Summary
Fat metabolism, reproduction, and aging are intertwined regulatory axes; however, the mechanism by which they are coupled remains poorly understood. We found that germline stem cells (GSCs) actively modulate lipid hydrolysis in Caenorhabditis elegans, which in turn regulates longevity. GSC arrest promotes systemic lipolysis via induction of a specific fat lipase. Subsequently, fat mobilization is promoted and life span is prolonged. Constitutive expression of this lipase in fat storage tissue generates lean and long-lived animals. This lipase is a key factor in the lipid hydrolysis and increased longevity that are induced by decreased insulin signaling. These results suggest a link between C. elegans fat metabolism and longevity.
Abalance of fat storage and mobilization is a universal feature of animal physiology (1). Reproduction is an energy-intensive process, which is modulated by the availability of nutrients and in turn influences lipid metabolism (2). Reproductive ability declines with age, and many organisms undergo reproductive senescence (3). Obesity increases with age and is also associated with the transition to menopause in women (4). Genetic studies have suggested endocrine roles of adipose tissue and the reproductive system in regulation of life span (5–8). Thus, understanding the mechanisms by which fat metabolism is coupled to reproductive cues may reveal systemic regulation of fat metabolism and provide insights into the control of aging.
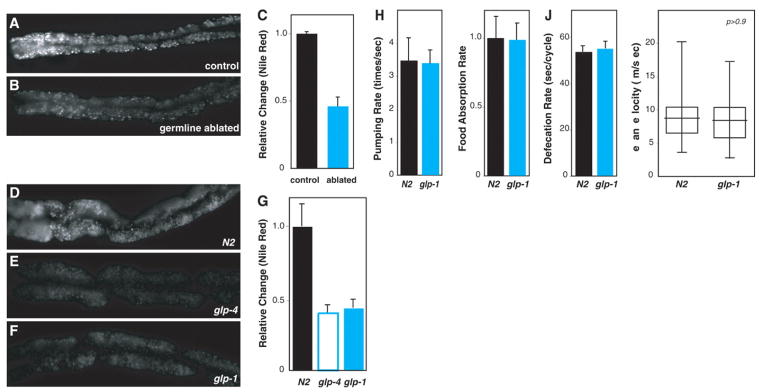
In C. elegans, the energetic demands of progeny production are profound. The gonad undergoes many more mitoses than does somatic tissue, and the biomass of the oocytes produced is approximately equal to the biomass increase from egg to adult. Thus, in the absence of reproduction, a surfeit of available energy could lead to an increase in fat storage. To test this idea, we ablated the precursor cells of the germ line in C. elegans with the use of a laser microbeam. The vital dye Nile Red was used to visualize fat storage droplets in living animals (9). Opposite to the expected increase in fat storage, germ line–ablated animals stored 50% as much fat as untreated animals (Fig. 1, A to C). This finding suggested a regulatory mechanism coupling reproduction and fat metabolism.
Fig. 1. Influence of reproductive activity on fat metabolism.
(A to C) Ablation of germline precursor cells resulted in a 50% reduction in adult fat storage (P < 0.0001) (n = 15). (D to G) The same degree of reduction was observed in glp-4(bn2) and glp-1(e2141) mutants defective in germline proliferation (P < 0.0001) (N2 and glp-4, n = 15; glp-1, n = 18). (H to J) Decreased fat in glp-1 was not due to less food intake (measured as pharyngeal pumping and food absorption rates) or food retention time (measured as defecation rate) (P > 0.5 for each) (pumping rate, N2, n = 12; glp-1, n = 18; food absorption rate, N2 and glp-1, n = 5; defecation rate, N2 and glp-1, n = 7). (K) Comparison of locomotory behavior showed that physical activity does not change in glp-1. Experiments were performed at the restrictive temperature (25°C) for both wild-type (N2) and glp mutants (n = 21 for each).
Fat storage is also aberrant in the sterile mutants glp-1(e2141ts) and glp-4(bn2ts), which are defective in germline proliferation (10, 11). The glp mutants showed a 50% decrease in fat storage at the nonpermissive temperature relative to the wild type (N2) (Fig. 1, D to G). A similar decrease was observed by staining with a BODIPY-labeled fatty acid analog (fig. S1) (12) or SudanBlack, a fat-specific dye (fig. S2) (13). At the permissive temperature, the glp mutants reproduced normally and their fat storage was similar to that of the wild type (fig. S3).
