN) complex in processing of DNA ends during meiosis [15-18]. The conserved MRX complex influences many aspects of chromosome break metabolism; it is a sensor of DSBs and physically localizes to sites of damage, it activates DNA damage checkpoint signaling cascades, it regulates resection of the DNA ends and also serves as DNA tether (Table 1). The existence of structural and functional homologues of Rad50 and Mre11 in all domains of life underscores their universal importance [19-25]. This review will focus on the nuclease activity of Mre11, after a brief description of the salient features of each member of the complex.
TABLE 1.
Proteins involved in DSB processing
| Protein | Homologues | Function | Sensitivity | Resection defect |
|---|---|---|---|---|
| Mre11 | H.sapiens Mre11 M.musculus Mre11 S.pombe Mre11 | Damage signalling Endo/exonuclease | CPT/IR/MMS | ++ |
| Rad50 | H.sapiens Rad50 M.musculus Rad50 S.pombe Rad50 | Damage signalling Tethering of DNA ends | CPT/IR/MMS | ++ |
| Xrs2 | H.sapiens Nbs1 M.musculus Nbs1 S.pombe Nbs1 | Damage signalling Tel1/ATM activation | CPT/IR/MMS | ++ |
| Tel1 | H.sapiens ATM M.musculus ATM S.pombe Tel1 | Checkpoint protein kinase | HU/MMS | + |
| Mec1 | H.sapiens ATR M.musculus ATR S.pombe Rad3 | Checkpoint protein kinase | HU/MMS | − |
| Sae2 | H.sapiens CtIP S.pombe Ctp1 | Endonuclease | CPT/MMS | + |
| Exo1 | H.sapiens Exo1 M.musculus Exo1 S.pombe Exo1 | 5′-3′ exonuclease Flap endonuclease | MMS | + |
| Sgs1 | H.sapiens BLM/WRN M.musculus BLM/WRN S.pombe Rqh1 | 3′-5′ helicase | HU/MMS | + |
| Dna2 | H.sapiens Dna2 M.musculus Dna2 S.pombe Dna2 | 5′-3′ helicase/endonuclease | HU/IR/MMS | + |
Mre11, Rad50 and Xrs2/NBS1 form a heterotrimeric complex that binds DNA. Mre11 has manganese dependent nuclease activities in vitro, including 3′-5′ ds exonuclease activity and ss endonuclease activity that acts on 5′ overhangs, 3′ flaps and 3′ branches. The observation that ssDNA homopolymers are resistant to cleavage suggests that Mre11 is structure specific and cleaves ssDNA by trapping transient secondary structures that allow the enzyme to cut at the preferred single/double-stranded transition [26]. Mre11 contains five conserved phosphoesterase motifs in the N-terminal region found in the superfamily of phosphodiesterases [7,21]. Conserved residues within the phosphodiesterase motifs, for example, Asp 16, Asp 56, His 125 and His 213, are required for Mre11 endo- and exonuclease activities in vitro [27-30]. Mutations that eliminate the Mre11 nuclease activity are generally referred to as mre11-nd (nuclease defective) alleles. Mre11-Rad50 was also found to have weak DNA unwinding (stem-loop opening) activity and to support endonucleolytic cleavage of hairpin ends [22,29,31-33]. Both Rad50 and Xrs2/Nbs1 enhance the nuclease activity of Mre11 in vitro [26,31,34,35]. The C-terminal region of Mre11 has two DNA-binding sites, one of which is essential for Spo11-dependent DSB formation [18,27,29].
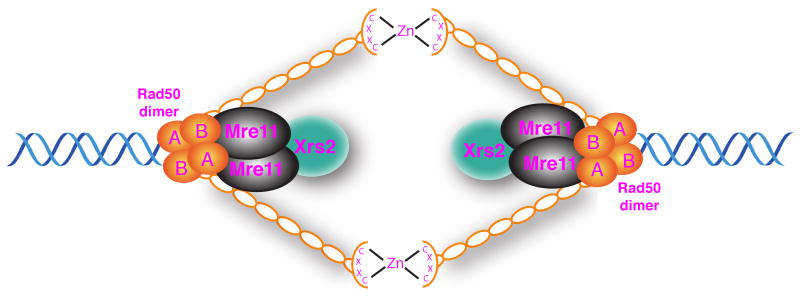
S. cerevisiae Rad50 is a 153 kDa protein with homology to E. coli SbcC and belongs to the structural maintenance of chromosomes (SMC) family of proteins [20,21,36]. Proteins in this family contain two heptad repeats in their center that fold into a coiled-coil, bringing the N-terminal Walker A and C-terminal Walker B ATPase motifs in close proximity [37]. Mre11 binds to the base of the coiled coil near the Rad50 DNA binding domain forming a Mre112Rad502 heterotetramer [38,39]. The apex of the Rad50 coiled-coil contains a conserved Cys-X-X-Cys motif in a hook-shaped domain that dimerizes with a second hook domain via cysteine-mediated zinc ion coordination (Figure 2) [39]. Crystallographic and scanning EM studies have shown that this hook domain allows dimerization between two MR complexes to tether DNA molecules [37,39,40]. Wiltzius et al. [41] showed that hook mutants of Rad50 were deficient in meiotic DSB formation and restoration of dimerization using a conditional dimerization module restored DSB formation. This suggests that the activity of MRX via the Rad50 coiled-coil might bridge sister chromatids in order to establish a proper architecture of the sisters to be cleaved.
Figure 2. Bridging of DSB ends by the Mre11-Rad50-Xrs2 complex.
Each pentameric complex is comprised of a Rad50 dimer, an Mre11 dimer and an Xrs2 monomer. The apex of the Rad50 coiled-coil, where the Cys-X-X-Cys motif resides, provides the dimerization interphase via cysteine-mediated zinc ion coordination. Mre11 binds to the base of the coiled coil near the Rad50 DNA binding region. The Xrs2 monomer associates with the complex via interaction with the Mre11 dimer. Intermolecular dimerization between two individual Mre112Rad502Xrs21 complexes tethers the two ends of a DSB.
The third member of the complex is less well conserved. Xrs2, the yeast component, and Xrs2, the human component, share some common motifs. These include the N-terminal FHA domain, which mediate protein-protein interaction and which binds to phosphorylated H2A, and the conserved C-terminal domain (CCD), which is involved in interaction with Mre11 and contains phosphorylation sites for the checkpoint protein ATM/Tel1 [42,43]. Mutant studies in S. cerevisiae have shown that the Mre11-Xrs2 interaction is crucial to all functions of the MRX complex [44].
2.3. Role of MRX and Sae2/Com1 in meiosis
The mre11 mutant was first described as meiotic recombination deficient and it was shown that deletion of any member of the MRX complex leads to an absence of meiotic recombination and extremely low spore viability [15,19,45]. All three members of the MRX complex are required in addition to the catalytic component Spo11, for the formation of meiotic DSBs in S. cerevisiae [10]. Although the MR complex appears to be universally required for DSB processing in eukaryotes, the function in meiotic DSB formation is not conserved [46]. A specific class of separation-of-function alleles of RAD50 and MRE11, rad50S and mre11S, respectively, were isolated that allow DSB formation by Spo11, but are completely defective for processing of Spo11-induced DSB and thus accumulate unprocessed DSBs with Spo11 covalently attached at their 5′ termini [13,15,18]. The mre11-nd alleles allow DSB formation, but prevent removal of Spo11 from DNA ends and subsequent repair demonstrating the importance of the Mre11 nuclease activity for meiotic recombination [17,27-29]. The mre11S allele has a mutation in the N-terminal part of the protein, but retains nuclease activity in vitro (E. Morgan and L. S. Symington, unpublished data) [18]. During meiotic prophase, Mre11 was found to transiently associate with DSB specific regions [47], raising the possibility that MRX plays an architectural role and ensures that the factors required for processing are present at the site of cleavage. All nine different mutations that resulted in the rad50S phenotype mapped to the N-terminus of Rad50 and were located in the vicinity of the Walker A-type ATPase domain of Rad50 [15]. Exactly how these mutations affect the in vitro functions of the Mre11 complex has not been determined. The isolation of null mutations in a different gene, SAE2/COM1, which conferred a phenotype identical to rad50S, suggested that rad50S could be defective in its interaction with the product of that gene.
SAE2/COM1 was discovered in two genetic screens that were designed to isolate meiotic mutants, defective at steps after the initiation of Spo11-induced DSBs, but before resolution of the recombination intermediates [48,49]. The phenotype of the isolated sae2 null mutant was shown to be identical to that conferred by rad50S and mre11-S/nd alleles; unresected DSBs, absence of homologous recombination, reduced homologous synapsis and weak sensitivity to MMS [48,49]. The genetic evidence provided the first suggestion that the role of Sae2 may intersect with that of the MRX complex. Since then Sae2 has been considered the unofficial ‘fourth member’ of the MRX complex.