Fat storage can be altered by changes in either energy input or expenditure. Food intake and retention in the gut are unchanged in glp-1 (Fig. 1, H and J); the food absorption rate is also normal (Fig. 1I). Normal locomotion in glp-1 suggests that less fat storage is not due to an increase in physical activity (Fig. 1K). Therefore, decreased fat storage in the germ line–defective mutants is unlikely to be the result of alterations in energy intake and/or physical activity, and more likely reveals an altered endocrine signaling axis.
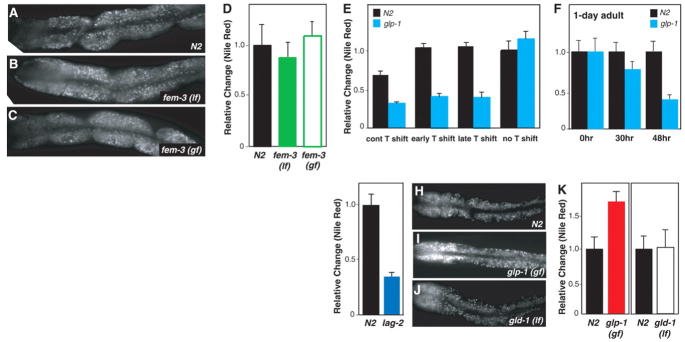
Production of vitellogenin-rich oocytes is the most energy-intensive reproductive function. We used fem-3 sterile mutants to examine whether gametogenesis influences fat storage. The gain-of-function allele fem-3(q20ts) produces only sperm, whereas the loss-of-function allele fem-3(e2006ts) produces only oocytes at the nonpermissive temperature (14, 15). Neither fem-3 mutant exhibited abnormal lipid accumulation (Fig. 2, A to D). This result excludes the possibility that gametogenesis regulates fat storage, and it also suggests that sterility per se does not cause a change in lipid accumulation.
Fig. 2. Regulation of fat metabolism by GSC proliferation.
(A to D) fem-3(e2006) loss-of-function mutants (lf) producing only oocytes or fem-3(q20) gain-of-function mutants (gf) generating only sperm showed the same fat storage as in the wild type (P > 0.1) (n = 15 for each). (E) Shift to 25°C during early or late larval development did not affect fat storage in the wild type (P > 0.1) but caused a 50% decrease in glp-1(e2141) mutants (P < 0.0001) (n = 15 for each genotype and treatment). (F) GSC arrest, caused by temperature shifting of 1-day-old glp-1 adults, caused a decrease in fat storage (0 hours, P > 0.5; 30 hours, P < 0.005; 48 hours, P < 0.0001)(n = 17 for each genotype and treatment). (G) lag-2(q420) showed reduced fat (P < 0.0001) (N2, n = 12; lag-2, n = 15). (H to K) GSC overproliferation in the glp-1(ar202) gain-of-function mutant causes increased fat (P < 0.0001). In contrast, the loss-of-function mutant of gld-1(q485), in which early-phase meiotic germ cells overproliferate, did not change fat storage (P > 0.1) (N2, n = 12; glp-1, n = 15; gld-1, n = 13).
To test whether germline proliferation regulates fat storage, we shifted glp-1 mutants to the restrictive temperature at different evelopmental stages to arrest germline proliferation at distinct points. Adults that are generated from L2 (early) temperature shifts carry few mitotic germ cells, whereas adults from L4 (late) temperature shifts form the germ line with essentially wild type– sized mitotic and meiotic germ cells and differentiated sperm. Despite a very different composition of the germ line, adult fat storage was decreased to a similar extent under all conditions (Fig. 2E). One process shared by all temperature shifts is germline stem cell (GSC) arrest (16), which could induce the decrease in fat storage. We therefore shifted temperature at 1 day of adulthood, after animals started to reproduce; this should affect adult GSCs but not the already proliferated germ line. By 30 hours at the restrictive temperature, fat storage in glp-1 started to decrease (Fig. 2F). Within 48 hours, lipid accumulation in glp-1 was reduced to an extent comparable to that seen with the developmental temperature shifts (Fig. 2F). This result suggests that GSCs regulate fat storage during adulthood.
The somatic distal tip cell forms the niche of GSCs. The Notch ligand LAG-2 expressed in the distal tip cell is required to maintain GSC identity (17). Like glp-1 mutants, lag-2(q420ts) mutants (18) showed a 50% decrease in fat storage (Fig. 2G). glp-1(ar202gf) mutants with a hyperactive GLP-1, in which entry into meiosis is prevented and GSCs overproliferate (19), showed a factor of 1.7 fat increase (Fig. 2, H, I, and K), which suggests that a deficit of GSCs signals low fat storage and that GSC overproliferation signals high fat storage. No change in fat content was detected in gld-1(q485) mutants, in which early-phase meiotic germ cells reenter into the mitotic cell cycle and overproliferate (20) (Fig. 2, H, J, and K). Thus, once germ cells undergo differentiation, they lose the ability to modulate fat storage.
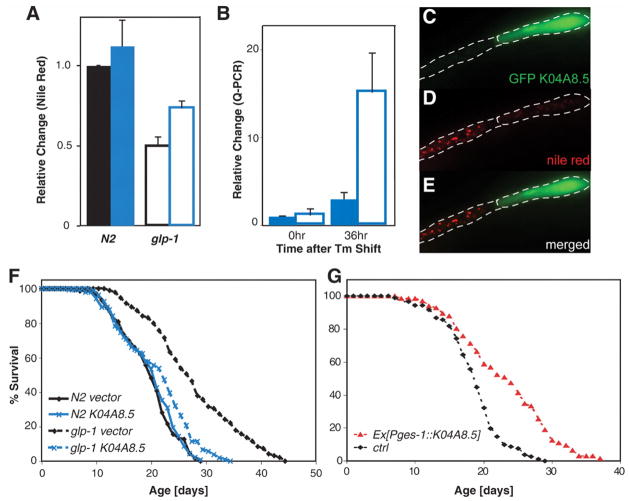
To understand the mechanisms by which GSCs regulate fat storage, we reduced the activities of 163 metabolic genes by RNA interference (RNAi) and screened for gene inactivations that increase fat storage in glp-1 (table S1). Among 16 potential candidate genes identified, K04A8.5 encoded a triglyceride lipase, which most strongly affected fat storage. Inactivation of K04A8.5 partially restored fat storage in glp-1 but had marginal effect on the wild type (Fig. 3A). GSC arrest caused a marked increase in the transcriptional levels of K04A8.5 (Fig. 3B), and a promoter–green fluorescent protein (GFP) reporter that was not detected under normal conditions became detectable in the glp-1 gut at the restrictive temperature (fig. S4). High gene dosage of K04A8.5 decreased fat storage in the wild type, and genetic mosaic animals showed that intestinal cells that constitutively express K04A8.5 had fewer lipid droplets than did neighboring nontransgenic cells (Fig. 3, C to E). These results imply that this lipase acts in fat storage tissue rather than in endocrine cells or GSCs. Thus, the decrease in fat storage upon GSC arrest is induced by increased lipid hydrolysis via up-regulation of K04A8.5.
Fig. 3. A role for triglyceride lipase in lipid hydrolysis and longevity.
(A) K04A8.5 RNAi partially restored fat storage in glp-1(e2141) (open bars, P < 0.001) but had a marginal effect in the wild type (solid bars, 10% increase in fat storage, P > 0.1)(n = 20 for each genotype and RNAi feeding). Blue, K04A8.5 RNAi; black, vector control. (B) K04A8.5 expression was up-regulated in glp-1 (P < 0.0001; solid bars, N2; open bars, glp-1). (C to E) Genetic mosaic analysis shows that the number and intensity of lipid droplets both decreased in the cell constitutively expressing K04A8.5, marked by GFP, relative to its sister cell. (F) K04A8.5 RNAi had no effect on the life span of wild-type animals (P > 0.01) but suppressed the increased longevity of glp-1 (P < 0.0001; 24% reduction in mean life span). (G) Constitutive expression of K04A8.5 in the intestine extended life span (P < 0.0001; 24% mean life-span extension).
GSC arrest caused by glp-1 loss of function resulted in extended life span (Fig. 3F and table S2) (8); K04A8.5 RNAi suppressed this increased longevity but did not reduce wild-type life span (Fig. 3F and table S2). Therefore, up-regulation of this lipase gene mediates both lipid hydrolysis and longevity in GSC-arrested animals. Constitutive expression of K04A8.5 specifically in the intestine led to life spans that were 24% longer than in control siblings (Fig. 3G and table S3). Thus, lipid hydrolysis in fat storage tissue prolongs life span, which connects the metabolic functions of adipose tissue to life-span control.