Recently, Sae2-related proteins were identified in the human, fission yeast, plants and worm. The Arabidopsis thaliana COM1 protein was identified through a sequence similarity search to a set of fungal Com1/Sae2 homologues and was shown to be essential for female and male meiosis. Similarly to S. cerevisiae Sae2, it was shown to act downstream of AtSPO11-1 and upstream of AtDMC1 during meiosis and it was found important for the turnover of AtSPO11-1 and the processing of meiotic DSBs [50]. Alleles of the Caenorhabditis elegans com-1 were first obtained in a screen for maternal-effect embryonic lethality [51]. Characterization of these mutants revealed that C. elegans Com1/Sae2 was required for the processing of meiotic DSBs as well as having a repair function in non-meiotic cells [52]. The fission yeast Ctp1 was identified in a screen for MBF-regulated genes that are periodically transcribed in S phase; ctp1Δ zygotes were found to display severe spore viablitiy defects [53].
Neale et al. [54] showed that removal of covalently bound Spo11 occurs by endonucleolytic cleavage rather than direct hydrolysis of the covalent protein-DNA linkage. This endonucleolytic cleavage was found to be asymmetric and a few bases away from the break site, releasing an oligonucleotide with Spo11 attached. Removal of Spo11 was shown to be defective in sae2 and rad50S mutants, consistent with the notion that Mre11 and/or Sae2 could mediate endonucleolytic processing.
In view of the behavior of the mre11-nd and rad50S alleles, one hypothesis suggested is that the MRX complex has a role during resection after the initial endonucleolytic removal of Spo11-linked oligonucleotides. This has been difficult to prove because all mre11-nd alleles, which would presumably be defective for resection, are also defective in removing Spo11. However, studies on the repair of an HO-endonuclease induced DSB in vegetative cells have shown that rad50S and mre11-nd strains are proficient in 5′-3′ resection, indicating that this function is likely to be redundant with other nucleases [17,55]. This does not exclude the possibility that the MRX complex could participate in resection (by recruiting other factors/nucleases for example), in a manner that is not dependent on its nuclease activities.
2.4. Role of Exo1 in meiotic processing
One of the strongest candidates for lengthening of 5′-3′ resection tracts during meiosis is Exo1. Exo1 is a member of the Rad2 family of structure-specific nucleases, which possess 5′-3′ exonuclease and 5′-flap endonuclease activities in vitro [56]. It is highly conserved throughout the eukaryotic domain and is implicated in recombination and mismatch repair in vivo [57-59]. Exo1 was first purified from S. pombe induced to undergo meiosis, when the Exo1 transcript is induced about 10-fold resulting in a corresponding increase in exonuclease activity [60]. Budding yeast Exo1 and Tosca, the Drosophila homologue, are also induced during meiosis [57,61]. Although exo1 mutants of S. cerevisiae exhibit reduced spore viability, the pattern of spore viability is similar to mlh1 mutants and is thought to be due to the function of Exo1 in the crossover control pathway [61-63].
Physical analysis of meiotic DSBs in exo1Δ mutants showed that they are formed and processed to heterogeneous fragments indicative of resection. In homologous pairing mutants (such as dmc1Δ) that accumulate extensively resected DSB fragments, the extent of degradation was reduced by the exo1Δ mutation, suggesting a possible role for Exo1 in resection of meiotic DSBs [61]. If so, an exo1Δ mutation would be expected to decrease the levels of gene conversion of markers distant from the initiating DSB. However, that was not the case in all studies carried out and a decrease was found at some loci, but not at others, suggesting that the Exo1 effect could be locus specific and/or that Exo1 is redundant with other nucleases [62,63].
3. DSB processing in mitotic DSBs
3.1. Formation of mitotic DSBs
Studies of DSB repair in S. cerevisiae have been greatly assisted by the use of mega-endonucleases that create a single DSB at a defined recognition site. The best characterized of these endonucleases are HO and I-SceI, which have long nonpalindromic recognition sequences and generate 4 bp staggered cuts with 3′-OH overhangs [64,65]. The DSB ends formed after the action of these endonuclease have 5′-P and 3′-OH groups suitable for ligation or priming DNA synthesis, and thus have been called ‘clean’ breaks. DSBs formed after exposure to ionizing radiation (IR), bleomycin or methylating agents can give rise to a variety of chemically heterogeneous ends, including those with modified terminal nucleotides or even protein-DNA adducts, referred to as ‘dirty’ ends. These ends require processing by nucleases or other DNA modifying enzymes to enable repair by HR or NHEJ.
3.2. The role of MRX in processing mitotic DSBs
The RAD50 and XRS2 genes were originally identified in screens for ionizing radiation sensitive mutants. Unlike other IR sensitive mutants, such as rad51, the mrx mutants show higher radiation resistance as diploids, particularly in the G1 phase of the cell cycle, and exhibit a hyper-recombination phenotype in assays that measure spontaneous recombination between chromosome homologues [45,66-70]. These phenotypes have been attributed to a defect in sister chromatid recombination. The mrx mutants also show greatly increased rates of spontaneous gross chromosome rearrangements, thought to be a consequence of defective DSBR and checkpoint activation [71]. After the identification and characterization of the rad50S alleles it logically followed that RAD50 might control the 5′-3′ processing of mitotic DSBs. The mrx null mutants exhibit significantly delayed resection of HO-induced DSBs at the MAT locus but do not completely abolish processing and the ability to complete repair by gene conversion [72,73]. Interestingly, the MRX complex is important for nucleosome eviction from chromatin flanking a DSB by activation of the INO80 remodeling activity, and also interacts directly with the RSC nucleosome-remodeling complex [74,75]. These interactions with chromatin remodeling complexes may contribute to the role of MRX in resection of DNA ends.
While all of the mre11-nd alleles block meiotic DSB resection, the mre11-D56N, mre11-H125N and mre11-3 alleles confer only mild to intermediate IR sensitivity and do not impair processing of an HO-induced DSB [17,27,28,76,77]. The mre11-nd alleles (mre11-2, mre11-4 and mre11-58S) that confer sensitivity to IR and MMS comparable to the null allele encode proteins that fail to interact with Rad50 or Xrs2 implying that for mitotic DSB resection, the integrity of the MRX complex is more important than the catalytic activity of the Mre11 nuclease [17,29,76,78]. Although the mre11-D56N mutant shows moderate sensitivity to high doses of IR there is no apparent defect in processing multiple HO-induced DSBs suggesting that the Mre11 nuclease activity is not required for resection of clean DSBs but rather for processing of ends with covalent adducts, such as those created by IR [55]. That the nuclease activity is not required for processive 5′-3′ resection but rather for an initial endonucleolytic clipping of ends is imagined to resemble the initial processing of meiotic DSBs in which Spo11 is released bound to an oligonucleotide [54]. Consistent with the idea that Mre11 is a specialized nuclease capable of removing bulky adducts from DNA ends, E. coli SbcC/SbcD efficiently removes biotin-avidin complexes from the 5′ ends of DNA duplexes in vitro [79]. Moreover, Mre11-dependent removal of terminal protein-DNA adducts was shown to be required for initiating viral concatemerization in adenoviruses, providing further evidence for the Mre11 nuclease in processing terminal DNA adducts [80].
In contrast to the phenotype in S. cerevisiae, the mre11-H134S (equivalent to Sc mre11-H125N) mutant of S. pombe displays sensitivity to clastogens comparable to that of the null mutant, suggesting that the endonuclease activity is more crucial for repair of DSBs in this organism [81]. However, it should be noted that the mre11Δ mutant of S. cerevisiae exhibits far greater IR sensitivity than the S. pombe mre11Δ mutant [28,81,82]. Buis et al. [83] engineered mouse alleles, which either abrogate the nuclease activities or inactivate the entire MRN complex, and found that Mre11 nuclease deficiency phenocopied the absence of MRN in DNA repair but not in ATM activation. The murine Mre11-H129N protein retained Nbs1 and Rad50 interactions, but showed impaired recruitment of RPA and Rad51 into IR-induced nuclear foci, reduced clonogenic survival to IR and etoposide, a 90% reduction in HR repair efficiency, impaired ATR activation, early embryonic lethality and dramatic genomic instability [83]. The strong phenotype caused by Mre11 nuclease deficiency in S. pombe and mammals as compared to S. cerevisiae raises intriguing evolutionary questions. Buis et al. proposed that the lack of endonuclease activity of the human CtIP [84] makes Mre11 the only nuclease of the complex, whereas in budding yeast Mre11 and Sae2 could act redundantly [83]. Alternatively, the initial MRN-CtIP cleavage step of resection might be more important in some organisms than others [85].
3.3. Role of Sae2 in processing mitotic DSBs
SAE2 was detected in a genetic screen aimed at identifying mutants with low fidelity DSB repair during vegetative growth [86]. In this assay, sae2Δ, rad50S and mre11-nd strains accumulate aberrant recombination products that included a large amplification of part of the genomic DNA between inverted repeats. It was suggested that during the course of break-induced replication, hairpin fold-back structures arise that cannot be processed in sae2Δ, rad50S or mre11-nd strains, leading to chromosomal amplification events [86,87]. Lobachev et al [88] also reported a role for Sae2 and the MRX complex in processing hairpin-capped ends formed by cleavage of inverted repeats that extrude to form a cruciform structure. sae2Δ, rad50s and mre11-nd strains accumulated chromosomes with hairpin-capped ends at the sites of the inverted repeats, as well as large amplifications of the entire chromosome arm adjacent to the repeats [88]. The similar results from these two studies suggested that Sae2 and the MRX complex, the Mre11 nuclease activity in particular, play a unique role in processing hairpin-containing DNA structures in S. cerevisiae, consistent with previous in vitro studies [26,33].
Driven by these observations Lengsfeld et al. [89] tried to address the role of S. cerevisiae Sae2 in hairpin processing using purified proteins in vitro. They made the surprisingly observation that Sae2 itself exhibits endonuclease activity on ssDNA and ssDNA/dsDNA transitions and nuclease activity on ssDNA adjacent to hairpin structures, with the latter being stimulated by the MRX complex [89]. That Sae2 itself possesses endonuclease activity suggested the possibility that Sae2 may play dual roles- as a regulator of Mre11 nuclease function and/or as a nuclease. Unfortunately, an active site for this activity could not be detected and there are no recognizable domains or functional motifs in Sae2. Direct protein-protein interactions between MRX and Sae2 could not be detected in the absence of DNA in vitro, in agreement with what was previously reported by yeast two hybrid studies [90]. Even so, the cooperative DNA binding exhibited by MRX and Sae2, and coimmunoprecipitation of Sae2 with components of the MRX complex only in the presence of DNA, suggest that the proteins might interact on DNA in vivo [89,91]. The N-terminus of Sae2 contains domains that control Sae2 homotypic interactions and was found to be essential for nuclease activity [89,91]. The product of a rad50S allele, Rad50-R20M, exhibited a partial deficiency in Sae2 cooperative nuclease activity consistent with the long-suggested notion that the MRX complexes in rad50S strains are impaired in their association with Sae2 [89]. Furthermore, over-expression of Sae2 resulted in partial suppression of the rad50S defect in a single-strand annealing (SSA) assay that requires extensive resection (∼25-kb) of an HO-induced break placed between two direct repeats [92]. In this assay, repair of the DSB was defective in sae2Δ and rad50S strains and formation of ssDNA intermediates was delayed in the sae2Δ mutant [92].
Lisby et al. investigated the recruitment and association of checkpoint and repair proteins to sites of damage by visualizing the cellular response to DNA damage in living cells, using marked DSBs and fluorescently tagged proteins [93,94]. They observed that deletion of SAE2 delayed disassembly of Mre11 foci and the subsequent recruitment of Rad52 [93]. Moreover, Sae2 foci appeared at the precise time when Mre11 foci disassemble and Rad52 foci form [93]. The timing and transient nature of Sae2 foci suggested that Sae2 plays a role in the transition from the initial damage recognition to the 5′-3′ resection and recruitment of downstream factors. The kinetics of Mre11 and Rad52 foci formation and disappearance in the nuclease defective mre11-H125N and mre11-D56N cells was similar to that of sae2Δ [93]. In cells treated with IR, the delay in the disassembly of Mre11-H125N foci was increased almost 2-fold compared to the delay observed after an I-SceI induced DSB, consistent with the intermediate sensitivity to IR displayed by the mre11-H125N mutant [28,93]. This suggests Mre11 is required to act as structure-specific nuclease on aberrant DSB ends generated by IR, which are not formed at I-SceI-induced DSBs. These observations are consistent with the view that the Mre11 nuclease activity is not required for processive resection of the break, but rather for an initial processing, and this processing step appears to be regulated by Sae2.
3.4. Sae2 homologs and their role in repair of mitotic lesions
Ctp1, the fission yeast Sae2-related protein, was identified in a screen for cell cycle regulated genes [53]. Unlike the budding yeast sae2Δ mutant, the ctp1Δ mutant phenocopied S. pombe mrn null mutants in sensitivity to DNA damaging agents. Ctp1 was found to localize at DSBs and was required for HR-dependent plasmid integration but not for NHEJ-dependent plasmid recircularization [53]. The vertebrate homologue, CtIP, was first isolated as a binding partner of CtBP (C-terminal region of adenovirus E1A) and BRCA1 [95,96]. The CtIP C-terminus, which is highly conserved in CtIP related proteins in other vertebrate organisms, and is required for CtIP function in human cells, is weakly conserved in S. cerevisiae Sae2 [84]. In a recent study in which the mitotic role of CtIP was addressed, CtIP depletion caused hypersensitivity to DNA damaging agents, reduced frequency DSB-induced recombination and was shown to physically and functionally interact with the MRN complex [84]. Moreover, it is recruited to DSBs and promotes RPA foci formation and ATR recruitment to the sites of damage [84]. These findings support the general notion that there are conserved roles for CtIP-like proteins in controlling DSB resection, checkpoint signaling and homologous recombination.
3.5. Cell cycle control of resection
An important factor governing the choice between DSB repair pathways is the phase of the cell cycle. HR is generally restricted to the S and G2 phases, when DNA has replicated and the sister chromatid is available as a repair template [97,98]. NHEJ, on the other hand, operates throughout the cell cycle but seems to be more important in G1 [99-102]. The choice between the two pathways is governed by cyclin dependent protein kinases (CDKs) and several lines of evidence suggested that Cdk1 regulates DSB resection [97,98]. Cell cycle arrest of budding yeast cells in G1 by mating type pheromone or by inhibition of an analog-sensitive allele of CDC28 (Cdk1) kinase blocks resection and prevents Rad53 phosphorylation in response to a single HO-induced DSB [97,98]. Sae2 was recently identified as one target of the Cdk1-dependent control of resection. Cdc28 phosphorylates Ser267 of Sae2 in unperturbed cycling cells [103,104]. Mutation of this site to a non-phosphorylatable residue, S267A, phenocopies sae2Δ, including hypersensitivity to camptothecin, defective sporulation, reduced hairpin-induced recombination, impaired DSB processing, persistent Mre11 foci and delayed Rad52 recruitment [104]. A sae2-S267E mutation that behaves as phosphomimic, not only complements these phenotypes but overcomes the necessity of CDK activity for DSB resection [104]. Both mutations modulate the balance between HR and NHEJ during the cell cycle, suggesting that the commitment to DSB resection is highly regulated to ensure that the cell engages the most appropriate DNA repair pathway [104].
3.6. Role of Exo1 in processing mitotic DSBs
The production of 3′ ssDNA after resection of a DSB would presumably require the activity of a 5′-3′ exonuclease and Exo1 as such, appeared as an excellent candidate for the resection nuclease in mitotic cells. As was already discussed, Exo1 was initially purified from S. pombe extracts induced to undergo meiosis, when Exo1 transcription and activity is increased [60]. Exo1 was also purified from mitotic extracts based on the ability to degrade duplex DNA to produce substrates for single-strand annealing [105,106]. Although the exo1Δ mutant does not show sensitivity to IR, spontaneous deletions between direct repeats are reduced in frequency [105]. Analysis of the kinetics of resection of HO-induced DSBs in the exo1Δ mutant showed there was less resection than in wild-type cells, but this did not impair homologous recombination by gene conversion [55,107].
Several studies have suggested that Mre11 and Exo1 have redundant roles in the processing of DSBs in mitotic cells. Over-expression of Exo1 suppresses the DNA repair defect of mre11, rad50 and xrs2 mutants and can partially overcome the mrx defect in resection of an HO-induced DSB [61,77,107-109]. This activity is dependent on the nuclease activity of Exo1, suggesting that Exo1 can take over some functions of the MRX complex [107,109]. Moreover, exo1Δ mre11Δ double mutants exhibit a synthetic growth defect, are more sensitive to MMS and IR, and display an even longer delay in the processing of an HO-induced DSB than mre11Δ single mutants [61,107]. In S. pombe, radiation-induced Rad51 foci are not detected in the exo1Δ rad50Δ double mutant consistent with the single-strand tails being very short [82]. While these studies suggest that Exo1 and the Mre11 complex have redundant roles in resection, the exo1Δ mre11-H125N double mutant is more IR resistance than the mre11Δ mutant and has no defect in DSB-induced gene conversion [107]. If Exo1 were the only activity redundant with Mre11, then the exo1Δ mre11Δ and exo1Δ mre11-H125N strains would be expected to exhibit similar phenotypes and this is not the case, suggesting that there is at least one other redundant nuclease required for processing of DSBs [107]. In the absence of Exo1 and Sae2, 5′-3′ resection of DSBs is reduced, but resection and homologous recombination still occur [85,110].
Null alleles of genes encoding other members of the Rad2 family of 5′-3′ exonucleases, Rad2, Rad27, Din7 and Yen1, and Pso2 a member of the metallo-lactamase family of proteins, have been examined for resection defects when combined with exo1 and mre11-H125N, but none of these combinations resulted in a further reduction in formation of ssDNA compared with the exo1 single mutant [55,111] (E. P. Mimitou and L. S. Symington, unpublished data). Moreover, exhaustive screens to look for single viable mutants hypersensitive to IR have been performed, but none of the mutants identified in this screen appeared to be good candidates for involvement in 5′ to 3′ resection [112].