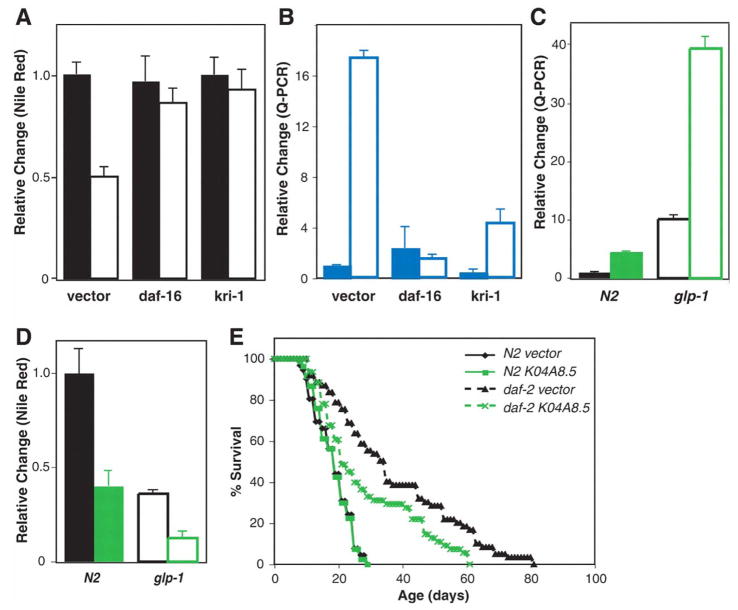
We investigated the signaling pathways regulating K04A8.5 expression in the intestine. The forkhead transcription factor DAF-16 is translocated into nuclei in the intestine upon GSC arrest (21). To test whether daf-16 is involved in regulation of fat storage by GSC proliferation, we inactivated daf-16 by RNAi in wild-type and glp-1 mutants and assayed fat storage. daf-16 inactivation restored fat storage in glp-1 but did not affect wild-type fat storage (Fig. 4A and fig. S5). K04A8.5 up-regulation in glp-1 was abolished in the absence of daf-16 but was not altered in wild-type animals subjected to daf-16 RNAi (Fig. 4B). Thus, upon GSC arrest, DAF-16 is activated in the intestine to promote lipid hydrolysis through induction of K04A8.5 expression. External stresses such as heat shock and oxidative stress activate daf-16 (22, 23). After heat shock and paraquat treatment, the DAF-16 targets hsp-16.1 and ctl-2 were up-regulated but K04A8.5 was not (fig. S6). These results suggest a specific regulation of K04A8.5 by the signal from the germ line.
Fig. 4. Synergistic regulation of fat metabolism by GSC proliferation and insulin signaling.
(A) Either daf-16 or kri-1 RNAi restored lipid accumulation in glp-1 (P < 0.0001); neither of them affected fat storage in the wild type (P > 0.05) (n = 17 for each genotype and RNAi feeding). Solid bars, N2; open bars, glp-1. (B) daf-16 and kri-1 were required to up-regulate K04A8.5 upon GSC arrest. daf-16 or kri-1 RNAi suppressed K04A8.5 induction in glp-1 (P < 0.001) (n = 15 for each genotype and RNAi feeding). Solid bars, N2; open bars, glp-1. (C) K04A8.5 was induced in animals subjected to daf-2 RNAi only at adulthood (P < 0.0001). This induction by daf-2 RNAi was enhanced in the glp-1 mutant (P < 0.0001). Green, daf-2 RNAi; black, vector control. Solid bars, N2; open bars, glp-1. (D) Adult-specific daf-2 RNAi decreased fat storage by 50% in the wild type (P < 0.0001). Loss of the germ line and reduction of daf-2 activity were synergistic in reducing fat storage (P < 0.0001). Green, daf-2 RNAi; black, vector control. Solid bars, N2; open bars, glp-1. (E) K04A8.5 RNAi partially suppressed the longevity of daf-2(e1370) mutants (P < 0.005; 24% decrease in mean life span) but had no effect on the life span of the wild type (P > 0.01).
KRI-1, the human KRIT 1 homolog, and DAF-12, the nuclear hormone receptor, are both required for the intestinal nuclear localization of DAF-16 in GSC-arrested animals (21). These factors could act upstream of DAF-16 to sense signals from GSC and, in response, regulate lipid accumulation. Like daf-16 RNAi, kri-1 RNAi significantly reduced K04A8.5 expression and increased the fat content in glp-1 (Fig. 4, A and B, and fig. S5). In contrast, reducing daf-12 function did not affect K04A8.5 levels and caused a slight decrease in lipid accumulation in both wild-type and glp-1 mutants (fig. S7). Therefore, GSC arrest promotes lipid hydrolysis in the intestine through activation of the kri-1/daf-16 signaling pathway, but independently of daf-12 lipophilic hormone signaling.