3.7. Exo1 and Sgs1-Dna2 function redundantly in 5′-3′ resection
One possible mechanism for DNA end resection would involve a helicase in conjunction with a single-strand specific endo- or exonuclease. In E. coli the RecQ helicase and the RecJ 5′-3′ exonuclease function in DSB resection in the absence of the dominant RecBCD activity [113], suggesting the possibility that the yeast RecQ helicase, Sgs1, might also function in DSB resection. An SSA assay (Figure 1), requiring extensive resection to allow repair of an I-SceI or HO induced DSB, was used address this hypothesis [85,114]. Although deletion of SGS1 resulted in only a slight delay in repair of repeats located 5-7 kb apart, a greater defect was observed for repeats separated by 25 kb [85,114]. However, the exo1Δ sgs1Δ double mutant was completely deficient in SSA [85,114]. In the exo1Δ sgs1Δ double mutant novel intermediates were observed with slightly faster mobility than the DSB cut fragments due to partial resection, and these became smeared over time suggesting slow processing [85,114]. The resection defect observed for sgs1 was also observed for mutants defective for the Sgs1-interacting proteins, Top3 and Rmi1, suggesting the entire STR complex is required for DSB processing [114]. Although the resection defect in the exo1Δ sgs1Δ double mutant completely abolished repair by SSA, repair by gene conversion was still observed, albeit with reduced efficiency, suggesting that short ssDNA tails are sufficient to initiate Rad51 dependent gene conversion [85]. The resection activity was shown to require the Exo1 nuclease and Sgs1 helicase activities [85,114,115]. A role for a RecQ family helicase in resection appears to be conserved in human cells as the mammalian Sgs1 homolog, BLM, was shown to function in a parallel pathway with Exo1 to promote DSB resection [115]. In vitro studies using purified proteins have shown that BLM stimulates Exo1 nuclease activity raising the possibility that Exo1 functions in both Sgs1/BLM dependent and independent pathways [116].
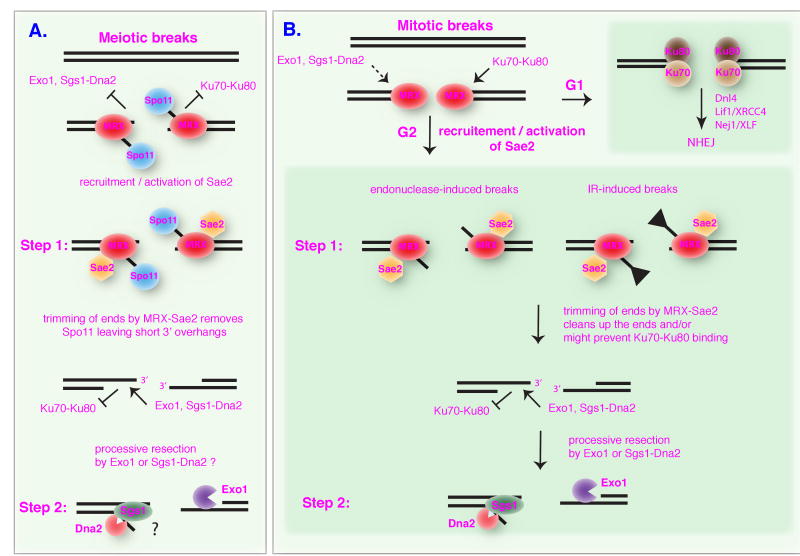
Because previous studies suggested MRX and Sae2 function in an early processing step, it was logical to ask whether the partial resection observed in the exo1Δ sgs1Δ double mutant requires these factors. The synthetic lethality of sgs1Δ with sae2Δ, and exo1Δ sgs1Δ with mre11Δ or mre11-H125N, necessitated the construction of a conditional allele of SAE2 [85]. Indeed, conditional loss of SAE2 in the exo1Δ sgs1Δ background led to a prominent stabilization of the cut fragment and no apparent gene conversion, confirming the complete block to resection and necessity of resection to initiate Rad51-mediated strand invasion. These observations led to the proposal of a two-step mechanism for DSB resection [85]. The first step is dependent on MRX and Sae2 and results in the endonucleolytic removal of about 50-100 nucleotides from the 5′ end, which gives rise to an intermediate with short 3′ ssDNA tails that is rapidly processed in a second step by either Exo1 or Sgs1 with a single-strand-specific nuclease (Figure 3). In support of this model, recent studies have shown Mre11-dependent processing of DSBs to form short ssDNA oligonucleotides that stimulate ATM activity [117]. It is likely that the extensive resection after MRX-Sae2 clipping is fast, explaining why the initial processing in mitotic cells had previously escaped detection.
Figure 3. Two step mechanism of DSB resection in meiosis and mitosis.
A. After meiotic DSBs are formed, Spo11-bound ends are poor substrates for Exo1 or Sgs1-Dna2, but when processed by MRX-Sae2 to remove Spo11 and form short 3′ overhangs they can be used by either Exo1 or Sgs1-Dna2. Whether Sgs1-Dna2 functions in processing meiotic DSBs remains to be determined. B. In G1, when Sae2 is not phosphorylated by CDK, the ends are preferentially bound by Ku70-Ku80 channeling repair to the NHEJ pathway. In G2, activation of Sae2 by CDK, leads to trimming of DSB ends, dissociation of the MRX complex from the DSB ends and generation of short RPA bound ssDNA overhangs. This trimming step could potentially inhibit Ku70-Ku80 binding and instead promote extensive resection by Sgs1-Dna2 or Exo1.
Zhu et al. [114] provided evidence that the nuclease acting with Sgs1 is Dna2. Dna2 is a conserved endonuclease/helicase implicated in Okazaki fragment processing and in DSB and post-replication repair pathways [118,119]. By monitoring the kinetics of disappearance of an HO cut fragment, or restriction fragments located 3, 10 or 28 kb from the HO cut site, Zhu et al. [114] found that MRX and Sae2 initiate DSB resection and either Sgs1 with Dna2 or Exo1 rapidly degrade 5′ strands to expose long 3′ ssDNA tails. Because the phenotype of the exo1Δ dna2Δ double mutant is similar to the exo1Δ sgs1Δ double mutant, and the sgs1Δ resection defect is indistinguishable from sgs1Δ dna2Δ, it appears that Sgs1-Dna2 function in a parallel pathway to Exo1. However, it remains possible that Sgs1 provides a substrate that can be utilized by either Dna2 or Exo1, as suggested by the in vitro studies [116]. The nuclease activity of Dna2, but not the helicase, was found to be required for DSB resection, consistent with the view that Sgs1 unwinds DNA ends and the Dna2 nuclease removes the 5′ strand. DNA2 has also been identified as a major DSB processing activity in Xenopus egg extracts and is required for an in vitro SSA reaction [120]. Whether Dna2 interacts directly with Sgs1 to promote resection, or is recruited via the interaction with RPA remains to be determined.
That Sgs1 acts at an early step of DSBR was unexpected because genetic studies had implied a role for it after Rad51 action, and biochemical studies have shown the human BLM protein to dissociate D-loops and double Holliday junction intermediates [121-124]. However, BLM localizes rapidly to sites of laser light-induced DSBs, and Sgs1 has been shown to interact with Mre11 in damaged cells, consistent with an early role in repair [125,126]. It has been reported that exo1Δ sgs1Δ double mutants show a synergistic increase in the frequency of gene targeting and in short repeat recombination [127,128]. This effect could be due to the late role of Sgs1 in unwinding recombination intermediates, and/or to the stabilization of linear DNA in the absence of extensive resection.
3.8. Regulation of Exo1 activity
Recent studies of human and yeast Exo1 provide insight into how Exo1 activity is regulated. Morin et al. [9] showed that Exo1 is phosphorylated in response to telomere uncapping and DNA damage in budding yeast. The phosphorylation was dependent on Rad9, Rad53, Mec1, the checkpoint clamp loader Rad24 and the Rad17 component of the clamp [9]. Consistent with this finding, Exo1 was identified as a Rad53 target in a proteome-wide screen of proteins phosphorylated in response to MMS [129]. Morin et al. [9] mapped four distinct major phosphorylation sites in the C-terminal half of the protein and constructed Exo1 mutants where all four serines were mutated to alanines (to prevent phosphorylation) or glutamic acids (to mimic phosphorylation). The behavior of the mutants was subtle but indicated that phosphorylation inhibits Exo1 activity in vivo by limiting the accumulation of ssDNA at telomeres and therefore participates in a negative feedback loop to limit DNA damage checkpoint activation [9]. Similar results were obtained from studies on the regulation of the human Exo1 activity. El-Shemerly et al. demonstrated that human Exo1 is phosphorylated by ATR and degraded after treatment of cells with the S phase inhibitor, hydroxyurea [130,131].
3.9. Rationale for a two-step processing mechanism
Spo11 bound to meiotic DSBs appears to function as a barrier to resection by Exo1 or Sgs1-Dna2, making the initial cleavage by MRX and Sae2 obligatory for DSB processing. Hence rad50S, sae2Δ and mre11-nd mutants, defective in the initial processing step, accumulate unprocessed DSBs and fail to complete meiosis [15,17,18,27,48,49]. It is also likely that Spo11-bound ends prevent binding by the Ku heterodimer (composed of Ku70 and Ku80) thus preventing NHEJ and promoting the HR pathway.
In budding yeast, the MRX-Sae2 cleavage step is not essential for processing DSBs generated by endonucleases because mre11Δ, mre11-nd and sae2Δ mutants are able to resect these ends, even though initiation of resection is delayed [17,85,92,93,100,114]. In mitotic cells, the Ku heterodimer is thought to bind to duplex DNA ends to protect them from degradation and mediate recruitment of downstream factors, such as the Lif1/XRCC4-Dnl4 complex, involved in NHEJ repair [132]. The MRX complex and Ku compete for DNA end binding because in the absence of Ku, Mre11 binding to ends increases in G1 cells and an increased number of ends initiate resection [100,102,133]. Conversely, over-expression of Ku can inhibit Mre11 binding to DSBs and initiation of resection in G2-arrested cells [102]. As mentioned previously, over-expression of Exo1 partially suppresses the IR sensitivity and resection defects of the mre11Δ mutant [61,77,107,109]. One possible mechanism for this suppression is that Exo1 competes with Ku for binding to DNA ends and when present in higher concentration is able to process ends for HR. In budding and fission yeast deletion of YKU70 results in suppression of the IR sensitivity of mre11Δ and ctp1Δ mutants that it is dependent on Exo1, consistent with the view that Exo1 is normally prevented from accessing 5′ ends at DSBs by Ku [53,67,82]. All these observations suggest that in the absence of the inhibitory action of Ku, Exo1 is able to access DNA ends without the initial clipping event by MRX-Sae2. The incomplete suppression of the IR sensitivity of mre11-nd and sae2Δ mutants by yku70Δ is most likely due to aberrant ends that require MRX-Sae2 cleavage. In S. pombe and mouse cells the initial processing step by MRN-Ctp1/CtIP appears to be more important for processing “clean” ends than in S. cerevisiae; one possible explanation is that Exo1 competes less effectively with Ku in these organisms. At present we do not know whether Sgs1-Dna2 can access ends made available in the absence of Ku, or whether the MRX-Sae2 processing step yields a substrate for processing by Sgs1-Dna2. Purified BLM and Sgs1 bind poorly to blunt-ended duplex DNA, but can unwind dsDNA with a 3′ tail [134,135]. The initial processing step by MRX-Sae2 may provide a substrate less suitable for Ku binding thus committing cells to extensive resection and homologous recombination.
4. Telomeres as DSBs
Telomeres are specialized DNA-protein complexes that define the physical ends of linear chromosomes. By protecting chromosomes from end degradation and end-to-end fusions, telomeres ensure genome stability and chromosome integrity. Their basic structure is conserved among eukaryotes and consists of short tandem DNA repeats, which are G-rich in the strand containing the 3′-end, the G strand. The G strand extends beyond its complementary strand to form a ssDNA overhang, called the G tail [136]. The 3′ overhangs of telomeres could be perceived as intermediates of DSB and if processed inappropriately by the DNA repair machinery would lead to chromosome fusion and/or degradation and cell cycle arrest. Cells therefore have to distinguish telomeres from DSBs by ensuring that different classes of protein bind to telomeric and DSB DNA ends. In S. cerevisiae, this is achieved by multiple telomere binding proteins, including Cdc13-Stn1-Ten1, a ssDNA-binding complex that binds specifically to the G-tails [137], and the Ku70-Ku80 heterodimer (reviewed in [138]).
4.1. Resection at normal telomeres
The protruding ssDNA G tails play a central role in modulating telomere length, since they serve as a substrate for extension by telomerase (reviewed in [139]. If they were only the result of semi-conservative DNA synthesis, G tails would be expected to be present at only one end of the chromosome, but they can be detected at both ends even in the absence of telomerase [140,141]. This suggests the C-strand of newly synthesized blunt-ended molecules is actively resected after completion of DNA synthesis. Null mutations in MRX genes result in short telomeres in most genetic backgrounds, and complete loss of telomeric DNA and senescence in one background, implicating the MRX complex in telomere maintenance [61,142-144]. MRX/N are required to maintain the wild type length of G tails in both S. cerevisiae and humans [145,146]. The disruption of the MRX complex does not completely abolish G-tail generation, suggesting the requirement for redundant nucleases [145,147]. In S. pombe, the dna2-C2 conditional mutant showed a decrease in the amount G-rich overhang in S-phase suggesting one of the redundant nucleases is Dna2 [148]. The MRX complex appears to be the primary factor responsible for single stranded tail generation in a de novo telomere reaction [149]. Diede and Gottschling showed that an HO-induced DSB adjacent to a telomere tract is resected to generate a 3′ ssDNA tail before Cdc13 binding and elongation by telomerase [149]. In this assay mrx null mutants as well as mre11-nd mutants were defective in formation of 3′ ssDNA tails and generation of telomeres in vivo [147,149]. However, mre11-nd mutants show normal telomere maintenance in cycling cells, suggesting that properties other than the nuclease activity are important to allow resection of telomeric ends [147]. The de novo telomere addition assay is performed in G2 arrested cells and this may explain the complete dependence on MRX for resection.
TEL1 was identified in a screen of yeast mutants that display short telomeres and subsequently shown to encode the yeast ortholog of the ATM kinase [150,151]. Tel1 is found at telomeres during S phase in an MRX dependent manner and contributes to maintainin proper telomere length via recruitment of telomerase [152-154]. The role of Sae2/CtIP at telomeres is not clear yet and it would be interesting to see whether it performs functions similar to those observed at DSBs. sae2Δ cells were found to exhibit slightly elongated telomeres and cause further elongation in combination with a mec1 mutation, similar to observations of rad50S cells [91,144]. In S. pombe, MRN complexes assemble and recruit Tel1 to telomeres in the absence of Ctp1 [53].
As with DSBs, generation of ssDNA at telomeres requires the activity of CDK. Inhibition of CDK was shown to completely block the addition of telomere repeats in a de novo telomere addition assay and to prevent generation of G strand overhangs [147,155]. Resection at telomeres, therefore, is limited to late S and G2/M phase, coinciding with a time frame in which the length of G-tails increases and telomerase can elongate telomeric DNA [156,157].
4.2. Resection at uncapped telomeres
Budding yeast mutants defective in telomere binding proteins are useful for addressing the mechanisms by which checkpoint pathways recognize damaged DNA because in these cells telomeres become potent activators of DNA damage checkpoint pathways. In the temperature sensitive cdc13-1 strain loss of Cdc13 function at the restrictive temperature leads to telomere uncapping, accumulation of ssDNA at the telomeres and Rad9 dependent cell cycle arrest [158,159]. The Ku complex is also important in telomere capping. Both yku70Δ and yku80Δ mutants display temperature sensitive growth, have short telomeres, accumulate ssDNA at telomeres and display altered telomeric silencing [160-165]. Furthermore, the Ku complex binds to the telomerase RNA directly and is localized at telomeres [166,167]. Maringele and Lydall demonstrated that the cell cycle arrest of yku mutants at restrictive temperature is associated with increased levels of ssDNA in subtelomeric Y' regions, which triggers both DNA damage (Chk1, Mec1 and Rad9) and spindle (Mad2) checkpoint pathways [159]. Rad9, therefore, seems to be critical for cell cycle arrest in many strains that have telomere defects. In a recent study Lazzaro et al. provided evidence for a mechanism governing the negative regulation of ssDNA accumulation by Rad9 at telomeres; they suggested that the binding of Rad9 to methylated histone H3-K79 inhibits a Rad50-dependent resection process at uncapped telomeres [168].
Studies of telomere uncapping revealed an important role for Exo1 in degradation of DNA ends. Although exo1Δ mutants show no telomere length defects [61,107], the exo1Δ mutation suppressed the temperature dependent growth defects and reduced ssDNA accumulation in yku70Δ and cdc13-1 mutants cultured at restrictive temperatures, suggesting that Exo1 creates ssDNA at telomeres in the absence of Cdc13 or the Ku heterodimer [159,169]. Moreover, Exo1 but not Mre11 was shown to be essential for generating ssDNA at telomeres, because exo1Δ suppressed the accumulation of telomeric ssDNA and relieved the cell cycle arrest in yku70Δ mre11Δ double mutants [159]. The short-telomere phenotype displayed by yku80Δ mutants is partially suppressed by exo1Δ suggesting Exo1 plays a direct role in telomere shortening [170]. However, studies in S. pombe showed that EXO1 deletion does not suppress accumulation of ssDNA in taz1Δ rad50Δ pku70Δ strains [82]. Instead, Rad50 and Dna2 are required for ssDNA generation at uncapped telomeres [148]. Recently, the role of Exo1 at uncapped telomeres in mice was demonstrated and it was found to be similar to the one observed in budding yeast. In particular, Exo1 was shown to participate in the formation of ssDNA, RPA recruitment and ATR activation in response to telomere dysfunction [171]. Notably, mice lacking Exo1 lived significantly longer than control mice expressing Exo1, suggesting that regulation of Exo1 activity can affect mammalian cell responses to uncapped telomeres and mammalian lifespan [171].