We examined K04A8.5 expression in other long-lived animals, such as worms with reducing function in insulin receptor/daf-2. daf-2 is crucial in regulation of fat metabolism during larval development (24). Therefore, we reduced daf-2 function only at adulthood by RNAi feeding. Reducing daf-2 activity at adulthood caused up-regulation of K04A8.5 and decreased fat storage (Fig. 4, C and D, and fig. S8). Loss of thegerm line and reduced daf-2 signaling synergistically induced K04A8.5 and decreased fat storage (Fig. 4, C and D, and fig. S8). We also found that K04A8.5 RNAi partially suppressed the longevity of daf-2 mutants (Fig. 4E and table S4). These results suggest that lipid hydrolysis is also connected to life-span control in the daf-2 long-lived animals.
Our findings reveal an endocrine signaling axis from GSCs to fat storage tissue, with feedback from the fat storage to the longevity of the animal. Somatic stem cells are thought to mediate tissue regeneration after wounding, and such regeneration is also known to decline with aging. How the proliferation of adult stem cells is coupled to the requirement for replacement cells during normal and pathological aging may be related to the metabolic pathways we have discovered between germline stem cells and the longevity of C. elegans.
Supplementary Material
Footnotes
This manuscript has been accepted for publication in Science. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior written permission of AAAS.
References and Notes
- 1.Zechner R, Strauss JG, Haemmerle G, Lass A, Zimmermann R. Curr Opin Lipidol. 2005;16:333. doi: 10.1097/01.mol.0000169354.20395.1c. [DOI] [PubMed] [Google Scholar]
- 2.Tissenbaum HA, Ruvkun G. Genetics. 1998;148:703. doi: 10.1093/genetics/148.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burks DJ, et al. Nature. 2000;407:377. doi: 10.1038/35030105. [DOI] [PubMed] [Google Scholar]
- 4.Carr MC. J Clin Endocrinol Metab. 2003;88:2404. doi: 10.1210/jc.2003-030242. [DOI] [PubMed] [Google Scholar]
- 5.Blüher M, Kahn BB, Kahn CR. Science. 2003;299:572. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 6.Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Nature. 2004;429:562. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 7.Giannakou ME, et al. Science. 2004;305:361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- 8.Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Science. 2002;295:502. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- 9.Ashrafi K, et al. Nature. 2003;421:268. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- 10.Beanan MJ, Strome S. Development. 1992;116:755. doi: 10.1242/dev.116.3.755. [DOI] [PubMed] [Google Scholar]
- 11.Priess JR, Schnabel H, Schnabel R. Cell. 1987;51:601. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- 12.Mak HY, Nelson LS, Basson M, Johnson CD, Ruvkun G. Nat Genet. 2006;38:363. doi: 10.1038/ng1739. [DOI] [PubMed] [Google Scholar]
- 13.Ogg S, et al. Nature. 1997;389:994. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 14.Hodgkin J. Genetics. 1986;114:15. doi: 10.1093/genetics/114.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barton MK, Schedl TB, Kimble J. Genetics. 1987;115:107. doi: 10.1093/genetics/115.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimble JE, White JG. Dev Biol. 1981;81:208. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- 17.Wong MD, Jin Z, Xie T. Annu Rev Genet. 2005;39:173. doi: 10.1146/annurev.genet.39.073003.105855. [DOI] [PubMed] [Google Scholar]
- 18.Henderson ST, Gao D, Lambie EJ, Kimble J. Development. 1994;120:2913. doi: 10.1242/dev.120.10.2913. [DOI] [PubMed] [Google Scholar]
- 19.Pepper AS, Killian DJ, Hubbard EJ. Genetics. 2003;163:115. doi: 10.1093/genetics/163.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francis R, Maine E, Schedl T. Genetics. 1995;139:607. doi: 10.1093/genetics/139.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berman JR, Kenyon C. Cell. 2006;124:1055. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 22.Lee SS, Kennedy S, Tolonen AC, Ruvkun G. Science. 2003;300:644. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- 23.Wolff S, et al. Cell. 2006;124:1039. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
- 24.Wolkow CA, Kimura KD, Lee MS, Ruvkun G. Science. 2000;290:147. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- 25.We thank H. Mak, J. Dittman, and J. Avruch for critical reading of the manuscript; N. Ringstad for laser ablation techniques; V. Rottiers, A. Antebi, and the Caenorhabditis Genetics Center for providing strains; A. Fire for GFP vectors; and members of the Ruvkun lab for discussions. Supported by Life Sciences Research Foundation and Ellison Medical Foundation fellowships (M.C.W.), a Human Frontier Science Program fellowship (E.J.O.), and NIH grants 5R01AG016636 and 5R37AG14161 (G.R.).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.