One intriguing feature of telomere resection is the more notable role played by checkpoint proteins, especially the Rad24/RFC clamp loader and the 9-1-1 checkpoint clamp [172,173]. Rad24, similarly to Exo1, is important for generating ssDNA at telomeres of cdc13-1 mutants, but unlike Exo1, Rad24 is not required for generating ssDNA at yku70Δ telomeres [159,169,173]. Thus, Rad24 contributes to ssDNA production at telomeres by regulating a nuclease (ExoX) other than Exo1 and genetic studies have excluded Mre11, Rad2, Din7, Yen1 and Nuc1 from this role [169]. To date, the identity of ExoX remains elusive. Considering the recently identified role of Sgs1-Dna2 in resection and the finding in S. pombe that Dna2 is involved in the generation of the G rich overhangs, it will be interesting to address whether Sgs1 and/or Dna2 are involved in processing uncapped telomeres in budding yeast [85,114,115,148]. In a recent study, sgs1Δ was shown to confer a similar phenotype to exo1Δ in suppression of the senescence phenotype of tlc1Δ (RNA component of telomerase) rad52Δ double mutants by promoting formation of large palindromes at chromosome ends [174,175]. This is thought be due to delayed resection of uncapped ends and generation of fold-back structures at single-stranded regions, similar to the mechanism proposed for palindrome formation next to DSBs in sae2Δ mutants [87], and is consistent with the notion that Sgs1 is able to access and degrade uncapped telomeres.
5. Concluding Remarks
Our understanding of how DNA ends are resected has advanced considerably with the discovery of Sgs1 and Dna2 as components of a redundant processing pathway. Current data suggest a model in which the MRX complex and Sae2 perform the initial processing step to remove an oligonucleotide from the 5′ ends; in a second step processive resection by either Sgs1-Dna2 or Exo1 results in formation of long 3′ ssDNA tails. Whether these alternative mechanisms are completely overlapping or have specialized roles at different phases of the cell cycle, or at different substrates remains to be determined. The recent identification of Sae2 as a target of CDK1 to limit resection to the S and G2 phases of the cell cycle, and Exo1 as a target for down-regulation of resection in response to checkpoint activation suggest these processes are highly regulated.
Acknowledgments
We are grateful to members of the Symington laboratory, past and present, for their contributions, and to W. K. Holloman for discussions and critical reading of the manuscript. Research performed in my laboratory that is cited in this review was supported by grants from the National Institutes for Health (GM41784 and GM54099).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 2.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson DO, Holloman WK. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc Natl Acad Sci U S A. 1996;93:5419–5424. doi: 10.1073/pnas.93.11.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 6.Li F, Dong J, Pan X, Oum JH, Boeke JD, Lee SE. Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Mol Cell. 2008;30:325–335. doi: 10.1016/j.molcel.2008.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 8.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 9.Morin I, Ngo HP, Greenall A, Zubko MK, Morrice N, Lydall D. Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J. 2008 doi: 10.1038/emboj.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 11.Keeney S, Neale MJ. Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem Soc Trans. 2006;34:523–525. doi: 10.1042/BST0340523. [DOI] [PubMed] [Google Scholar]
- 12.Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 13.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 14.Lichten M. Meiotic recombination: breaking the genome to save it. Curr Biol. 2001;11:R253–256. doi: 10.1016/s0960-9822(01)00131-2. [DOI] [PubMed] [Google Scholar]
- 15.Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 16.Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 17.Tsubouchi H, Ogawa H. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol Cell Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nairz K, Klein F. mre11S--a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 1997;11:2272–2290. doi: 10.1101/gad.11.17.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ajimura M, Leem SH, Ogawa H. Identification of new genes required for meiotic recombination in Saccharomyces cerevisiae. Genetics. 1993;133:51–66. doi: 10.1093/genetics/133.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alani E, Subbiah S, Kleckner N. The yeast RAD50 gene encodes a predicted 153-kD protein containing a purine nucleotide-binding domain and two large heptad-repeat regions. Genetics. 1989;122:47–57. doi: 10.1093/genetics/122.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharples GJ, Leach DR. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol Microbiol. 1995;17:1215–1217. doi: 10.1111/j.1365-2958.1995.mmi_17061215_1.x. [DOI] [PubMed] [Google Scholar]
- 22.D'Amours D, Jackson SP. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 23.Hopfner KP, Karcher A, Shin D, Fairley C, Tainer JA, Carney JP. Mre11 and Rad50 from Pyrococcus furiosus: cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J Bacteriol. 2000;182:6036–6041. doi: 10.1128/jb.182.21.6036-6041.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrini JH, Walsh ME, DiMare C, Chen XN, Korenberg JR, Weaver DT. Isolation and characterization of the human MRE11 homologue. Genomics. 1995;29:80–86. doi: 10.1006/geno.1995.1217. [DOI] [PubMed] [Google Scholar]
- 25.Dolganov GM, Maser RS, Novikov A, Tosto L, Chong S, Bressan DA, Petrini JH. Human Rad50 is physically associated with human Mre11: identification of a conserved multiprotein complex implicated in recombinational DNA repair. Mol Cell Biol. 1996;16:4832–4841. doi: 10.1128/mcb.16.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trujillo KM, Sung P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J Biol Chem. 2001;276:35458–35464. doi: 10.1074/jbc.M105482200. [DOI] [PubMed] [Google Scholar]
- 27.Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- 30.Arthur LM, Gustausson K, Hopfner KP, Carson CT, Stracker TH, Karcher A, Felton D, Weitzman MD, Tainer J, Carney JP. Structural and functional analysis of Mre11-3. Nucleic Acids Res. 2004;32:1886–1893. doi: 10.1093/nar/gkh343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trujillo KM, Roh DH, Chen L, Van Komen S, Tomkinson A, Sung P. Yeast xrs2 binds DNA and helps target rad50 and mre11 to DNA ends. J Biol Chem. 2003;278:48957–48964. doi: 10.1074/jbc.M309877200. [DOI] [PubMed] [Google Scholar]
- 33.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 34.Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 35.Desai-Mehta A, Cerosaletti KM, Concannon P. Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol Cell Biol. 2001;21:2184–2191. doi: 10.1128/MCB.21.6.2184-2191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raymond WE, Kleckner N. RAD50 protein of S.cerevisiae exhibits ATP-dependent DNA binding. Nucleic Acids Res. 1993;21:3851–3856. doi: 10.1093/nar/21.16.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell. 2001;8:1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- 38.Anderson DE, Trujillo KM, Sung P, Erickson HP. Structure of the Rad50 × Mre11 DNA repair complex from Saccharomyces cerevisiae by electron microscopy. J Biol Chem. 2001;276:37027–37033. doi: 10.1074/jbc.M106179200. [DOI] [PubMed] [Google Scholar]
- 39.Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, Carney JP, Petrini JH, Tainer JA. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 40.Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- 41.Wiltzius JJ, Hohl M, Fleming JC, Petrini JH. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat Struct Mol Biol. 2005;12:403–407. doi: 10.1038/nsmb928. [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi J, Tauchi H, Sakamoto S, Nakamura A, Morishima K, Matsuura S, Kobayashi T, Tamai K, Tanimoto K, Komatsu K. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr Biol. 2002;12:1846–1851. doi: 10.1016/s0960-9822(02)01259-9. [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi J, Antoccia A, Tauchi H, Matsuura S, Komatsu K. NBS1 and its functional role in the DNA damage response. DNA Repair (Amst) 2004;3:855–861. doi: 10.1016/j.dnarep.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 44.Tsukamoto Y, Mitsuoka C, Terasawa M, Ogawa H, Ogawa T. Xrs2p regulates Mre11p translocation to the nucleus and plays a role in telomere elongation and meiotic recombination. Mol Biol Cell. 2005;16:597–608. doi: 10.1091/mbc.E04-09-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivanov EL, Korolev VG, Fabre F. XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics. 1992;132:651–664. doi: 10.1093/genetics/132.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Borde V. The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res. 2007;15:551–563. doi: 10.1007/s10577-007-1147-9. [DOI] [PubMed] [Google Scholar]
- 47.Borde V, Lin W, Novikov E, Petrini JH, Lichten M, Nicolas A. Association of Mre11p with double-strand break sites during yeast meiosis. Mol Cell. 2004;13:389–401. doi: 10.1016/s1097-2765(04)00034-6. [DOI] [PubMed] [Google Scholar]
- 48.McKee AH, Kleckner N. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics. 1997;146:797–816. doi: 10.1093/genetics/146.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prinz S, Amon A, Klein F. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics. 1997;146:781–795. doi: 10.1093/genetics/146.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uanschou C, Siwiec T, Pedrosa-Harand A, Kerzendorfer C, Sanchez-Moran E, Novatchkova M, Akimcheva S, Woglar A, Klein F, Schlogelhofer P. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 2007;26:5061–5070. doi: 10.1038/sj.emboj.7601913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonczy P, Schnabel H, Kaletta T, Amores AD, Hyman T, Schnabel R. Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J Cell Biol. 1999;144:927–946. doi: 10.1083/jcb.144.5.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Penkner A, Portik-Dobos Z, Tang L, Schnabel R, Novatchkova M, Jantsch V, Loidl J. A conserved function for a Caenorhabditis elegans Com1/Sae2/CtIP protein homolog in meiotic recombination. EMBO J. 2007;26:5071–5082. doi: 10.1038/sj.emboj.7601916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Llorente B, Symington LS. The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol Cell Biol. 2004;24:9682–9694. doi: 10.1128/MCB.24.21.9682-9694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lieber MR. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 57.Digilio FA, Pannuti A, Lucchesi JC, Furia M, Polito LC. Tosca: a Drosophila gene encoding a nuclease specifically expressed in the female germline. Dev Biol. 1996;178:90–100. doi: 10.1006/dbio.1996.0200. [DOI] [PubMed] [Google Scholar]
- 58.Tishkoff DX, Boerger AL, Bertrand P, Filosi N, Gaida GM, Kane MF, Kolodner RD. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci U S A. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qiu J, Qian Y, Chen V, Guan MX, Shen B. Human exonuclease 1 functionally complements its yeast homologues in DNA recombination, RNA primer removal, and mutation avoidance. J Biol Chem. 1999;274:17893–17900. doi: 10.1074/jbc.274.25.17893. [DOI] [PubMed] [Google Scholar]
- 60.Szankasi P, Smith GR. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- 61.Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khazanehdari KA, Borts RH. EXO1 and MSH4 differentially affect crossing-over and segregation. Chromosoma. 2000;109:94–102. doi: 10.1007/s004120050416. [DOI] [PubMed] [Google Scholar]
- 63.Kirkpatrick DT, Ferguson JR, Petes TD, Symington LS. Decreased meiotic intergenic recombination and increased meiosis I nondisjunction in exo1 mutants of Saccharomyces cerevisiae. Genetics. 2000;156:1549–1557. doi: 10.1093/genetics/156.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Colleaux L, D'Auriol L, Galibert F, Dujon B. Recognition and cleavage site of the intron-encoded omega transposase. Proc Natl Acad Sci U S A. 1988;85:6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kostriken R, Heffron F. The product of the HO gene is a nuclease: purification and characterization of the enzyme. Cold Spring Harb Symp Quant Biol. 1984;49:89–96. doi: 10.1101/sqb.1984.049.01.012. [DOI] [PubMed] [Google Scholar]
- 66.Saeki T, Machida I, Nakai S. Genetic control of diploid recovery after gamma-irradiation in the yeast Saccharomyces cerevisiae. Mutat Res. 1980;73:251–265. doi: 10.1016/0027-5107(80)90192-x. [DOI] [PubMed] [Google Scholar]
- 67.Bressan DA, Baxter BK, Petrini JH. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7681–7687. doi: 10.1128/mcb.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johzuka K, Ogawa H. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hartsuiker E, Vaessen E, Carr AM, Kohli J. Fission yeast Rad50 stimulates sister chromatid recombination and links cohesion with repair. EMBO J. 2001;20:6660–6671. doi: 10.1093/emboj/20.23.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malone RE, Ward T, Lin S, Waring J. The RAD50 gene, a member of the double strand break repair epistasis group, is not required for spontaneous mitotic recombination in yeast. Curr Genet. 1990;18:111–116. doi: 10.1007/BF00312598. [DOI] [PubMed] [Google Scholar]
- 71.Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- 72.Ivanov EL, Sugawara N, White CI, Fabre F, Haber JE. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sugawara N, Haber JE. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shim EY, Ma JL, Oum JH, Yanez Y, Lee SE. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol Cell Biol. 2005;25:3934–3944. doi: 10.1128/MCB.25.10.3934-3944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bressan DA, Olivares HA, Nelms BE, Petrini JH. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics. 1998;150:591–600. doi: 10.1093/genetics/150.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee SE, Bressan DA, Petrini JH, Haber JE. Complementation between N-terminal Saccharomyces cerevisiae mre11 alleles in DNA repair and telomere length maintenance. DNA Repair (Amst) 2002;1:27–40. doi: 10.1016/s1568-7864(01)00003-9. [DOI] [PubMed] [Google Scholar]
- 78.Krogh BO, Llorente B, Lam A, Symington LS. Mutations in Mre11 phosphoesterase motif I that impair Saccharomyces cerevisiae Mre11-Rad50-Xrs2 complex stability in addition to nuclease activity. Genetics. 2005;171:1561–1570. doi: 10.1534/genetics.105.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Connelly JC, de Leau ES, Leach DR. Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair (Amst) 2003;2:795–807. doi: 10.1016/s1568-7864(03)00063-6. [DOI] [PubMed] [Google Scholar]
- 80.Stracker TH, Carson CT, Weitzman MD. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature. 2002;418:348–352. doi: 10.1038/nature00863. [DOI] [PubMed] [Google Scholar]
- 81.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, Moiani D, Carney JP, Russell P, Tainer JA. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, Iwasaki H, Mizuno K, Ohta K, Uritani M, Ushimaru T, Yoshinaga K, Ueno M. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol. 2003;23:5186–5197. doi: 10.1128/MCB.23.15.5186-5197.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rattray AJ, McGill CB, Shafer BK, Strathern JN. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics. 2001;158:109–122. doi: 10.1093/genetics/158.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rattray AJ, Shafer BK, Neelam B, Strathern JN. A mechanism of palindromic gene amplification in Saccharomyces cerevisiae. Genes Dev. 2005;19:1390–1399. doi: 10.1101/gad.1315805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 89.Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 91.Kim HS, Vijayakumar S, Reger M, Harrison JC, Haber JE, Weil C, Petrini JH. Functional interactions between Sae2 and the Mre11 complex. Genetics. 2008;178:711–723. doi: 10.1534/genetics.107.081331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem. 2005;280:38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- 93.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 94.Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol. 2003;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- 95.Schaeper U, Subramanian T, Lim L, Boyd JM, Chinnadurai G. Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J Biol Chem. 1998;273:8549–8552. doi: 10.1074/jbc.273.15.8549. [DOI] [PubMed] [Google Scholar]
- 96.Yu X, Wu LC, Bowcock AM, Aronheim A, Baer R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- 97.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Karathanasis E, Wilson TE. Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics. 2002;161:1015–1027. doi: 10.1093/genetics/161.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barlow JH, Lisby M, Rothstein R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell. 2008;30:73–85. doi: 10.1016/j.molcel.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zierhut C, Diffley JF. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 2008;27:1875–1885. doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Clerici M, Mantiero D, Guerini I, Lucchini G, Longhese MP. The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep. 2008;9:810–818. doi: 10.1038/embor.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Baroni E, Viscardi V, Cartagena-Lirola H, Lucchini G, Longhese MP. The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol Cell Biol. 2004;24:4151–4165. doi: 10.1128/MCB.24.10.4151-4165.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008 doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fiorentini P, Huang KN, Tishkoff DX, Kolodner RD, Symington LS. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang KN, Symington LS. A 5′-3′ exonuclease from Saccharomyces cerevisiae is required for in vitro recombination between linear DNA molecules with overlapping homology. Mol Cell Biol. 1993;13:3125–3134. doi: 10.1128/mcb.13.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Moreau S, Morgan EA, Symington LS. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics. 2001;159:1423–1433. doi: 10.1093/genetics/159.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chamankhah M, Fontanie T, Xiao W. The Saccharomyces cerevisiae mre11(ts) allele confers a separation of DNA repair and telomere maintenance functions. Genetics. 2000;155:569–576. doi: 10.1093/genetics/155.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lewis LK, Karthikeyan G, Westmoreland JW, Resnick MA. Differential suppression of DNA repair deficiencies of Yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase) Genetics. 2002;160:49–62. doi: 10.1093/genetics/160.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep. 2006;7:212–218. doi: 10.1038/sj.embor.7400593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lam AF, Krogh BO, Symington LS. Unique and overlapping functions of the Exo1, Mre11 and Pso2 nucleases in DNA repair. DNA Repair (Amst) 2008;7:655–662. doi: 10.1016/j.dnarep.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bennett CB, Lewis LK, Karthikeyan G, Lobachev KS, Jin YH, Sterling JF, Snipe JR, Resnick MA. Genes required for ionizing radiation resistance in yeast. Nat Genet. 2001;29:426–434. doi: 10.1038/ng778. [DOI] [PubMed] [Google Scholar]
- 113.Amundsen SK, Smith GR. Interchangeable parts of the Escherichia coli recombination machinery. Cell. 2003;112:741–744. doi: 10.1016/s0092-8674(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 114.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci U S A. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V. Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J. 2008;27:1953–1962. doi: 10.1038/emboj.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bae SH, Choi E, Lee KH, Park JS, Lee SH, Seo YS. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J Biol Chem. 1998;273:26880–26890. doi: 10.1074/jbc.273.41.26880. [DOI] [PubMed] [Google Scholar]
- 119.Budd ME, Campbell JL. The pattern of sensitivity of yeast dna2 mutants to DNA damaging agents suggests a role in DSB and postreplication repair pathways. Mutat Res. 2000;459:173–186. doi: 10.1016/s0921-8777(99)00072-5. [DOI] [PubMed] [Google Scholar]
- 120.Liao S, Toczylowski T, Yan H. Identification of the Xenopus DNA2 protein as a major nuclease for the 5′->3′ strand-specific processing of DNA ends. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 122.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.van Brabant AJ, Ye T, Sanz M, German IJ, Ellis NA, Holloman WK. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- 124.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 125.Chiolo I, Carotenuto W, Maffioletti G, Petrini JH, Foiani M, Liberi G. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol Cell Biol. 2005;25:5738–5751. doi: 10.1128/MCB.25.13.5738-5751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Karmakar P, Seki M, Kanamori M, Hashiguchi K, Ohtsuki M, Murata E, Inoue E, Tada S, Lan L, Yasui A, Enomoto T. BLM is an early responder to DNA double-strand breaks. Biochem Biophys Res Commun. 2006;348:62–69. doi: 10.1016/j.bbrc.2006.07.037. [DOI] [PubMed] [Google Scholar]
- 127.Nag DK, Cavallo SJ. Effects of mutations in SGS1 and in genes functionally related to SGS1 on inverted repeat-stimulated spontaneous unequal sister-chromatid exchange in yeast. BMC Mol Biol. 2007;8:120. doi: 10.1186/1471-2199-8-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stafa A, Svetec IK, Zgaga Z. Inactivation of the SGS1 and EXO1 genes synergistically stimulates plasmid integration in yeast. Food Technol Biotechnol. 2005;43:103–108. [Google Scholar]
- 129.Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci U S A. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.El-Shemerly M, Janscak P, Hess D, Jiricny J, Ferrari S. Degradation of human exonuclease 1b upon DNA synthesis inhibition. Cancer Res. 2005;65:3604–3609. doi: 10.1158/0008-5472.CAN-04-4069. [DOI] [PubMed] [Google Scholar]
- 131.El-Shemerly M, Hess D, Pyakurel AK, Moselhy S, Ferrari S. ATR-dependent pathways control hEXO1 stability in response to stalled forks. Nucleic Acids Res. 2008;36:511–519. doi: 10.1093/nar/gkm1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Featherstone C, Jackson SP. Ku, a DNA repair protein with multiple cellular functions? Mutat Res. 1999;434:3–15. doi: 10.1016/s0921-8777(99)00006-3. [DOI] [PubMed] [Google Scholar]
- 133.Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 134.Bennett RJ, Keck JL, Wang JC. Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J Mol Biol. 1999;289:235–248. doi: 10.1006/jmbi.1999.2739. [DOI] [PubMed] [Google Scholar]
- 135.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Henderson ER, Blackburn EH. An overhanging 3′ terminus is a conserved feature of telomeres. Mol Cell Biol. 1989;9:345–348. doi: 10.1128/mcb.9.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 138.Longhese MP. DNA damage response at functional and dysfunctional telomeres. Genes Dev. 2008;22:125–140. doi: 10.1101/gad.1626908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gilson E, Geli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 140.Wellinger RJ, Ethier K, Labrecque P, Zakian VA. Evidence for a new step in telomere maintenance. Cell. 1996;85:423–433. doi: 10.1016/s0092-8674(00)81120-4. [DOI] [PubMed] [Google Scholar]
- 141.Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 142.Ritchie KB, Petes TD. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics. 2000;155:475–479. doi: 10.1093/genetics/155.1.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wilson S, Warr N, Taylor DL, Watts FZ. The role of Schizosaccharomyces pombe Rad32, the Mre11 homologue, and other DNA damage response proteins in non-homologous end joining and telomere length maintenance. Nucleic Acids Res. 1999;27:2655–2661. doi: 10.1093/nar/27.13.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kironmai KM, Muniyappa K. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells. 1997;2:443–455. doi: 10.1046/j.1365-2443.1997.1330331.x. [DOI] [PubMed] [Google Scholar]
- 145.Larrivee M, LeBel C, Wellinger RJ. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 2004;18:1391–1396. doi: 10.1101/gad.1199404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chai W, Sfeir AJ, Hoshiyama H, Shay JW, Wright WE. The involvement of the Mre11/Rad50/Nbs1 complex in the generation of G-overhangs at human telomeres. EMBO Rep. 2006;7:225–230. doi: 10.1038/sj.embor.7400600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Frank CJ, Hyde M, Greider CW. Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol Cell. 2006;24:423–432. doi: 10.1016/j.molcel.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 148.Tomita K, Kibe T, Kang HY, Seo YS, Uritani M, Ushimaru T, Ueno M. Fission yeast Dna2 is required for generation of the telomeric single-strand overhang. Mol Cell Biol. 2004;24:9557–9567. doi: 10.1128/MCB.24.21.9557-9567.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Diede SJ, Gottschling DE. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr Biol. 2001;11:1336–1340. doi: 10.1016/s0960-9822(01)00400-6. [DOI] [PubMed] [Google Scholar]
- 150.Lustig AJ, Petes TD. Identification of yeast mutants with altered telomere structure. Proc Natl Acad Sci U S A. 1986;83:1398–1402. doi: 10.1073/pnas.83.5.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Greenwell PW, Kronmal SL, Porter SE, Gassenhuber J, Obermaier B, Petes TD. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- 152.Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol Cell. 2007;27:851–858. doi: 10.1016/j.molcel.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 153.Ritchie KB, Mallory JC, Petes TD. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6065–6075. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sabourin M, Tuzon CT, Zakian VA. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol Cell. 2007;27:550–561. doi: 10.1016/j.molcel.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Vodenicharov MD, Wellinger RJ. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol Cell. 2006;24:127–137. doi: 10.1016/j.molcel.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 156.Wellinger RJ, Wolf AJ, Zakian VA. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell. 1993;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- 157.Marcand S, Brevet V, Mann C, Gilson E. Cell cycle restriction of telomere elongation. Curr Biol. 2000;10:487–490. doi: 10.1016/s0960-9822(00)00450-4. [DOI] [PubMed] [Google Scholar]
- 158.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Maringele L, Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Feldmann H, Winnacker EL. A putative homologue of the human autoantigen Ku from Saccharomyces cerevisiae. J Biol Chem. 1993;268:12895–12900. [PubMed] [Google Scholar]
- 161.Barnes G, Rio D. DNA double-strand-break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94:867–872. doi: 10.1073/pnas.94.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Boulton SJ, Jackson SP. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gravel S, Larrivee M, Labrecque P, Wellinger RJ. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- 165.Porter SE, Greenwell PW, Ritchie KB, Petes TD. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- 167.Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- 168.Lazzaro F, Sapountzi V, Granata M, Pellicioli A, Vaze M, Haber JE, Plevani P, Lydall D, Muzi-Falconi M. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J. 2008;27:1502–1512. doi: 10.1038/emboj.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zubko MK, Guillard S, Lydall D. Exo1 and Rad24 differentially regulate generation of ssDNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants. Genetics. 2004;168:103–115. doi: 10.1534/genetics.104.027904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bertuch AA, Lundblad V. EXO1 contributes to telomere maintenance in both telomerase-proficient and telomerase-deficient Saccharomyces cerevisiae. Genetics. 2004;166:1651–1659. doi: 10.1534/genetics.166.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Schaetzlein S, Kodandaramireddy NR, Ju Z, Lechel A, Stepczynska A, Lilli DR, Clark AB, Rudolph C, Kuhnel F, Wei K, Schlegelberger B, Schirmacher P, Kunkel TA, Greenberg RA, Edelmann W, Rudolph KL. Exonuclease-1 deletion impairs DNA damage signaling and prolongs lifespan of telomere-dysfunctional mice. Cell. 2007;130:863–877. doi: 10.1016/j.cell.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jia X, Weinert T, Lydall D. Mec1 and Rad53 inhibit formation of single-stranded DNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants. Genetics. 2004;166:753–764. doi: 10.1534/genetics.166.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- 174.Lee JY, Mogen JL, Chavez A, Johnson FB. Sgs1 RecQ Helicase Inhibits Survival of Saccharomyces cerevisiae Cells Lacking Telomerase and Homologous Recombination. J Biol Chem. 2008;283:29847–29858. doi: 10.1074/jbc.M804760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Maringele L, Lydall D. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 2004;18:2663–2675. doi: 10.1101/gad.316504. [DOI] [PMC free article] [PubMed] [Google Scholar]